b98810724f35e2d9d8f50ac3a9262a5a.ppt
- Количество слайдов: 1
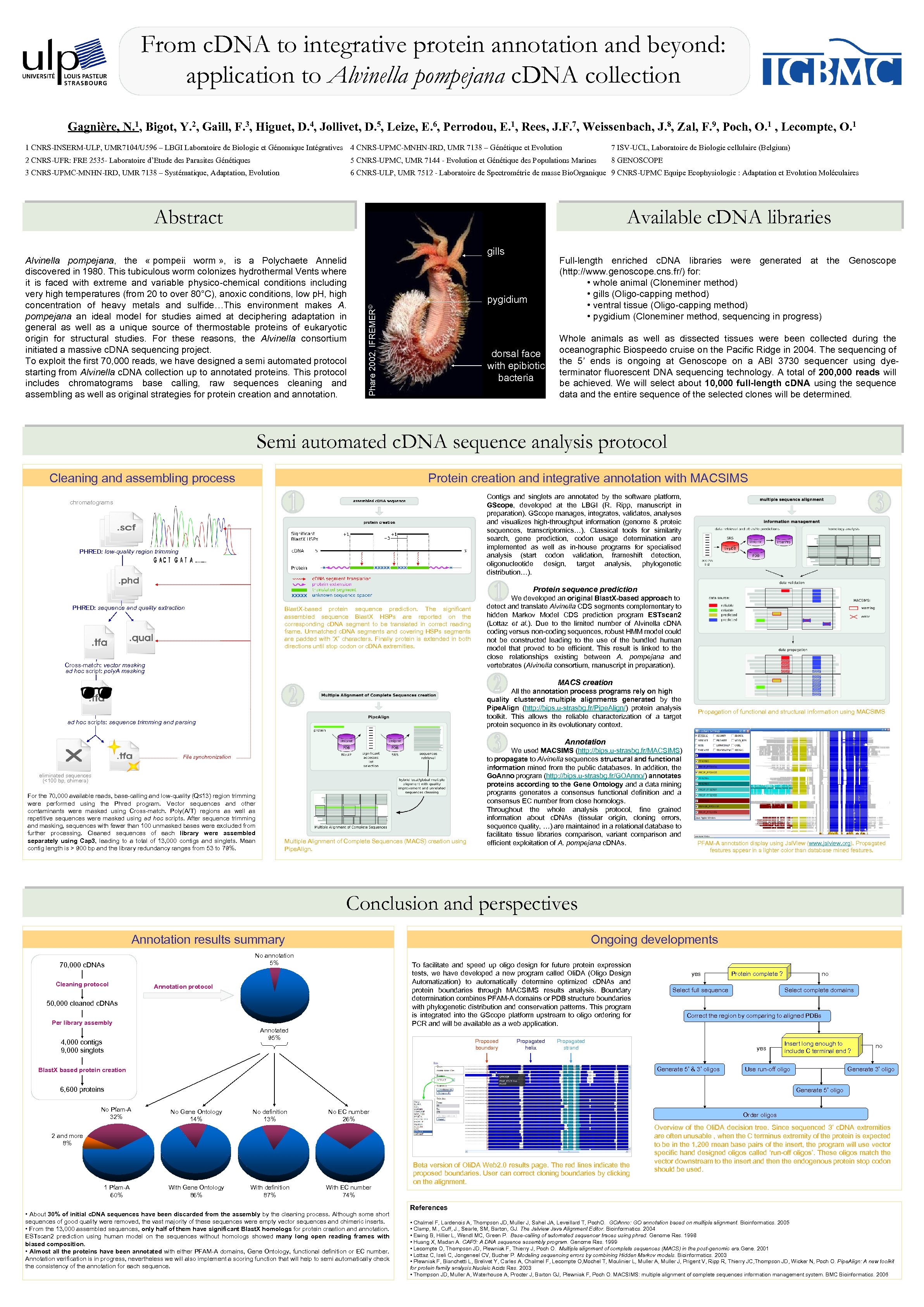 From c. DNA to integrative protein annotation and beyond: application to Alvinella pompejana c. DNA collection Gagnière, N. 1, Bigot, Y. 2, Gaill, F. 3, Higuet, D. 4, Jollivet, D. 5, Leize, E. 6, Perrodou, E. 1, Rees, J. F. 7, Weissenbach, J. 8, Zal, F. 9, Poch, O. 1 , Lecompte, O. 1 1 CNRS-INSERM-ULP, UMR 7104/U 596 – LBGI Laboratoire de Biologie et Génomique Intégratives 4 CNRS-UPMC-MNHN-IRD, UMR 7138 – Génétique et Evolution 7 ISV-UCL, Laboratoire de Biologie cellulaire (Belgium) 2 CNRS-UFR: FRE 2535 - Laboratoire d’Etude des Parasites Génétiques 5 CNRS-UPMC, UMR 7144 - Evolution et Génétique des Populations Marines 8 GENOSCOPE 3 CNRS-UPMC-MNHN-IRD, UMR 7138 – Systématique, Adaptation, Evolution 6 CNRS-ULP, UMR 7512 - Laboratoire de Spectrométrie de masse Bio. Organique 9 CNRS-UPMC Equipe Ecophysiologie : Adaptation et Evolution Moléculaires Abstract Available c. DNA libraries Full-length enriched c. DNA libraries were generated at the Genoscope (http: //www. genoscope. cns. fr/) for: • whole animal (Cloneminer method) • gills (Oligo-capping method) • ventral tissue (Oligo-capping method) • pygidium (Cloneminer method, sequencing in progress) pygidium Phare 2002, IFREMER© Alvinella pompejana, the « pompeii worm » , is a Polychaete Annelid discovered in 1980. This tubiculous worm colonizes hydrothermal Vents where it is faced with extreme and variable physico-chemical conditions including very high temperatures (from 20 to over 80°C), anoxic conditions, low p. H, high concentration of heavy metals and sulfide…This environment makes A. pompejana an ideal model for studies aimed at deciphering adaptation in general as well as a unique source of thermostable proteins of eukaryotic origin for structural studies. For these reasons, the Alvinella consortium initiated a massive c. DNA sequencing project. To exploit the first 70, 000 reads, we have designed a semi automated protocol starting from Alvinella c. DNA collection up to annotated proteins. This protocol includes chromatograms base calling, raw sequences cleaning and assembling as well as original strategies for protein creation and annotation. gills dorsal face with epibiotic bacteria Whole animals as well as dissected tissues were been collected during the oceanographic Biospeedo cruise on the Pacific Ridge in 2004. The sequencing of the 5’ ends is ongoing at Genoscope on a ABI 3730 sequencer using dyeterminator fluorescent DNA sequencing technology. A total of 200, 000 reads will be achieved. We will select about 10, 000 full-length c. DNA using the sequence data and the entire sequence of the selected clones will be determined. Semi automated c. DNA sequence analysis protocol Cleaning and assembling process Protein creation and integrative annotation with MACSIMS Contigs and singlets are annotated