bc34eb2f8bcc555990803ac47d20d9bb.ppt
- Количество слайдов: 41
Function-process links The theory
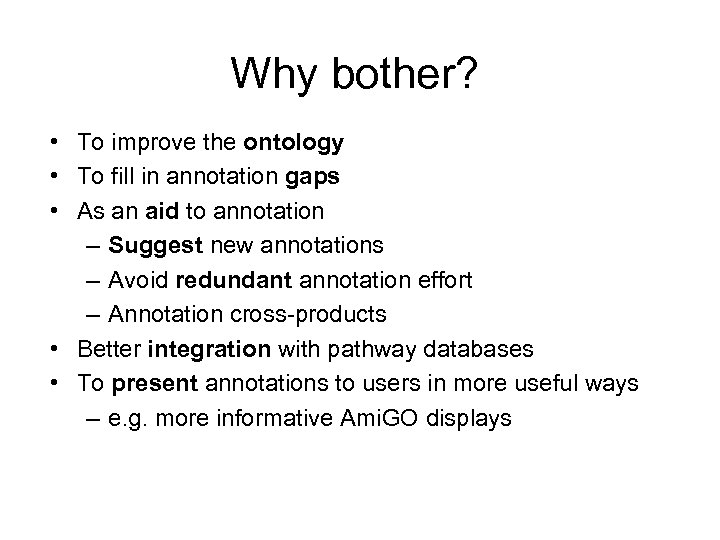
Why bother? • To improve the ontology • To fill in annotation gaps • As an aid to annotation – Suggest new annotations – Avoid redundant annotation effort – Annotation cross-products • Better integration with pathway databases • To present annotations to users in more useful ways – e. g. more informative Ami. GO displays
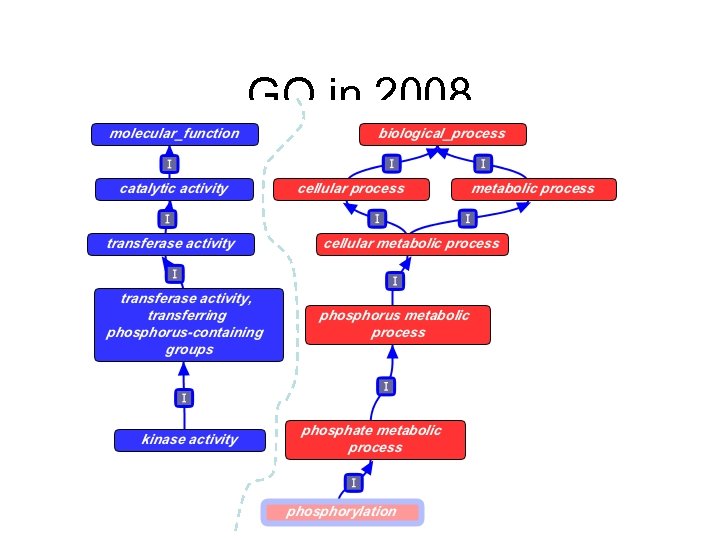
GO in 2008
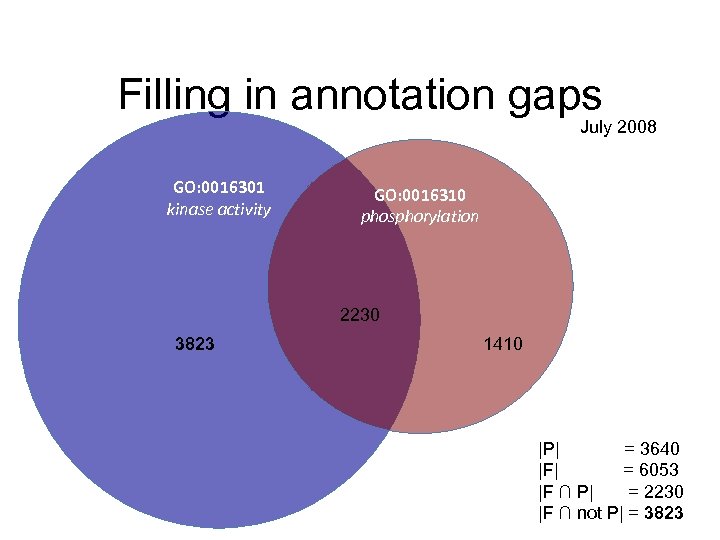
Filling in annotation gaps July 2008 GO: 0016301 kinase activity GO: 0016310 phosphorylation 2230 3823 1410 |P| = 3640 |F| = 6053 |F ∩ P| = 2230 |F ∩ not P| = 3823
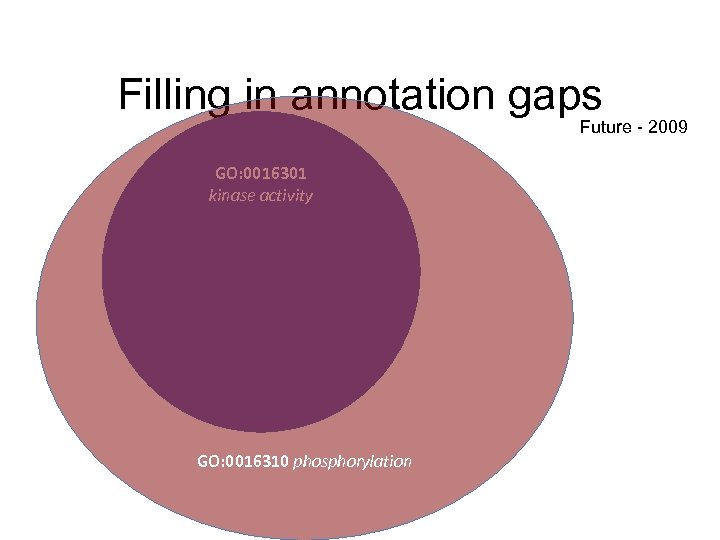
Filling in annotation gaps Future - 2009 GO: 0016301 kinase activity GO: 0016310 phosphorylation

Improved presentation to users
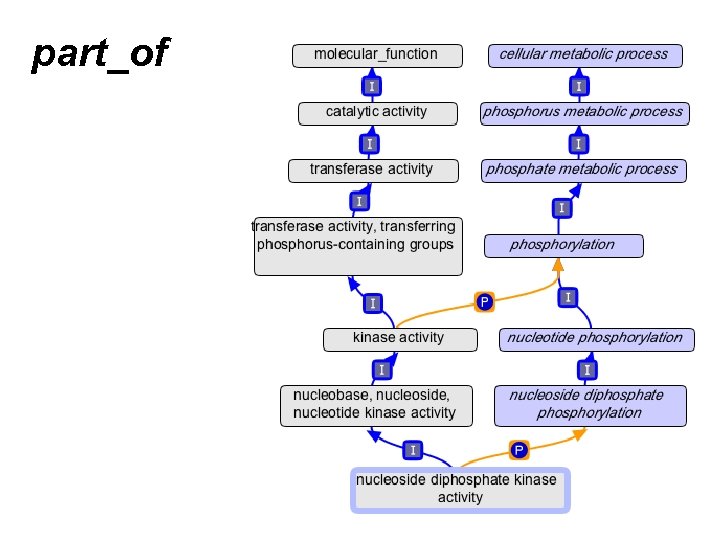
part_of
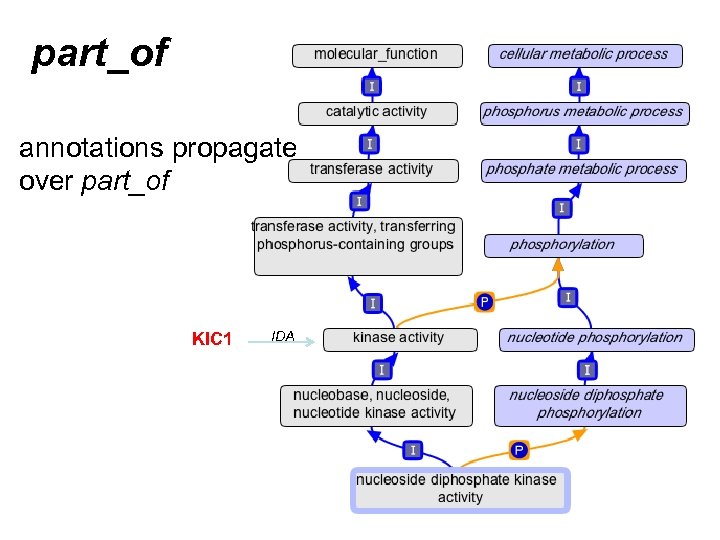
part_of annotations propagate over part_of KIC 1 IDA
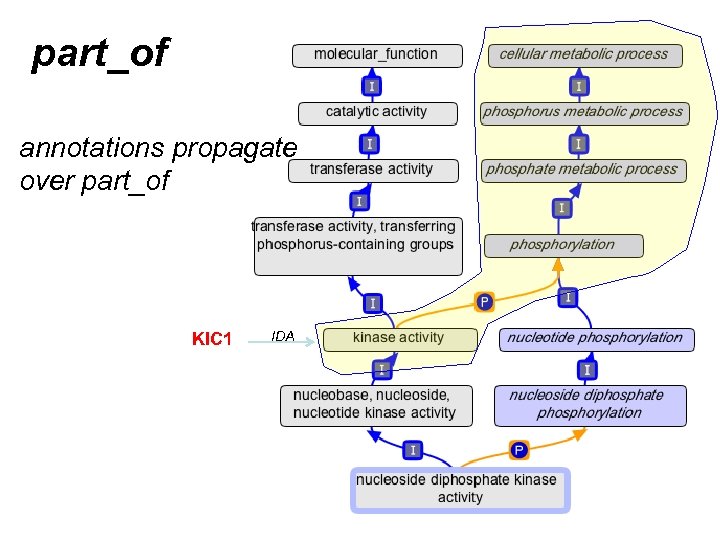
part_of annotations propagate over part_of KIC 1 IDA
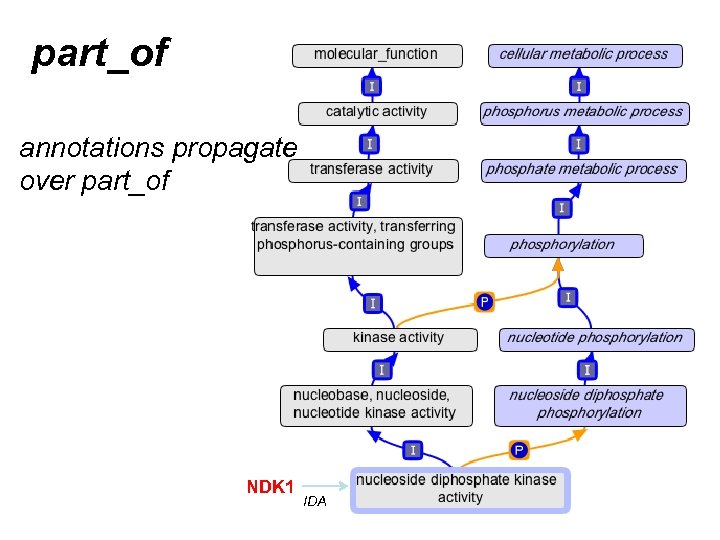
