d056f18e3b78a9bd3fa01ad776fdcd26.ppt
- Количество слайдов: 64
 From Gene to Protein
From Gene to Protein
 Question? u. How does DNA control a cell? u. By controlling Protein Synthesis. u. Proteins are the link between genotype and phenotype.
Question? u. How does DNA control a cell? u. By controlling Protein Synthesis. u. Proteins are the link between genotype and phenotype.
 Central Dogma DNA Transcription RNA Translation Polypeptide
Central Dogma DNA Transcription RNA Translation Polypeptide
 Explanation u. DNA - the Genetic code or genotype. u. RNA - the message or instructions. u. Polypeptide - the product for the phenotype.
Explanation u. DNA - the Genetic code or genotype. u. RNA - the message or instructions. u. Polypeptide - the product for the phenotype.
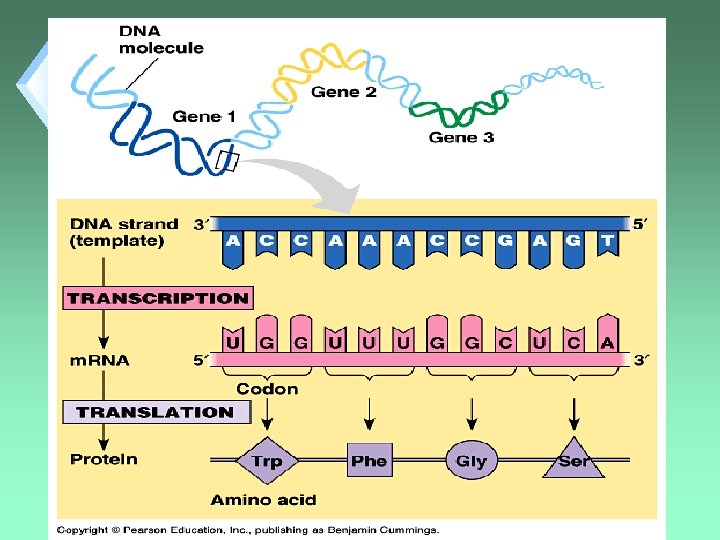
 Genetic Code u. Sequence of DNA bases that describe which Amino Acid to place in what order in a polypeptide. u. The genetic code gives the primary protein structure.
Genetic Code u. Sequence of DNA bases that describe which Amino Acid to place in what order in a polypeptide. u. The genetic code gives the primary protein structure.
 Genetic Code u. Is based on triplets of bases. u. Has redundancy; some AA's have more than 1 code. u 20 amino acids: 64 codons
Genetic Code u. Is based on triplets of bases. u. Has redundancy; some AA's have more than 1 code. u 20 amino acids: 64 codons
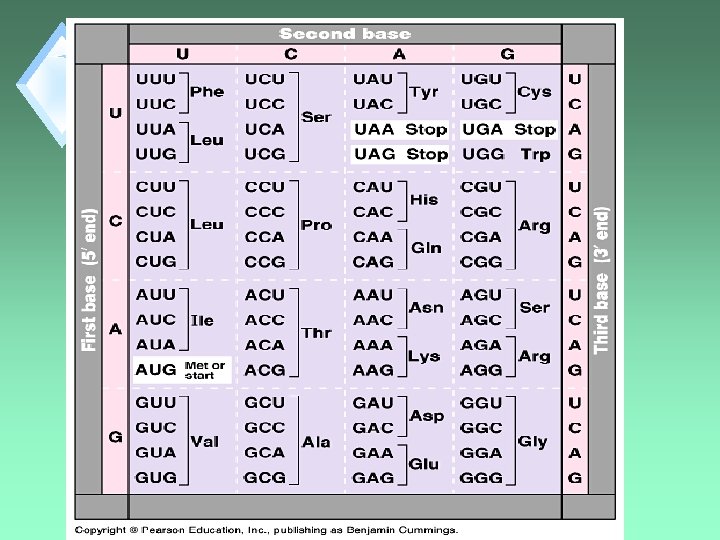
 Code Redundancy u. Third base in a codon shows "wobble”. u. First two bases are the most important in reading the code and giving the correct AA. The third base often doesn’t matter.
