5387e9e8aee5bc693020c319a5a01f3d.ppt
- Количество слайдов: 25
 FUGE - Satellite meeting 2005 Structural Genomics Consortium at Karolinska Institute Susanne Gräslund, Head Biotechnology
FUGE - Satellite meeting 2005 Structural Genomics Consortium at Karolinska Institute Susanne Gräslund, Head Biotechnology
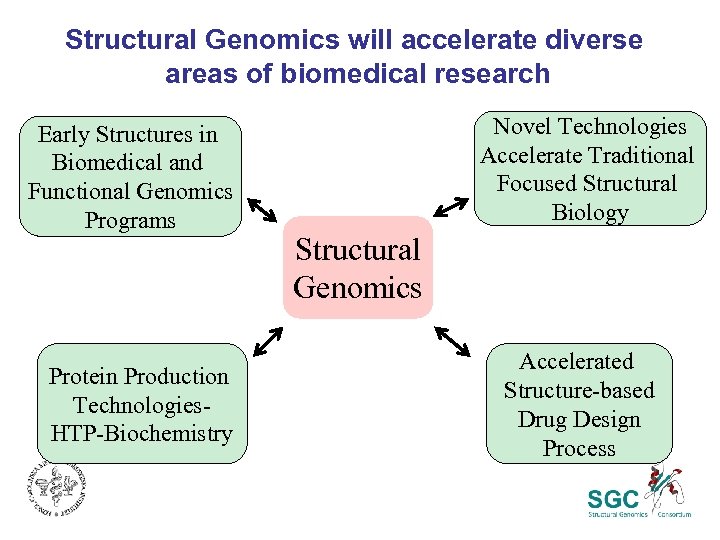 Structural Genomics will accelerate diverse areas of biomedical research Early Structures in Biomedical and Functional Genomics Programs Protein Production Technologies. HTP-Biochemistry Novel Technologies Accelerate Traditional Focused Structural Biology Structural Genomics Accelerated Structure-based Drug Design Process
Structural Genomics will accelerate diverse areas of biomedical research Early Structures in Biomedical and Functional Genomics Programs Protein Production Technologies. HTP-Biochemistry Novel Technologies Accelerate Traditional Focused Structural Biology Structural Genomics Accelerated Structure-based Drug Design Process
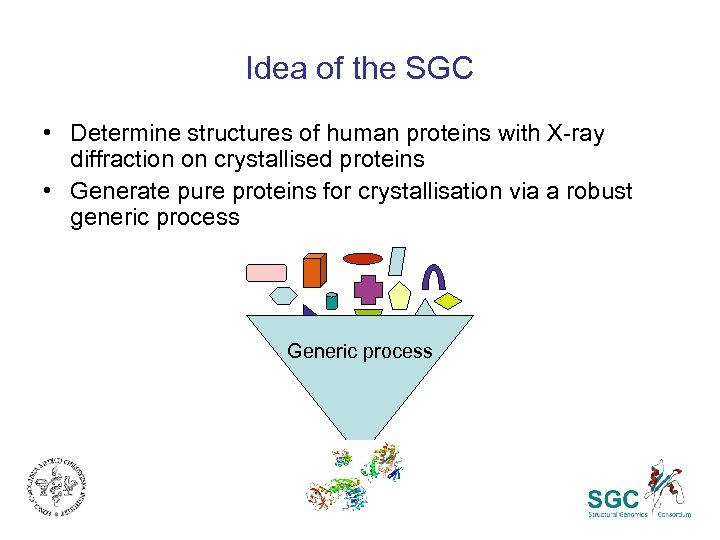 Idea of the SGC • Determine structures of human proteins with X-ray diffraction on crystallised proteins • Generate pure proteins for crystallisation via a robust generic process Generic process
Idea of the SGC • Determine structures of human proteins with X-ray diffraction on crystallised proteins • Generate pure proteins for crystallisation via a robust generic process Generic process
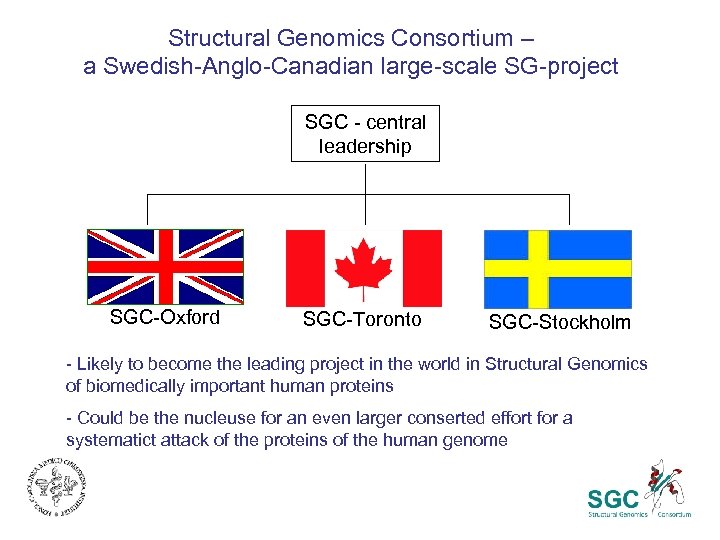 Structural Genomics Consortium – a Swedish-Anglo-Canadian large-scale SG-project SGC - central leadership SGC-Oxford SGC-Toronto SGC-Stockholm - Likely to become the leading project in the world in Structural Genomics of biomedically important human proteins - Could be the nucleuse for an even larger conserted effort for a systematict attack of the proteins of the human genome
Structural Genomics Consortium – a Swedish-Anglo-Canadian large-scale SG-project SGC - central leadership SGC-Oxford SGC-Toronto SGC-Stockholm - Likely to become the leading project in the world in Structural Genomics of biomedically important human proteins - Could be the nucleuse for an even larger conserted effort for a systematict attack of the proteins of the human genome