part_of annotations propagate over part_of NDK 1 IDA
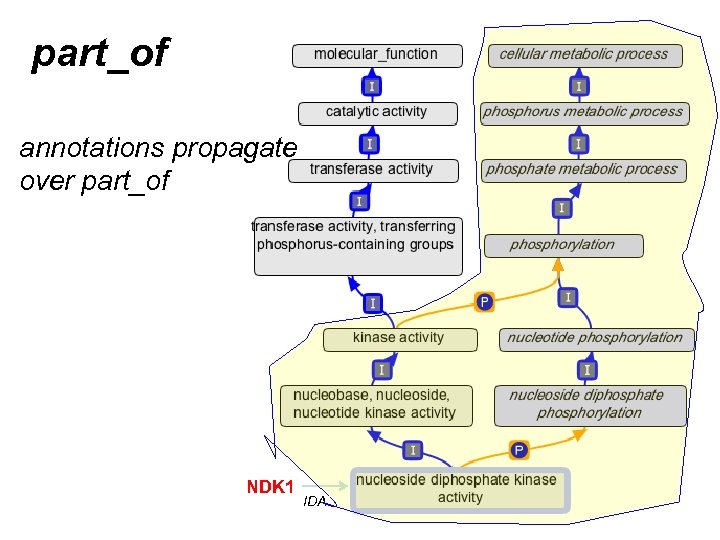
part_of annotations propagate over part_of NDK 1 IDA
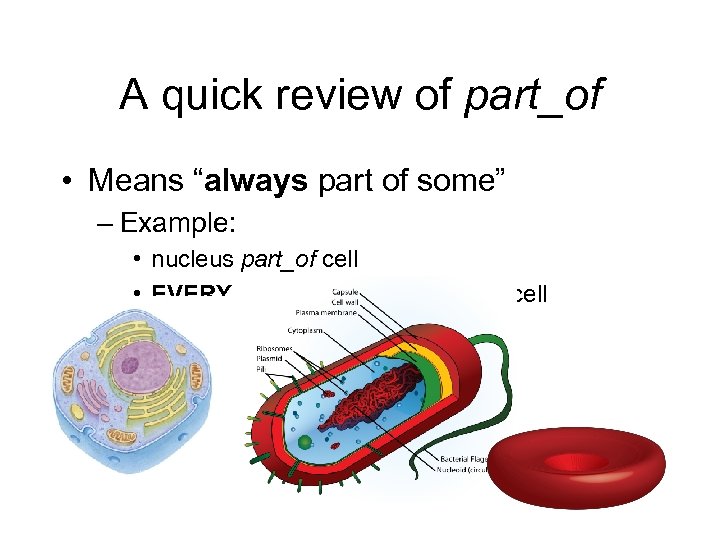
A quick review of part_of • Means “always part of some” – Example: • nucleus part_of cell • EVERY nucleus is part_of SOME cell
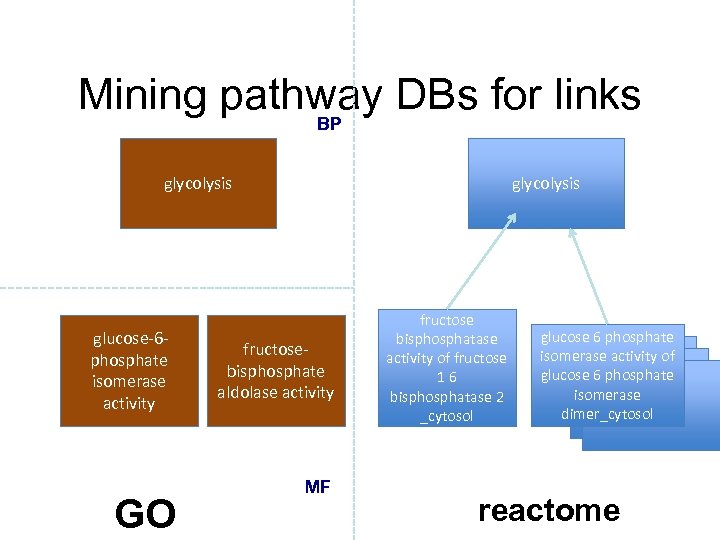
Mining pathway DBs for links BP glycolysis glucose-6 phosphate isomerase activity GO glycolysis fructosebisphosphate aldolase activity MF fructose bisphosphatase activity of fructose 16 bisphosphatase 2 _cytosol glucose 6 phosphate isomerase activity of glucose 6 phosphate isomerase dimer_cytosol reactome
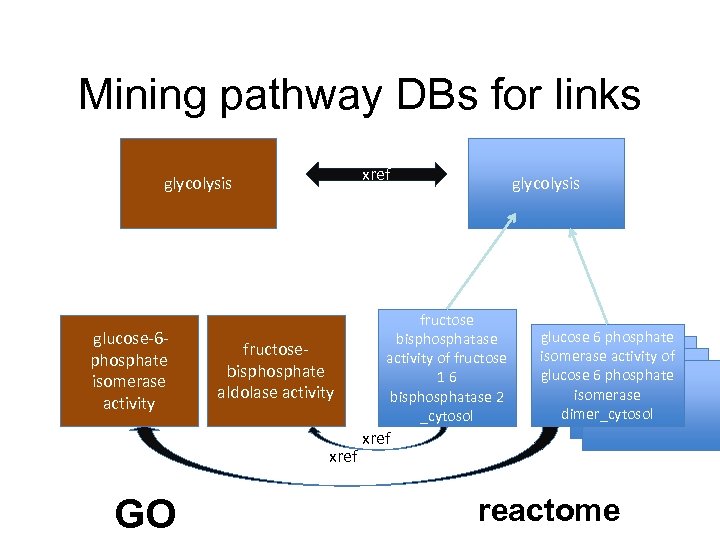
Mining pathway DBs for links xref glycolysis glucose-6 phosphate isomerase activity fructosebisphosphate aldolase activity xref GO glycolysis fructose bisphosphatase activity of fructose 16 bisphosphatase 2 _cytosol glucose 6 phosphate isomerase activity of glucose 6 phosphate isomerase dimer_cytosol xref reactome
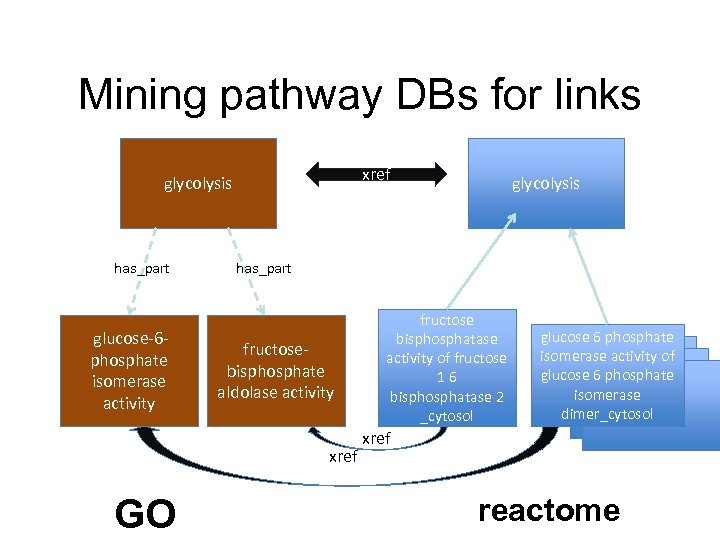