Code Redundancy u. Third base in a codon shows "wobble”. u. First two bases are the most important in reading the code and giving the correct AA. The third base often doesn’t matter.
 Code Evolution u. The genetic code is nearly universal. u. Ex: CCG = proline (all life) u. Reason - The code must have evolved very early. Life on earth must share a common ancestor.
Code Evolution u. The genetic code is nearly universal. u. Ex: CCG = proline (all life) u. Reason - The code must have evolved very early. Life on earth must share a common ancestor.
 Reading Frame and Frame Shift u. The “reading” of the code is every three bases (Reading Frame) u. Ex: the red cat ate the rat u. Frame shift – improper groupings of the bases u. Ex: thr edc ata tat her at u. The “words” only make sense if “read” in this grouping of three.
Reading Frame and Frame Shift u. The “reading” of the code is every three bases (Reading Frame) u. Ex: the red cat ate the rat u. Frame shift – improper groupings of the bases u. Ex: thr edc ata tat her at u. The “words” only make sense if “read” in this grouping of three.
 Transcription u. Process of making RNA from a DNA template. u. Only one strand is used as a template.
Transcription u. Process of making RNA from a DNA template. u. Only one strand is used as a template.
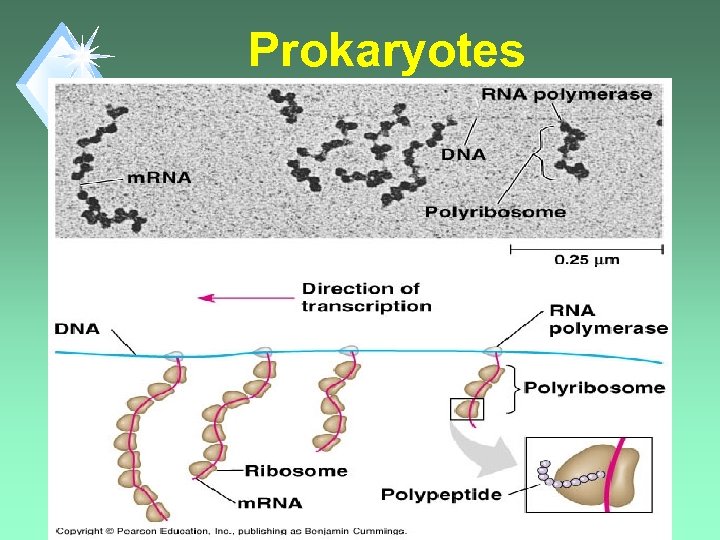
 Where in the cell does transcription take place? u. Eukaryotes: nucleus u. Prokaryotes: cytoplasm
Where in the cell does transcription take place? u. Eukaryotes: nucleus u. Prokaryotes: cytoplasm
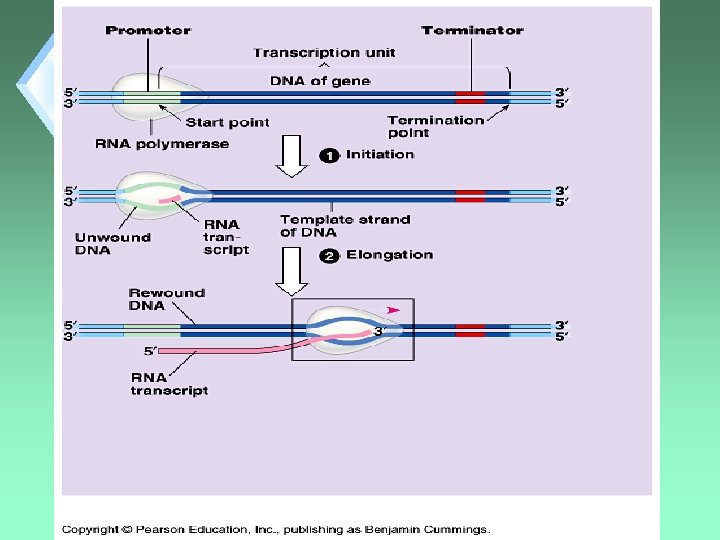
 Transcription Steps 1. 2. 3. 4. RNA Polymerase Binding Initiation Elongation Termination
Transcription Steps 1. 2. 3. 4. RNA Polymerase Binding Initiation Elongation Termination
 RNA Polymerase u. Enzyme for building RNA from RNA nucleotides.