 The SGC 1999: • Discussions initiated by Rob Cooke @GSK and colleagues • Major goal was to join forces and launch a pre-competitive organisation devoted to “early targets” of medical relevance • >10 Pharma’s and Biotech’s + Wellcome Trust in early discussions, built on SNP consortium model 2001 - 2003: • 2001: Business plan prepared, GSK (~10%) and WT (~90%) committed • 2003: Al Edwards recruited. • 2003: Consortium of Canadian investors committed to project • 2003: Strategic planning initiated in August and Ramp-Up in October
The SGC 1999: • Discussions initiated by Rob Cooke @GSK and colleagues • Major goal was to join forces and launch a pre-competitive organisation devoted to “early targets” of medical relevance • >10 Pharma’s and Biotech’s + Wellcome Trust in early discussions, built on SNP consortium model 2001 - 2003: • 2001: Business plan prepared, GSK (~10%) and WT (~90%) committed • 2003: Al Edwards recruited. • 2003: Consortium of Canadian investors committed to project • 2003: Strategic planning initiated in August and Ramp-Up in October
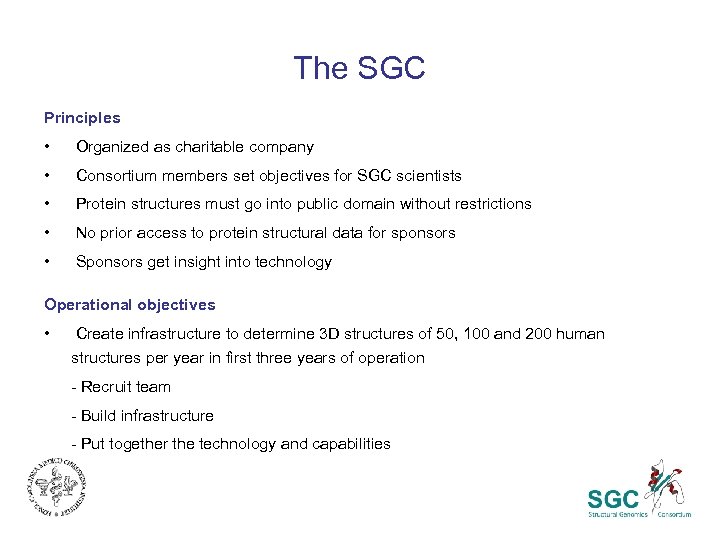 The SGC Principles • Organized as charitable company • Consortium members set objectives for SGC scientists • Protein structures must go into public domain without restrictions • No prior access to protein structural data for sponsors • Sponsors get insight into technology Operational objectives • Create infrastructure to determine 3 D structures of 50, 100 and 200 human structures per year in first three years of operation - Recruit team - Build infrastructure - Put together the technology and capabilities
The SGC Principles • Organized as charitable company • Consortium members set objectives for SGC scientists • Protein structures must go into public domain without restrictions • No prior access to protein structural data for sponsors • Sponsors get insight into technology Operational objectives • Create infrastructure to determine 3 D structures of 50, 100 and 200 human structures per year in first three years of operation - Recruit team - Build infrastructure - Put together the technology and capabilities
 SGC: The first 9 months
SGC: The first 9 months
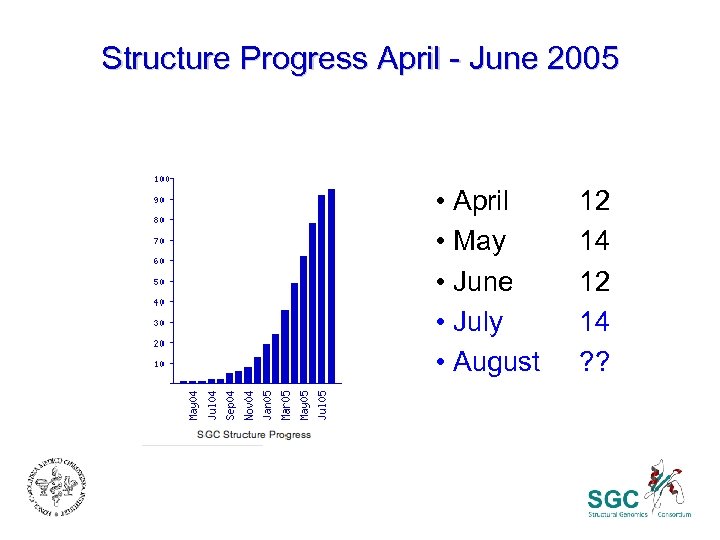 Structure Progress April - June 2005 • April • May • June • July • August 12 14 ? ?