Mining pathway DBs for links xref glycolysis has_part glucose-6 phosphate isomerase activity has_part fructosebisphosphate aldolase activity xref GO glycolysis fructose bisphosphatase activity of fructose 16 bisphosphatase 2 _cytosol glucose 6 phosphate isomerase activity of glucose 6 phosphate isomerase dimer_cytosol xref reactome
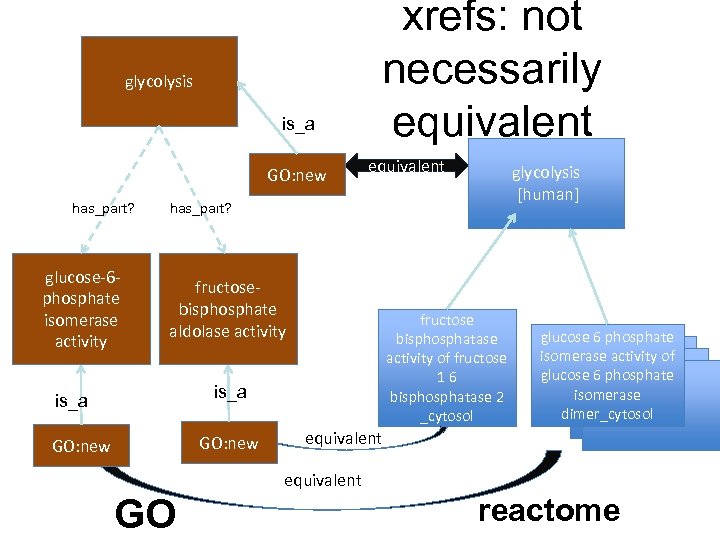
glycolysis is_a GO: new has_part? glucose-6 phosphate isomerase activity xrefs: not necessarily equivalent glycolysis [human] has_part? fructosebisphosphate aldolase activity fructose bisphosphatase activity of fructose 16 bisphosphatase 2 _cytosol is_a GO: new glucose 6 phosphate isomerase activity of glucose 6 phosphate isomerase dimer_cytosol equivalent GO reactome
![glycolysis is_a some_has_part GO: new xrefs: not necessarily equivalent glycolysis [human] some_has_part glucose-6 phosphate glycolysis is_a some_has_part GO: new xrefs: not necessarily equivalent glycolysis [human] some_has_part glucose-6 phosphate](https://present5.com/presentation/bc34eb2f8bcc555990803ac47d20d9bb/image-17.jpg)
glycolysis is_a some_has_part GO: new xrefs: not necessarily equivalent glycolysis [human] some_has_part glucose-6 phosphate isomerase activity fructosebisphosphate aldolase activity fructose bisphosphatase activity of fructose 16 bisphosphatase 2 _cytosol is_a GO: new glucose 6 phosphate isomerase activity of glucose 6 phosphate isomerase dimer_cytosol equivalent GO reactome
![xrefs: not necessarily equivalent glycolysis xref some_has_part glycolysis [human] some_has_part glucose-6 phosphate isomerase activity xrefs: not necessarily equivalent glycolysis xref some_has_part glycolysis [human] some_has_part glucose-6 phosphate isomerase activity](https://present5.com/presentation/bc34eb2f8bcc555990803ac47d20d9bb/image-18.jpg)
xrefs: not necessarily equivalent glycolysis xref some_has_part glycolysis [human] some_has_part glucose-6 phosphate isomerase activity fructosebisphosphate aldolase activity fructose bisphosphatase activity of fructose 16 bisphosphatase 2 _cytosol glucose 6 phosphate isomerase activity of glucose 6 phosphate isomerase dimer_cytosol xref GO reactome

Specifics • Low Hanging Fruit – Function to process links • Mostly part_of links • Some regulates links • Pathways – Process to function • has_part – Mining from pathways databases & curation
Function-process links Conclusions of the electron transport working group.