RNA Polymerase u. Enzyme for building RNA from RNA nucleotides.
 Binding u. Requires that the enzyme find the “proper” place on the DNA to attach and start transcription.
Binding u. Requires that the enzyme find the “proper” place on the DNA to attach and start transcription.
 Binding u. Is a complicated process u. Uses Promoter Regions (start region) on the DNA (upstream from the information for the protein)
Binding u. Is a complicated process u. Uses Promoter Regions (start region) on the DNA (upstream from the information for the protein)
 Initiation u. Actual unwinding of DNA to start RNA synthesis.
Initiation u. Actual unwinding of DNA to start RNA synthesis.
 Elongation u. RNA Polymerase untwists DNA 1 turn at a time. u. Exposes 10 DNA bases for pairing with RNA nucleotides.
Elongation u. RNA Polymerase untwists DNA 1 turn at a time. u. Exposes 10 DNA bases for pairing with RNA nucleotides.
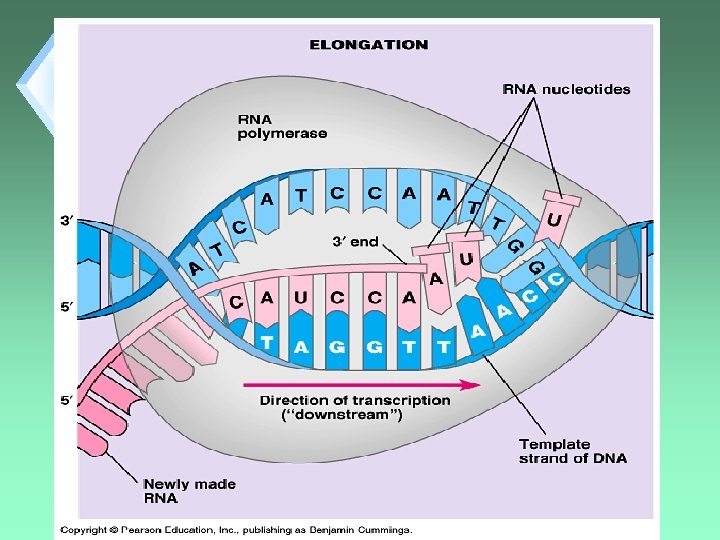
 Elongation u. Enzyme moves 5’ 3’. u. Rate is about 60 nucleotides per second.
Elongation u. Enzyme moves 5’ 3’. u. Rate is about 60 nucleotides per second.
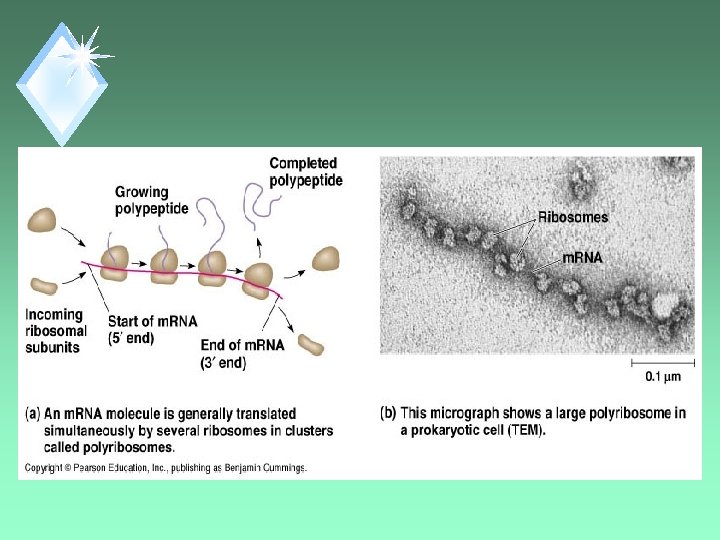
 Comment u. Each gene can be read by sequential RNA Polymerases giving several copies of RNA. u. Result - several copies of the protein can be made.