Structure Progress April - June 2005 • April • May • June • July • August 12 14 ? ?
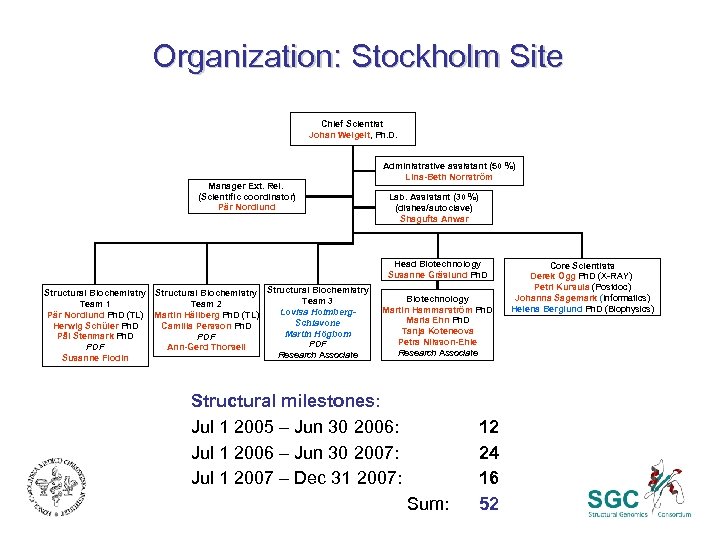 Organization: Stockholm Site Chief Scientist Johan Weigelt, Ph. D. Manager Ext. Rel. (Scientific coordinator) Pär Nordlund Administrative assistant (50 %) Lina-Beth Norrström Lab. Assistant (30 %) (dishes/autoclave) Shagufta Anwar Head Biotechnology Susanne Gräslund Ph. D Structural Biochemistry Team 3 Team 1 Team 2 Lovisa Holmberg. Pär Nordlund Ph. D (TL) Martin Hällberg Ph. D (TL) Schiavone Herwig Schüler Ph. D Camilla Persson Ph. D Martin Högbom Pål Stenmark Ph. D PDF PDF Ann-Gerd Thorsell Research Associate Susanne Flodin Biotechnology Martin Hammarström Ph. D Maria Ehn Ph. D Tanja Koteneova Petra Nilsson-Ehle Research Associate Structural milestones: Jul 1 2005 – Jun 30 2006: Jul 1 2006 – Jun 30 2007: Jul 1 2007 – Dec 31 2007: Sum: 12 24 16 52 Core Scientists Derek Ogg Ph. D (X-RAY) Petri Kursula (Postdoc) Johanna Sagemark (Informatics) Helena Berglund Ph. D (Biophysics)
Organization: Stockholm Site Chief Scientist Johan Weigelt, Ph. D. Manager Ext. Rel. (Scientific coordinator) Pär Nordlund Administrative assistant (50 %) Lina-Beth Norrström Lab. Assistant (30 %) (dishes/autoclave) Shagufta Anwar Head Biotechnology Susanne Gräslund Ph. D Structural Biochemistry Team 3 Team 1 Team 2 Lovisa Holmberg. Pär Nordlund Ph. D (TL) Martin Hällberg Ph. D (TL) Schiavone Herwig Schüler Ph. D Camilla Persson Ph. D Martin Högbom Pål Stenmark Ph. D PDF PDF Ann-Gerd Thorsell Research Associate Susanne Flodin Biotechnology Martin Hammarström Ph. D Maria Ehn Ph. D Tanja Koteneova Petra Nilsson-Ehle Research Associate Structural milestones: Jul 1 2005 – Jun 30 2006: Jul 1 2006 – Jun 30 2007: Jul 1 2007 – Dec 31 2007: Sum: 12 24 16 52 Core Scientists Derek Ogg Ph. D (X-RAY) Petri Kursula (Postdoc) Johanna Sagemark (Informatics) Helena Berglund Ph. D (Biophysics)
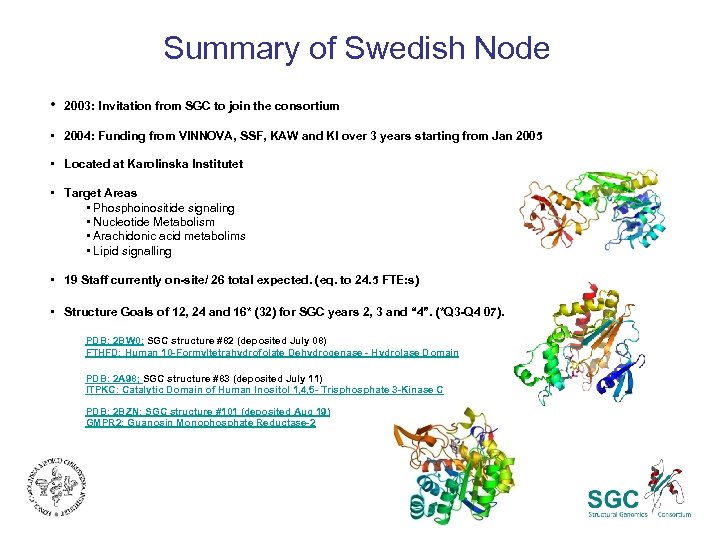 Summary of Swedish Node • 2003: Invitation from SGC to join the consortium • 2004: Funding from VINNOVA, SSF, KAW and KI over 3 years starting from Jan 2005 • Located at Karolinska Institutet • Target Areas • Phosphoinositide signaling • Nucleotide Metabolism • Arachidonic acid metabolims • Lipid signalling • 19 Staff currently on-site/ 26 total expected. (eq. to 24. 5 FTE: s) • Structure Goals of 12, 24 and 16* (32) for SGC years 2, 3 and “ 4”. (*Q 3 -Q 4 07). PDB: 2 BW 0; SGC structure #82 (deposited July 08) FTHFD: Human 10 -Formyltetrahydrofolate Dehydrogenase - Hydrolase Domain PDB: 2 A 98; SGC structure #83 (deposited July 11) ITPKC: Catalytic Domain of Human Inositol 1, 4, 5 - Trisphosphate 3 -Kinase C PDB: 2 BZN; SGC structure #101 (deposited Aug 19) GMPR 2: Guanosin Monophosphate Reductase-2