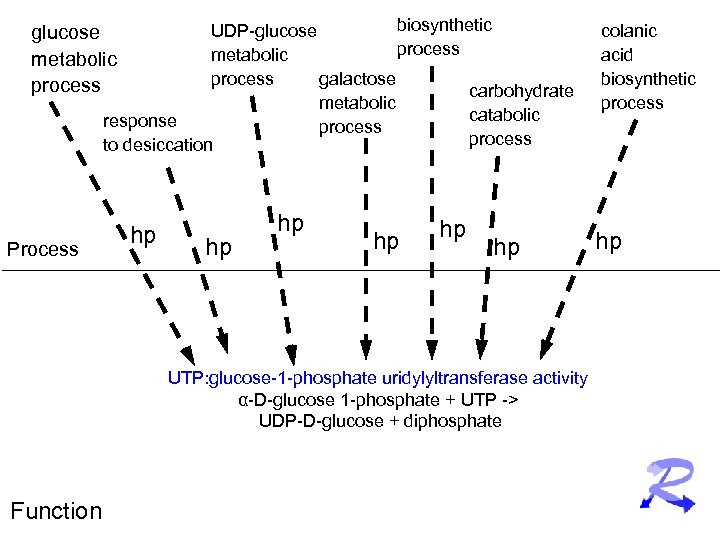
biosynthetic UDP-glucose process metabolic process galactose carbohydrate metabolic catabolic response process to desiccation glucose metabolic process Process hp hp hp UTP: glucose-1 -phosphate uridylyltransferase activity α-D-glucose 1 -phosphate + UTP -> UDP-D-glucose + diphosphate Function colanic acid biosynthetic process hp
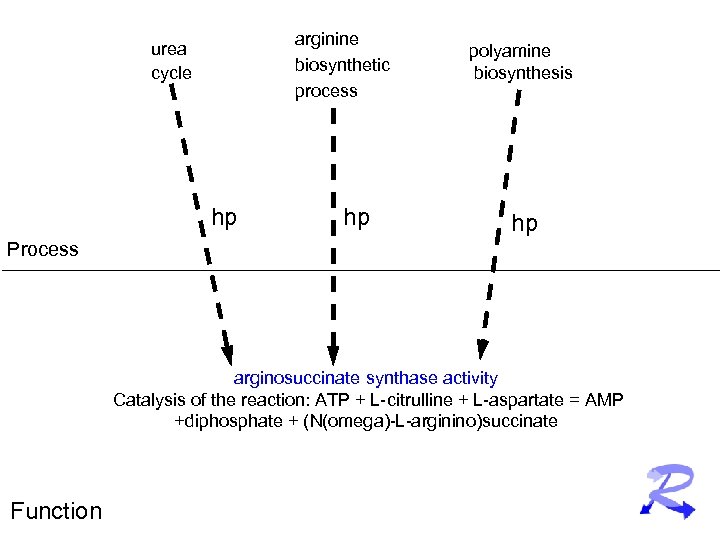
arginine biosynthetic process urea cycle hp hp polyamine biosynthesis hp Process arginosuccinate synthase activity Catalysis of the reaction: ATP + L-citrulline + L-aspartate = AMP +diphosphate + (N(omega)-L-arginino)succinate Function
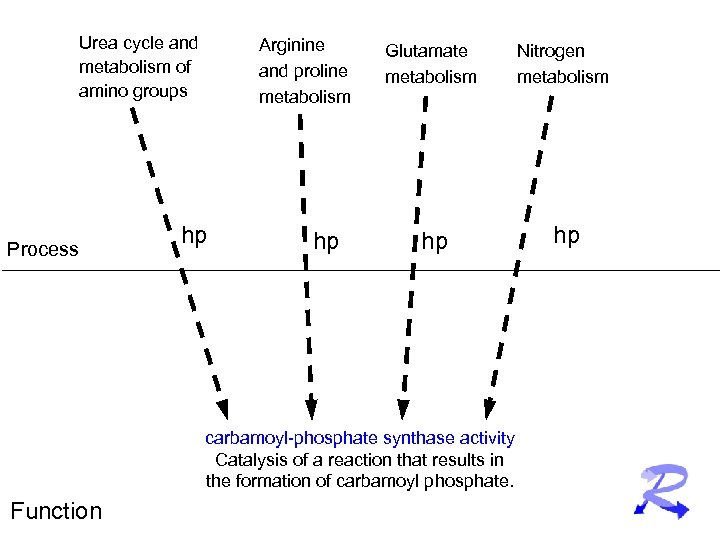
Urea cycle and metabolism of amino groups Process Arginine and proline metabolism hp hp Glutamate metabolism Nitrogen metabolism hp hp carbamoyl-phosphate synthase activity Catalysis of a reaction that results in the formation of carbamoyl phosphate. Function