Comment u. Each gene can be read by sequential RNA Polymerases giving several copies of RNA. u. Result - several copies of the protein can be made.
 Termination u. DNA sequence that tells RNA Polymerase to stop. u. Ex: AATAAA u. RNA Polymerase detaches from DNA after closing the helix.
Termination u. DNA sequence that tells RNA Polymerase to stop. u. Ex: AATAAA u. RNA Polymerase detaches from DNA after closing the helix.
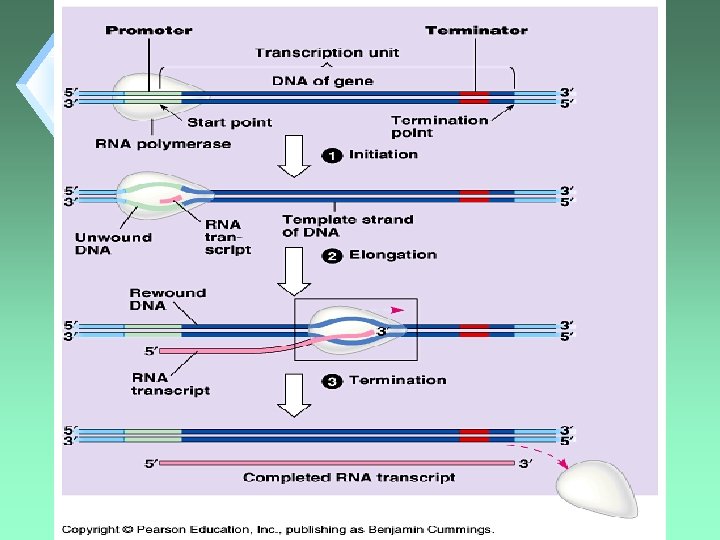
 Final Product u. Pre-m. RNA u. This is a “raw” RNA that will need processing.
Final Product u. Pre-m. RNA u. This is a “raw” RNA that will need processing.
 Modifications of RNA 1. 5’ Cap 2. Poly-A Tail 3. Splicing
Modifications of RNA 1. 5’ Cap 2. Poly-A Tail 3. Splicing
 5' Cap u. Modified Guanine nucleotide added to the 5' end. u. Protects m. RNA from digestive enzymes. u. Recognition sign for ribosome attachment.
5' Cap u. Modified Guanine nucleotide added to the 5' end. u. Protects m. RNA from digestive enzymes. u. Recognition sign for ribosome attachment.
 Poly-A Tail u 150 -200 Adenine nucleotides added to the 3' tail u. Protects m. RNA from digestive enzymes. u. Aids in m. RNA transport from nucleus.
Poly-A Tail u 150 -200 Adenine nucleotides added to the 3' tail u. Protects m. RNA from digestive enzymes. u. Aids in m. RNA transport from nucleus.
 RNA Splicing u. Removal of non-protein coding regions of RNA. u. Coding regions are then spliced back together.
RNA Splicing u. Removal of non-protein coding regions of RNA. u. Coding regions are then spliced back together.
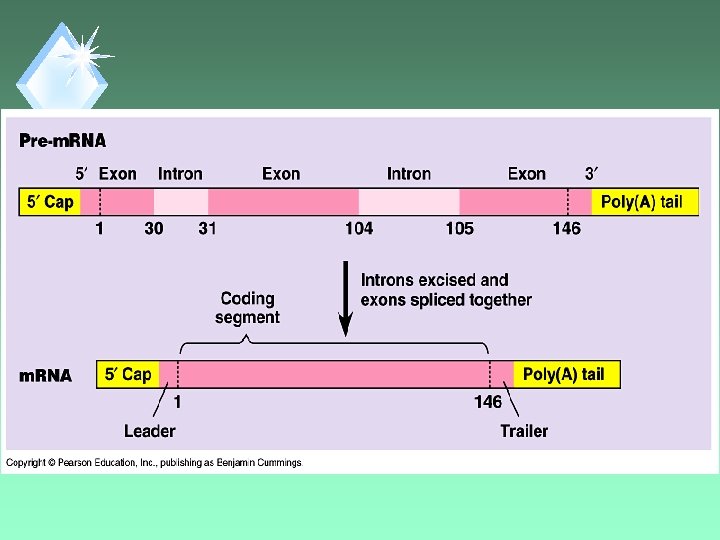
 Introns u. Intervening sequences. u. Removed from RNA.