Summary of Swedish Node • 2003: Invitation from SGC to join the consortium • 2004: Funding from VINNOVA, SSF, KAW and KI over 3 years starting from Jan 2005 • Located at Karolinska Institutet • Target Areas • Phosphoinositide signaling • Nucleotide Metabolism • Arachidonic acid metabolims • Lipid signalling • 19 Staff currently on-site/ 26 total expected. (eq. to 24. 5 FTE: s) • Structure Goals of 12, 24 and 16* (32) for SGC years 2, 3 and “ 4”. (*Q 3 -Q 4 07). PDB: 2 BW 0; SGC structure #82 (deposited July 08) FTHFD: Human 10 -Formyltetrahydrofolate Dehydrogenase - Hydrolase Domain PDB: 2 A 98; SGC structure #83 (deposited July 11) ITPKC: Catalytic Domain of Human Inositol 1, 4, 5 - Trisphosphate 3 -Kinase C PDB: 2 BZN; SGC structure #101 (deposited Aug 19) GMPR 2: Guanosin Monophosphate Reductase-2
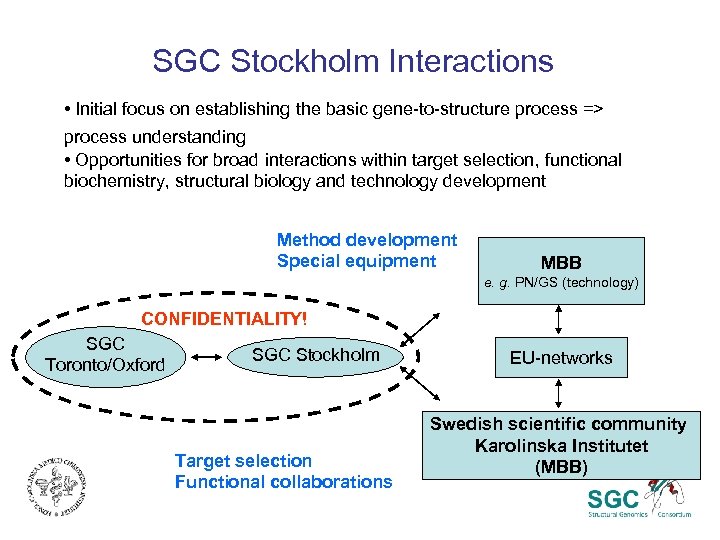 SGC Stockholm Interactions • Initial focus on establishing the basic gene-to-structure process => process understanding • Opportunities for broad interactions within target selection, functional biochemistry, structural biology and technology development Method development Special equipment MBB e. g. PN/GS (technology) CONFIDENTIALITY! SGC Toronto/Oxford SGC Stockholm Target selection Functional collaborations EU-networks Swedish scientific community Karolinska Institutet (MBB)
SGC Stockholm Interactions • Initial focus on establishing the basic gene-to-structure process => process understanding • Opportunities for broad interactions within target selection, functional biochemistry, structural biology and technology development Method development Special equipment MBB e. g. PN/GS (technology) CONFIDENTIALITY! SGC Toronto/Oxford SGC Stockholm Target selection Functional collaborations EU-networks Swedish scientific community Karolinska Institutet (MBB)
 SGC-Stockholm target selection process - Human proteins with high biomedical relevance - No overlap with targets selected in Toronto and Oxford (However, within Target areas is OK) - The SGC Scientific Committee approves/rejects Target area proposals and borderline targets. - Target selection process delegated to the SGC researchers. In Sweden the SAC will guide this work. - SGC-target list confidential until structures are deposited.