Lysine biosynthesis pathways
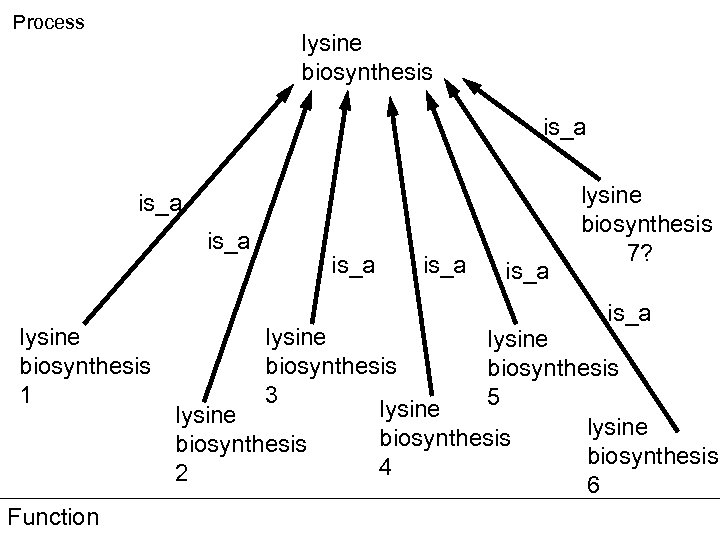
Process lysine biosynthesis is_a lysine biosynthesis 1 Function is_a lysine biosynthesis 7? is_a lysine biosynthesis 3 5 lysine biosynthesis 4 2 6
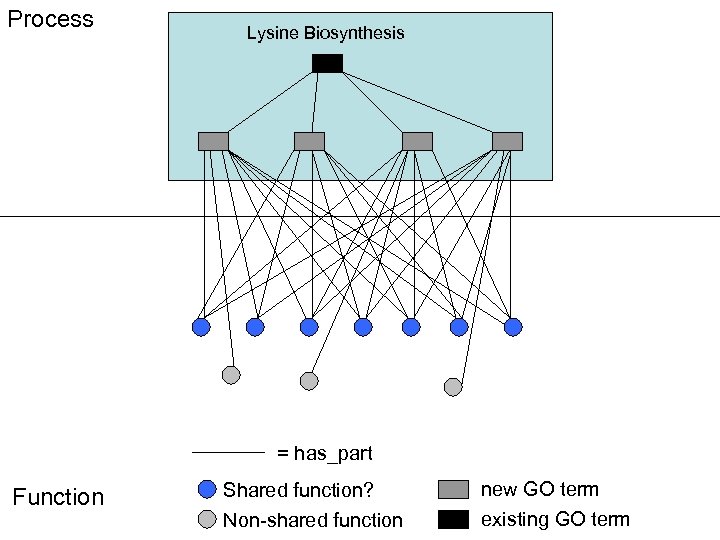
Process Lysine Biosynthesis = has_part Function Shared function? new GO term Non-shared function existing GO term
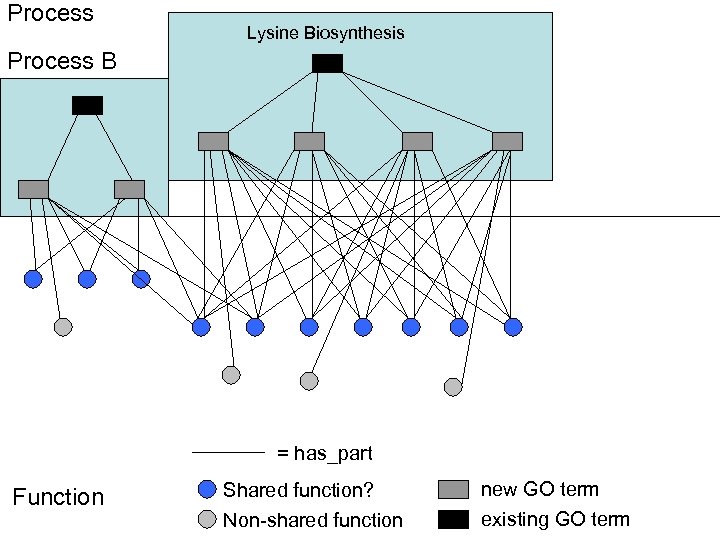
Process Lysine Biosynthesis Process B = has_part Function Shared function? new GO term Non-shared function existing GO term
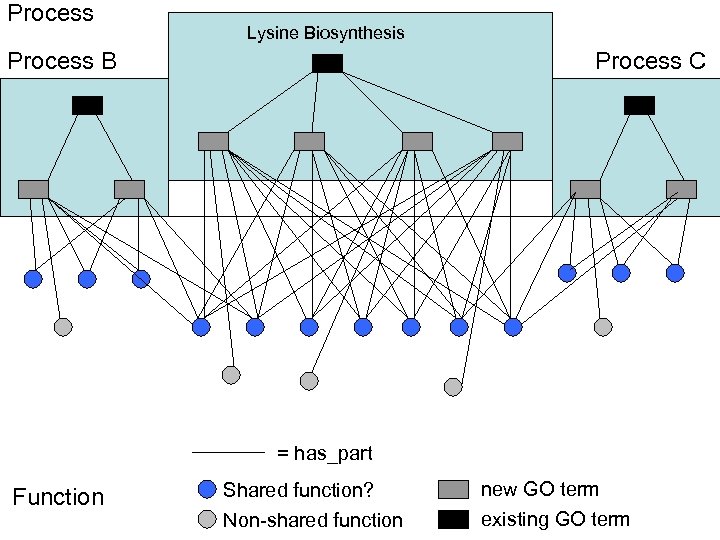
Process Lysine Biosynthesis Process B Process C = has_part Function Shared function? new GO term Non-shared function existing GO term

Process B Function Lysine Biosynthesis Relationship explosion (or Editorial office explosion) Process C
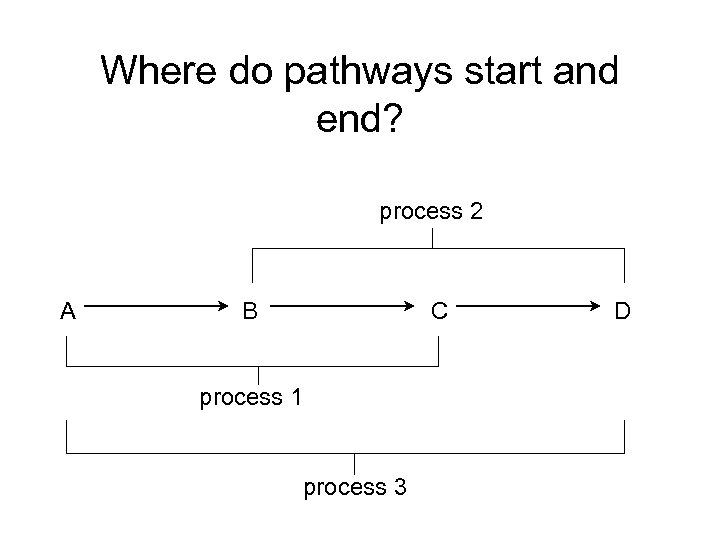
Where do pathways start and end? process 2 A B C process 1 process 3 D