by the software platform, GScope, developed at the LBGI (R. Ripp, manuscript in preparation). GScope manages, integrates, validates, analyses and visualizes high-throughput information (genome & proteic sequences, transcriptomics…). Classical tools for similarity search, gene prediction, codon usage determination are implemented as well as in-house programs for specialised analysis (start codon validation, frameshift detection, oligonucleotide design, target analysis, phylogenetic distribution…). chromatograms PHRED: low-quality region trimming Protein sequence prediction PHRED: sequence and quality extraction Blast. X-based protein sequence prediction. The significant assembled sequence Blast. X HSPs are reported on the corresponding c. DNA segment to be translated in correct reading frame. Unmatched c. DNA segments and covering HSPs segments are padded with ‘X’ characters. Finally protein is extended in both directions until stop codon or c. DNA extremities. Cross-match: vector masking ad hoc script: poly. A masking We developed an original Blast. X-based approach to detect and translate Alvinella CDS segments complementary to hidden Markov Model CDS prediction program ESTscan 2 (Lottaz et al. ). Due to the limited number of Alvinella c. DNA coding versus non-coding sequences, robust HMM model could not be constructed leading to the use of the bundled human model that proved to be efficient. This result is linked to the close relationships existing between A. pompejana and vertebrates (Alvinella consortium, manuscript in preparation). MACS creation All the annotation process programs rely on high quality clustered multiple alignments generated by the Pipe. Align (http: //bips. u-strasbg. fr/Pipe. Align/) protein analysis toolkit. This allows the reliable characterization of a target protein sequence in its evolutionary context. ad hoc scripts: sequence trimming and parsing Propagation of functional and structural information using MACSIMS Annotation File synchronization eliminated sequences (<100 bp, chimera) For the 70, 000 available reads, base-calling and low-quality (Q≤ 13) region trimming were performed using the Phred program. Vector sequences and other contaminants were masked using Cross-match. Poly(A/T) regions as well as repetitive sequences were masked using ad hoc scripts. After sequence trimming and masking, sequences with fewer than 100 unmasked bases were excluded from further processing. Cleaned sequences of each library were assembled separately using Cap 3, leading to a total of 13, 000 contigs and singlets. Mean contig length is > 900 bp and the library redundancy ranges from 53 to 79%. Multiple Alignment of Complete Sequences (MACS) creation using Pipe. Align. We used MACSIMS (http: //bips. u-strasbg. fr/MACSIMS) to propagate to Alvinella sequences structural and functional information mined from the public databases. In addition, the