Introns u. Intervening sequences. u. Removed from RNA.
 Exons u. Expressed sequences of RNA. u. Translated into AAs.
Exons u. Expressed sequences of RNA. u. Translated into AAs.
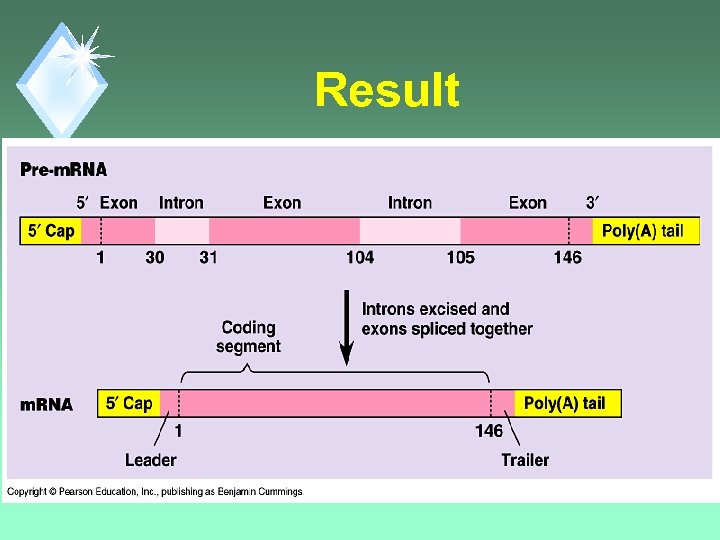 Result
Result
 Introns - Function u. Left-over DNA (? ) u. Way to lengthen genetic message. u. Old virus inserts (? ) u. Way to create new proteins.
Introns - Function u. Left-over DNA (? ) u. Way to lengthen genetic message. u. Old virus inserts (? ) u. Way to create new proteins.
 Translation u. Process by which a cell interprets a genetic message and builds a polypeptide.
Translation u. Process by which a cell interprets a genetic message and builds a polypeptide.
 Where in the cell does translation take place? u. Eukaryotes and prokaryotes: cytoplasm because that’s where the ribosomes are located.
Where in the cell does translation take place? u. Eukaryotes and prokaryotes: cytoplasm because that’s where the ribosomes are located.
 Materials Required ut. RNA u. Ribosomes um. RNA
Materials Required ut. RNA u. Ribosomes um. RNA
 Transfer RNA = t. RNA u. Made by transcription. u. About 80 nucleotides long. u Carries AA for polypeptide synthesis.
Transfer RNA = t. RNA u. Made by transcription. u. About 80 nucleotides long. u Carries AA for polypeptide synthesis.
 Structure of t. RNA u. Has double stranded regions and 3 loops. u. AA attachment site at the 3' end. u 1 loop serves as the Anticodon.
Structure of t. RNA u. Has double stranded regions and 3 loops. u. AA attachment site at the 3' end. u 1 loop serves as the Anticodon.
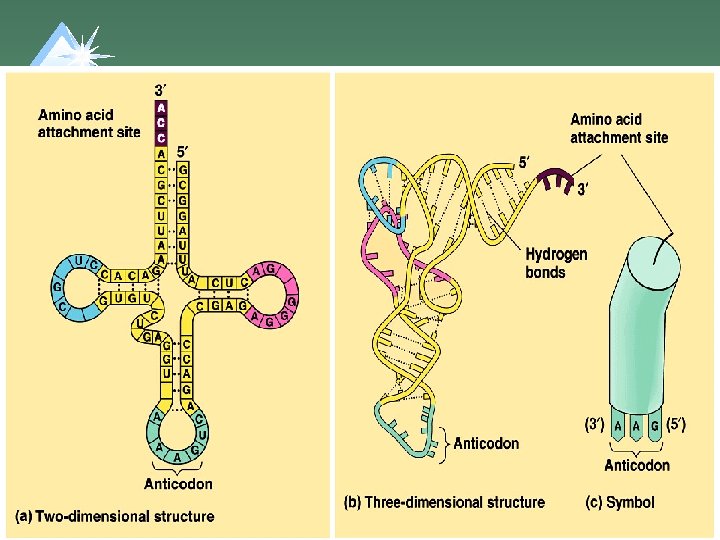
 Anticodon u. Region of t. RNA that base pairs to m. RNA codon. u. Usually is a compliment to the m. RNA bases, so reads the same as the DNA codon.