Use cases • Can we slim from function up to process? • Can we infer annotations to process from those to function?
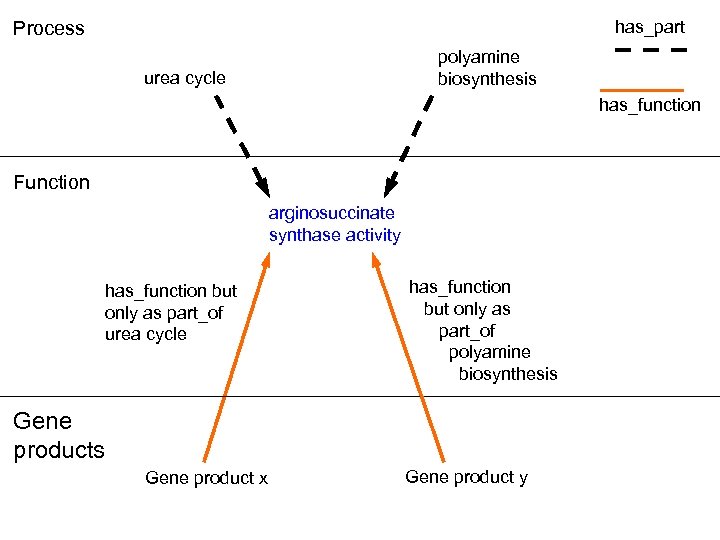
has_part Process polyamine biosynthesis urea cycle has_function Function arginosuccinate synthase activity has_function but only as part_of urea cycle has_function but only as part_of polyamine biosynthesis Gene product x Gene product y
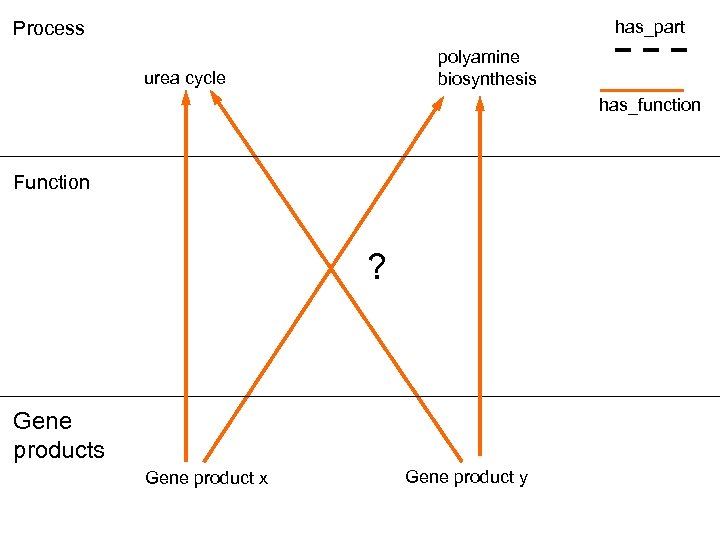
has_part Process polyamine biosynthesis urea cycle has_function Function ? Gene products Gene product x Gene product y
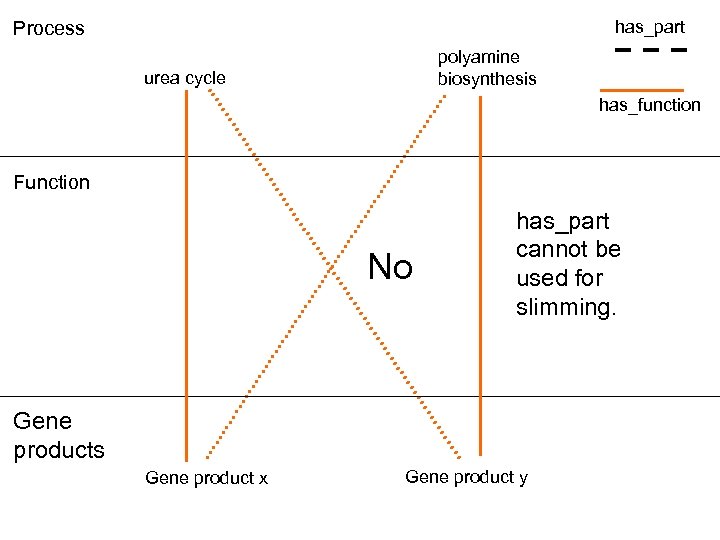
has_part Process polyamine biosynthesis urea cycle has_function Function No has_part cannot be used for slimming. Gene products Gene product x Gene product y

Can we infer annotations to process from those to function? • No. There is too much variation in process details, and too many functions are shared.