SGC-Stockholm target selection process - Human proteins with high biomedical relevance - No overlap with targets selected in Toronto and Oxford (However, within Target areas is OK) - The SGC Scientific Committee approves/rejects Target area proposals and borderline targets. - Target selection process delegated to the SGC researchers. In Sweden the SAC will guide this work. - SGC-target list confidential until structures are deposited.
 Recommendations for target selection - Targets should be selected as structural families or targets with “common chemistry”. - Structural families with members being potential drug targets (“druggable families”) should have priority. - Targets should have a reasonable feasibility for structural studies. - Structural families and biology areas where an SGC in-house expertise is available should have priority.
Recommendations for target selection - Targets should be selected as structural families or targets with “common chemistry”. - Structural families with members being potential drug targets (“druggable families”) should have priority. - Targets should have a reasonable feasibility for structural studies. - Structural families and biology areas where an SGC in-house expertise is available should have priority.
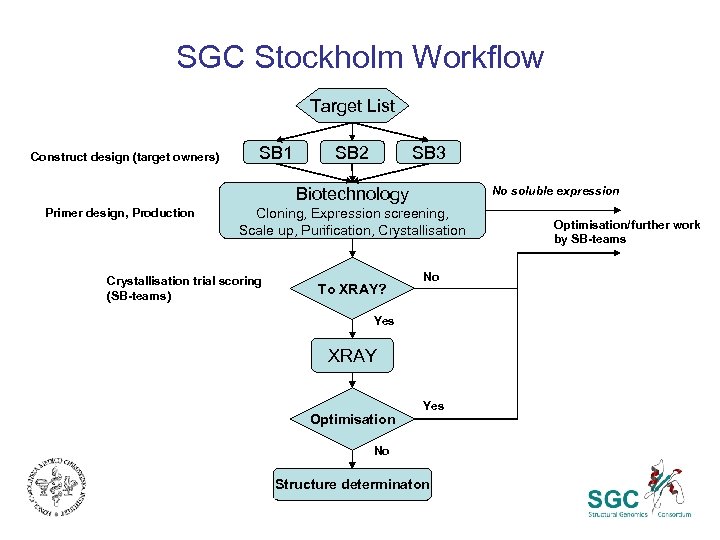 SGC Stockholm Workflow Target List Construct design (target owners) SB 1 SB 2 SB 3 No soluble expression Biotechnology Primer design, Production Cloning, Expression screening, Scale up, Purification, Crystallisation trial scoring (SB-teams) To XRAY? No Yes XRAY Optimisation Yes No Structure determinaton Optimisation/further work by SB-teams
SGC Stockholm Workflow Target List Construct design (target owners) SB 1 SB 2 SB 3 No soluble expression Biotechnology Primer design, Production Cloning, Expression screening, Scale up, Purification, Crystallisation trial scoring (SB-teams) To XRAY? No Yes XRAY Optimisation Yes No Structure determinaton Optimisation/further work by SB-teams
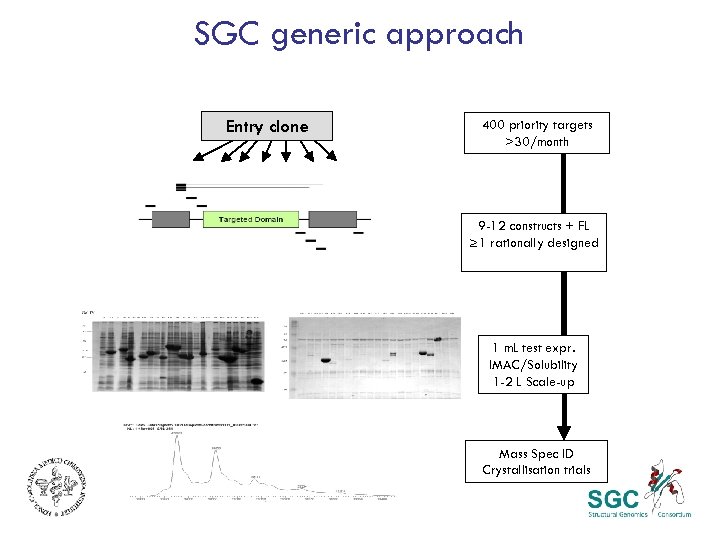 SGC generic approach Entry clone 400 priority targets >30/month 9 -12 constructs + FL ≥ 1 rationally designed 1 m. L test expr. IMAC/Solubility 1 -2 L Scale-up Mass Spec ID Crystallisation trials
SGC generic approach Entry clone 400 priority targets >30/month 9 -12 constructs + FL ≥ 1 rationally designed 1 m. L test expr. IMAC/Solubility 1 -2 L Scale-up Mass Spec ID Crystallisation trials