Anticodon u. Region of t. RNA that base pairs to m. RNA codon. u. Usually is a compliment to the m. RNA bases, so reads the same as the DNA codon.
 Example u. DNA - GAC um. RNA - CUG ut. RNA anticodon - GAC
Example u. DNA - GAC um. RNA - CUG ut. RNA anticodon - GAC
 Ribosomes u. Two subunits made in the nucleolus. u. Made of r. RNA (60%)and protein (40%). ur. RNA is the most abundant type of RNA in a cell.
Ribosomes u. Two subunits made in the nucleolus. u. Made of r. RNA (60%)and protein (40%). ur. RNA is the most abundant type of RNA in a cell.
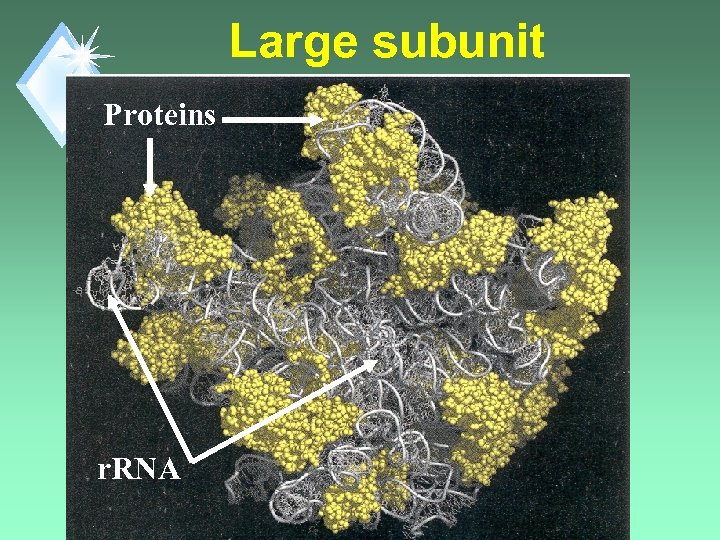 Large subunit Proteins r. RNA
Large subunit Proteins r. RNA
 Both sununits
Both sununits
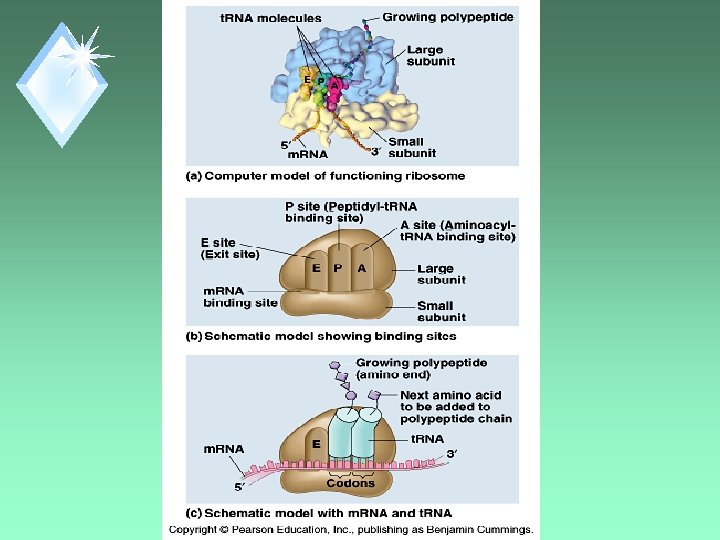
 Large Subunit u. Has 3 sites for t. RNA. u. P site: Peptidyl-t. RNA site carries the growing polypeptide chain. u A site: Aminoacyl-t. RNA site holds the t. RNA carrying the next AA to be added. u. E site: Exit site
Large Subunit u. Has 3 sites for t. RNA. u. P site: Peptidyl-t. RNA site carries the growing polypeptide chain. u A site: Aminoacyl-t. RNA site holds the t. RNA carrying the next AA to be added. u. E site: Exit site