Go. Anno program (http: //bips. u-strasbg. fr/GOAnno/) annotates proteins according to the Gene Ontology and a data mining programs generates a consensus functional definition and a consensus EC number from close homologs. Throughout the whole analysis protocol, fine grained information about c. DNAs (tissular origin, cloning errors, sequence quality, …) are maintained in a relational database to facilitate tissue libraries comparison, variant comparison and efficient exploitation of A. pompejana c. DNAs. PFAM-A annotation display using Jal. View (www. jalview. org). Propagated features appear in a lighter color than database mined features. Conclusion and perspectives Annotation results summary No annotation 5% 70, 000 c. DNAs Cleaning protocol Ongoing developments To facilitate and speed up oligo design for future protein expression tests, we have developed a new program called Oli. DA (Oligo Design Automatization) to automatically determine optimized c. DNAs and protein boundaries through MACSIMS results analysis. Boundary determination combines PFAM-A domains or PDB structure boundaries with phylogenetic distribution and conservation patterns. This program is integrated into the GScope platform upstream to oligo ordering for PCR and will be available as a web application. Annotation protocol 50, 000 cleaned c. DNAs Per library assembly Annotated 95% 4, 000 contigs 9, 000 singlets Proposed boundary Propagated helix yes Select full sequence Select complete domains yes Generate 5’ & 3’ oligos no Generate 3’ oligo Generate 5’ oligo No Gene Ontology 14% No definition 13% No EC number 26% Order oligos 2 and more 8% 1 Pfam-A 60% Insert long enough to include C terminal end ? Use run-off oligo 6, 600 proteins No Pfam-A 32% no Correct the region by comparing to aligned PDBs Propagated strand Blast. X based protein creation Protein complete ? With Gene Ontology 86% With definition 87% With EC number 74% • About 30% of initial c. DNA sequences have been discarded from the assembly by the cleaning process. Although some short sequences of good quality were removed, the vast majority of these sequences were empty vector sequences and chimeric inserts. • From the 13, 000 assembled sequences, only half of them have significant Blast. X homologs for protein creation and annotation. ESTscan 2 prediction using human model on the sequences without homologs showed many long open reading frames with biased composition. • Almost all the proteins have been annotated with either PFAM-A domains, Gene Ontology, functional definition or EC number. Annotation verification is in progress, nevertheless we will also implement a scoring function that will help to semi automatically check the consistency of the annotation for each sequence. Beta version of Oli. DA Web 2. 