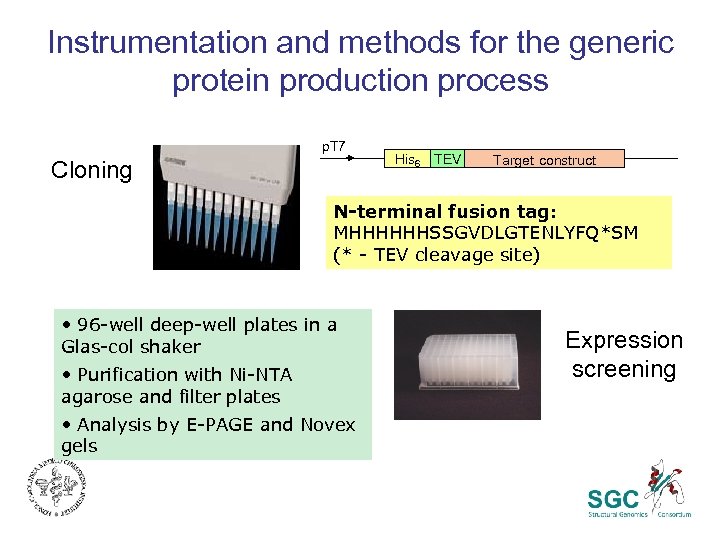 Instrumentation and methods for the generic protein production process p. T 7 Cloning His 6 TEV Target construct N-terminal fusion tag: MHHHHHHSSGVDLGTENLYFQ*SM (* - TEV cleavage site) • 96 -well deep-well plates in a Glas-col shaker • Purification with Ni-NTA agarose and filter plates • Analysis by E-PAGE and Novex gels Expression screening
Instrumentation and methods for the generic protein production process p. T 7 Cloning His 6 TEV Target construct N-terminal fusion tag: MHHHHHHSSGVDLGTENLYFQ*SM (* - TEV cleavage site) • 96 -well deep-well plates in a Glas-col shaker • Purification with Ni-NTA agarose and filter plates • Analysis by E-PAGE and Novex gels Expression screening
 Scale-up methods and instrumentation Cultivation • Scale-up cultivations performed in Tunair flasks • 2*750 ml Terrific Broth per construct • Two-step purification of 12 sampes per run, IMAC + GF • Phosphate buffers in IMAC step and HEPES in GF • Analysis by SDS-PAGE, pooling and concentration Purification
Scale-up methods and instrumentation Cultivation • Scale-up cultivations performed in Tunair flasks • 2*750 ml Terrific Broth per construct • Two-step purification of 12 sampes per run, IMAC + GF • Phosphate buffers in IMAC step and HEPES in GF • Analysis by SDS-PAGE, pooling and concentration Purification
 Crystallisation and X-ray • Crystallisation is performed either manually or with a robotic system (Phoenix) • Rock. Maker: keep track, view and score the crystallisation experiments that are placed into the crystal plate hotels/imaging system • Inhouse X-ray source: screening of crystals and collecting of datasets • High-resulotion datasets will also be collected at different synchrotrons
Crystallisation and X-ray • Crystallisation is performed either manually or with a robotic system (Phoenix) • Rock. Maker: keep track, view and score the crystallisation experiments that are placed into the crystal plate hotels/imaging system • Inhouse X-ray source: screening of crystals and collecting of datasets • High-resulotion datasets will also be collected at different synchrotrons
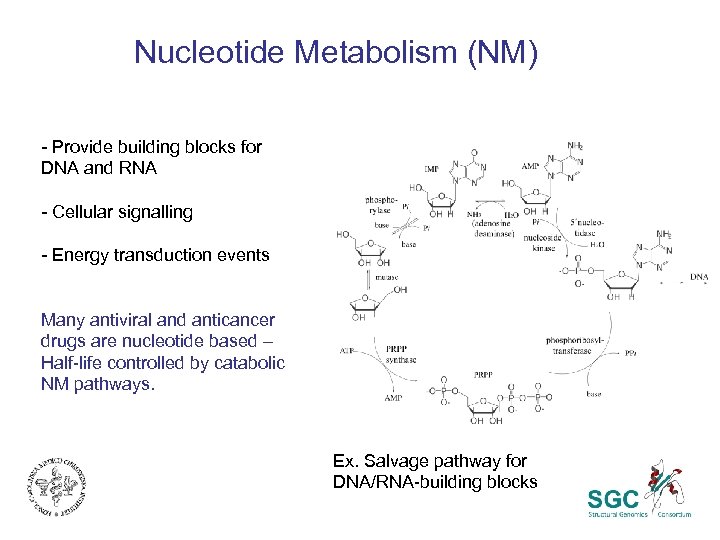 Nucleotide Metabolism (NM) - Provide building blocks for DNA and RNA - Cellular signalling - Energy transduction events Many antiviral and anticancer drugs are nucleotide based – Half-life controlled by catabolic NM pathways. Ex. Salvage pathway for DNA/RNA-building blocks