 Translation Steps 1. Initiation 2. Elongation 3. Termination
Translation Steps 1. Initiation 2. Elongation 3. Termination
 Initiation u. Brings together: um. RNA u. A t. RNA carrying the 1 st AA u 2 subunits of the ribosome
Initiation u. Brings together: um. RNA u. A t. RNA carrying the 1 st AA u 2 subunits of the ribosome
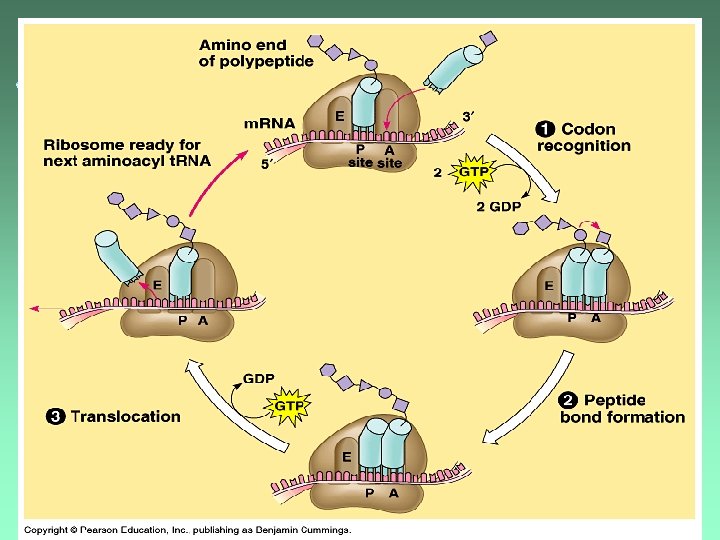
 Elongation Steps: 1. Codon Recognition 2. Peptide Bond Formation 3. Translocation
Elongation Steps: 1. Codon Recognition 2. Peptide Bond Formation 3. Translocation
 Codon Recognition ut. RNA anticodon matched to m. RNA codon in the A site.
Codon Recognition ut. RNA anticodon matched to m. RNA codon in the A site.
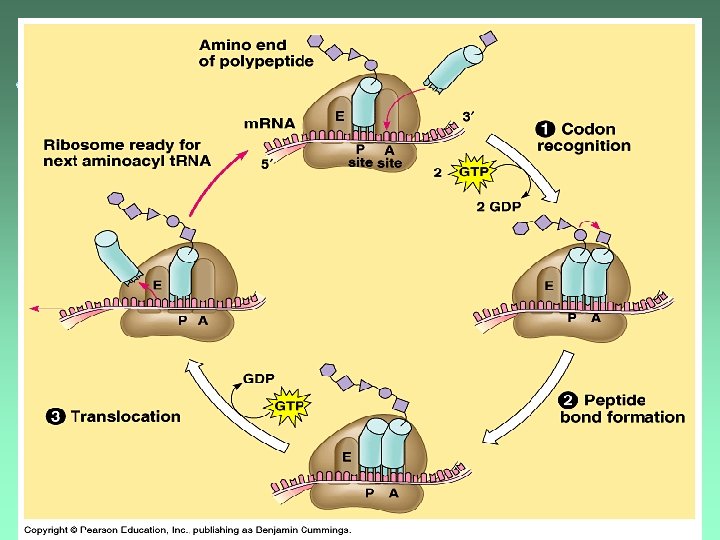
 Peptide Bond Formation u. A peptide bond is formed between the new AA and the polypeptide chain in the P -site.
Peptide Bond Formation u. A peptide bond is formed between the new AA and the polypeptide chain in the P -site.