So what can we do?
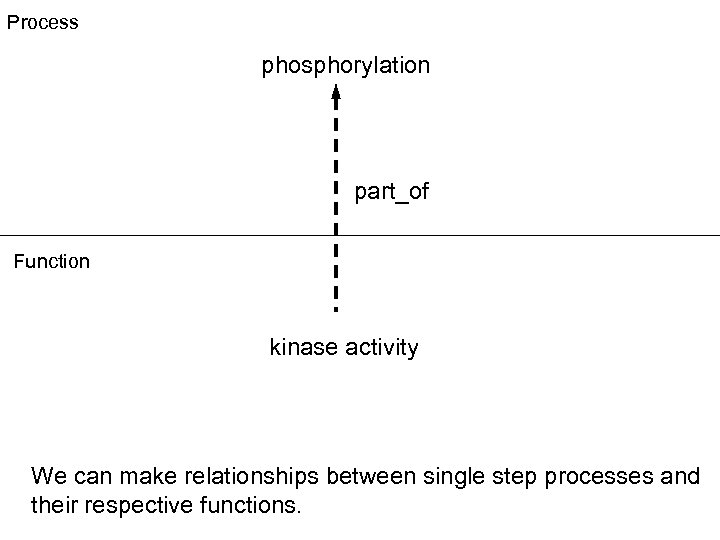
Process phosphorylation part_of Function kinase activity We can make relationships between single step processes and their respective functions.
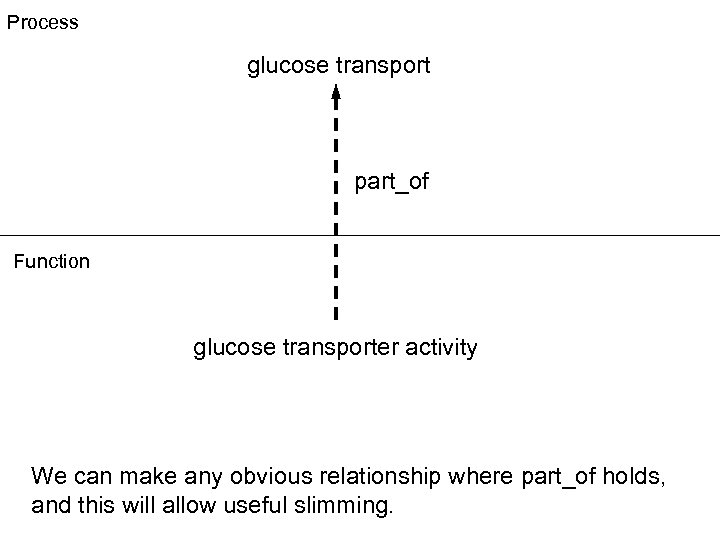
Process glucose transport part_of Function glucose transporter activity We can make any obvious relationship where part_of holds, and this will allow useful slimming.
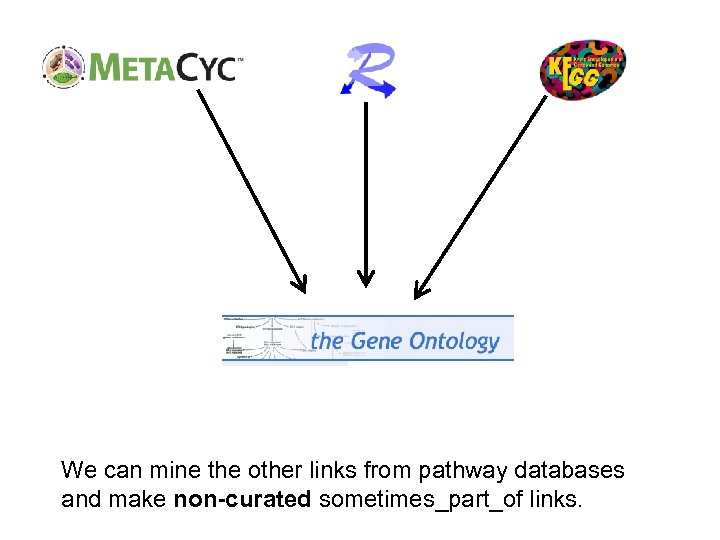
We can mine the other links from pathway databases and make non-curated sometimes_part_of links.

sometimes_part_of What does this buy us? • Very full coverage of function-process links. • No manual link curation. What work does it involve? • We maintain the mapping files e. g. reactome 2 go. • We write the mining scripts. • Work with pathway dbs to unify exchange formats and make data interoperable

Acknowledgements Michelle Gwinn-Giglio Debbie Siegele Ingrid Keseler Harold Drabkin Jennifer Deegan Chris Mungall Peifen Zhang
bc34eb2f8bcc555990803ac47d20d9bb.ppt