Nucleotide Metabolism (NM) - Provide building blocks for DNA and RNA - Cellular signalling - Energy transduction events Many antiviral and anticancer drugs are nucleotide based – Half-life controlled by catabolic NM pathways. Ex. Salvage pathway for DNA/RNA-building blocks
 Nucleotide Metabolism – Biomedical relevance - Deoxyribonucleotide synthesis pathways – e. g. ribonucleotide reductase, dihydrofolate reductase or thymidine synthase. - Enzymes in NM major cause of degradation of nucleotide based anti-cancer and anti-HIV drugs. - Mutations in a number of NM enzymes case diseases such as neurogastrointestinal encephalomyopathy (MNGIE) causing severe combined immuno-deficiency (SCID)
Nucleotide Metabolism – Biomedical relevance - Deoxyribonucleotide synthesis pathways – e. g. ribonucleotide reductase, dihydrofolate reductase or thymidine synthase. - Enzymes in NM major cause of degradation of nucleotide based anti-cancer and anti-HIV drugs. - Mutations in a number of NM enzymes case diseases such as neurogastrointestinal encephalomyopathy (MNGIE) causing severe combined immuno-deficiency (SCID)
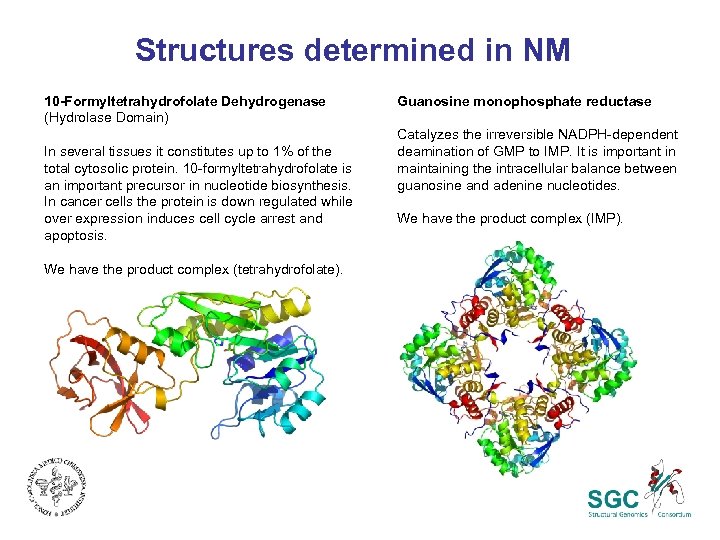 Structures determined in NM 10 -Formyltetrahydrofolate Dehydrogenase (Hydrolase Domain) In several tissues it constitutes up to 1% of the total cytosolic protein. 10 -formyltetrahydrofolate is an important precursor in nucleotide biosynthesis. In cancer cells the protein is down regulated while over expression induces cell cycle arrest and apoptosis. We have the product complex (tetrahydrofolate). Guanosine monophosphate reductase Catalyzes the irreversible NADPH-dependent deamination of GMP to IMP. It is important in maintaining the intracellular balance between guanosine and adenine nucleotides. We have the product complex (IMP).
Structures determined in NM 10 -Formyltetrahydrofolate Dehydrogenase (Hydrolase Domain) In several tissues it constitutes up to 1% of the total cytosolic protein. 10 -formyltetrahydrofolate is an important precursor in nucleotide biosynthesis. In cancer cells the protein is down regulated while over expression induces cell cycle arrest and apoptosis. We have the product complex (tetrahydrofolate). Guanosine monophosphate reductase Catalyzes the irreversible NADPH-dependent deamination of GMP to IMP. It is important in maintaining the intracellular balance between guanosine and adenine nucleotides. We have the product complex (IMP).