 After bond formation u. The polypeptide is now transferred from the t. RNA in the P-site to the t. RNA in the A -site.
After bond formation u. The polypeptide is now transferred from the t. RNA in the P-site to the t. RNA in the A -site.
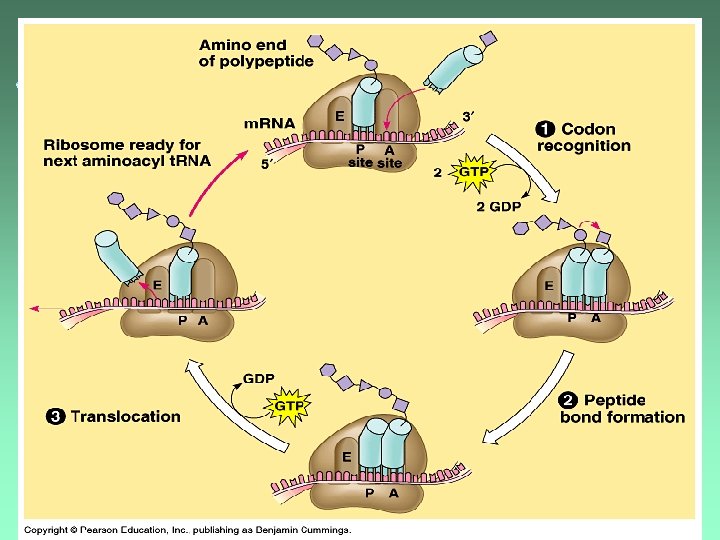
 Translocation ut. RNA in P-site is released. u. Ribosome advances 1 codon ut. RNA in A-site is now in the P -site. u. Process repeats with the next codon.
Translocation ut. RNA in P-site is released. u. Ribosome advances 1 codon ut. RNA in A-site is now in the P -site. u. Process repeats with the next codon.
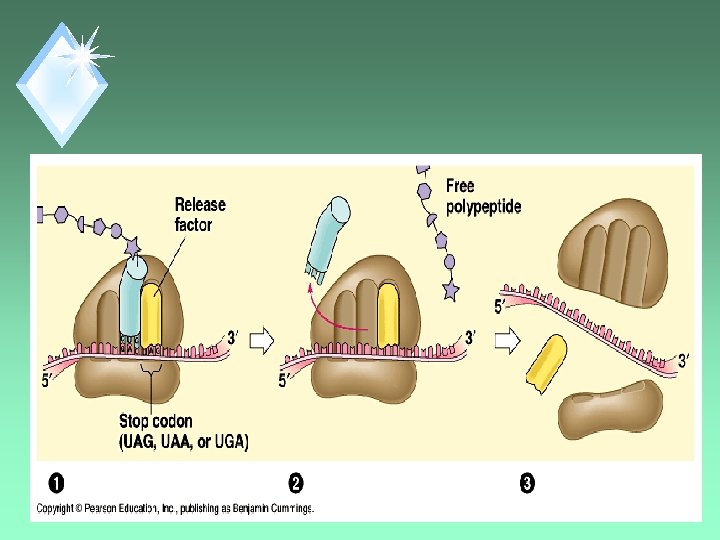
 Termination u. Triggered by stop codons. u. Release factor binds in the A-site instead of a t. RNA. u H 2 O is added instead of AA, freeing the polypeptide. u. Ribosome separates.
Termination u. Triggered by stop codons. u. Release factor binds in the A-site instead of a t. RNA. u H 2 O is added instead of AA, freeing the polypeptide. u. Ribosome separates.
 Prokaryotes
Prokaryotes
 Comment u. Polypeptide usually needs to be modified before it becomes functional.
Comment u. Polypeptide usually needs to be modified before it becomes functional.
 Examples u. Sugars, lipids, phosphate groups added. u. Some AAs removed. u. Protein may be cleaved. u. Join polypeptides together (Quaternary Structure).
Examples u. Sugars, lipids, phosphate groups added. u. Some AAs removed. u. Protein may be cleaved. u. Join polypeptides together (Quaternary Structure).