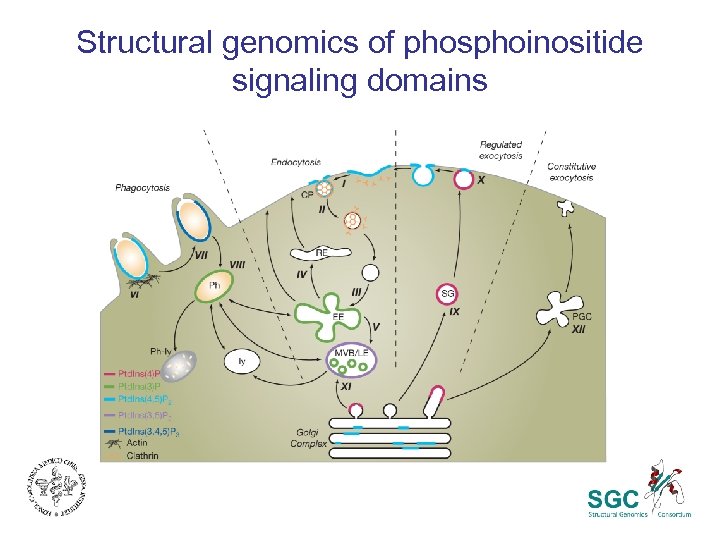 Structural genomics of phosphoinositide signaling domains
Structural genomics of phosphoinositide signaling domains
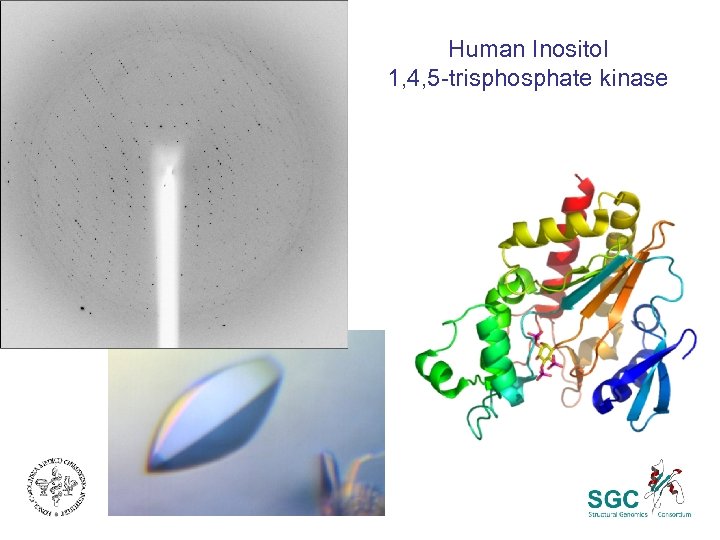 Human Inositol 1, 4, 5 -trisphosphate kinase
Human Inositol 1, 4, 5 -trisphosphate kinase
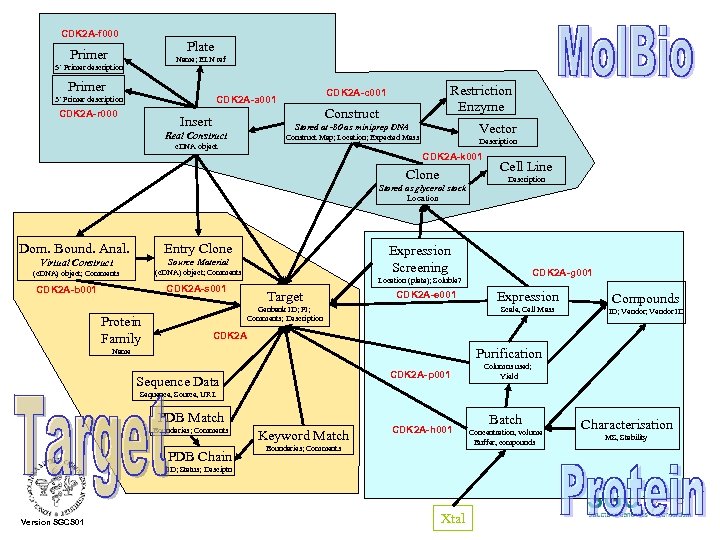 CDK 2 A-f 000 Plate Primer Name; ELN ref 5’ Primer description Primer CDK 2 A-r 000 Restriction Enzyme CDK 2 A-c 001 CDK 2 A-a 001 3’ Primer description Construct Insert Vector Stored at -80 as miniprep DNA Real Construct Map; Location; Expected Mass c. DNA object Description CDK 2 A-k 001 Clone Stored as glycerol stock Location Dom. Bound. Anal. Entry Clone Virtual Construct Source Material Protein Family CDK 2 A-g 001 Location (plate); Soluble? CDK 2 A-s 001 CDK 2 A-b 001 Description Expression Screening (c. DNA) object; Comments Cell Line Target CDK 2 A-e 001 Genbank ID; PI; Comments; Description Expression Scale, Cell Mass Compounds ID; Vendor ID CDK 2 A Purification Name CDK 2 A-p 001 Sequence Data Columns used; Yield Sequence, Source, URL PDB Match Boundaries; Comments PDB Chain Keyword Match CDK 2 A-h 001 Boundaries; Comments ID; Status; Desciptn Version SGCS 01 Xtal Batch Concentration, volume Buffer, compounds Characterisation MS, Stability
CDK 2 A-f 000 Plate Primer Name; ELN ref 5’ Primer description Primer CDK 2 A-r 000 Restriction Enzyme CDK 2 A-c 001 CDK 2 A-a 001 3’ Primer description Construct Insert Vector Stored at -80 as miniprep DNA Real Construct Map; Location; Expected Mass c. DNA object Description CDK 2 A-k 001 Clone Stored as glycerol stock Location Dom. Bound. Anal. Entry Clone Virtual Construct Source Material Protein Family CDK 2 A-g 001 Location (plate); Soluble? CDK 2 A-s 001 CDK 2 A-b 001 Description Expression Screening (c. DNA) object; Comments Cell Line Target CDK 2 A-e 001 Genbank ID; PI; Comments; Description Expression Scale, Cell Mass Compounds ID; Vendor ID CDK 2 A Purification Name CDK 2 A-p 001 Sequence Data Columns used; Yield Sequence, Source, URL PDB Match Boundaries; Comments PDB Chain Keyword Match CDK 2 A-h 001 Boundaries; Comments ID; Status; Desciptn Version SGCS 01 Xtal Batch Concentration, volume Buffer, compounds Characterisation MS, Stability
 Thank you for your attention ! And thanks to VINNOVA, SSF, KAW and KI for funding
Thank you for your attention ! And thanks to VINNOVA, SSF, KAW and KI for funding