0 results page. The red lines indicate the proposed boundaries. User can correct cloning boundaries by clicking on the alignment. Overview of the Oli. DA decision tree. Since sequenced 3’ c. DNA extremities are often unusable , when the C terminus extremity of the protein is expected to be in the 1, 200 mean base pairs of the insert, the program will use vector specific hand designed oligos called ‘run-off oligos’. These oligos match the vector downstream to the insert and then the endogenous protein stop codon should be used. References • Chalmel F, Lardenois A, Thompson JD, Muller J, Sahel JA, Leveillard T, Poch. O. GOAnno: GO annotation based on multiple alignment. Bioinformatics. 2005 • Clamp, M. , Cuff, J. , Searle, SM, Barton, GJ. The Jalview Java Alignment Editor. Bioinformatics. 2004 • Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. Genome Res. 1998 • Huang X, Madan A. CAP 3: A DNA sequence assembly program. Genome Res. 1999 • Lecompte O, Thompson JD, Plewniak F, Thierry J, Poch O. Multiple alignment of complete sequences (MACS) in the post-genomic era. Gene. 2001 • Lottaz C, Iseli C, Jongeneel CV, Bucher P. Modeling sequencing errors by combining Hidden Markov models. Bioinformatics. 2003 • Plewniak F, Bianchetti L, Brelivet Y, Carles A, Chalmel F, Lecompte O, Mochel T, Moulinier L, Muller A, Muller J, Prigent V, Ripp R, Thierry JC, Thompson JD, Wicker N, Poch O. Pipe. Align: A new toolkit for protein family analysis. Nucleic Acids Res. 2003 • Thompson JD, Muller A, Waterhouse A, Procter J, Barton GJ, Plewniak F, Poch O. MACSIMS: multiple alignment of complete sequences information management system. BMC Bioinformatics. 2006
From c. DNA to integrative protein annotation and beyond: application to Alvinella pompejana c. DNA collection Gagnière, N. 1, Bigot, Y. 2, Gaill, F. 3, Higuet, D. 4, Jollivet, D. 5, Leize, E. 6, Perrodou, E. 1, Rees, J. F. 7, Weissenbach, J. 8, Zal, F. 9, Poch, O. 1 , Lecompte, O. 1 1 CNRS-INSERM-ULP, UMR 7104/U 596 – LBGI Laboratoire de Biologie et Génomique Intégratives 4 CNRS-UPMC-MNHN-IRD, UMR 7138 – Génétique et Evolution 7 ISV-UCL, Laboratoire de Biologie cellulaire (Belgium) 2 CNRS-UFR: FRE 2535 - Laboratoire d’Etude des Parasites Génétiques 5 CNRS-UPMC, UMR 7144 - Evolution et Génétique des Populations Marines 8 GENOSCOPE 3 CNRS-UPMC-MNHN-IRD, UMR 7138 – Systématique, Adaptation, Evolution 6 CNRS-ULP, UMR 7512 - Laboratoire de Spectrométrie de masse Bio. Organique 9 CNRS-UPMC Equipe Ecophysiologie : Adaptation et Evolution Moléculaires Abstract Available c. DNA libraries Full-length enriched c. DNA libraries were generated at the Genoscope (http: //www. genoscope. cns. fr/) for: • whole animal (Cloneminer method) • gills (Oligo-capping method) • ventral tissue (Oligo-capping method) • pygidium (Cloneminer method, sequencing in progress) pygidium Phare 2002, IFREMER© Alvinella pompejana, the « pompeii worm » , is a Polychaete Annelid discovered in 1980. This tubiculous worm colonizes hydrothermal Vents where it is faced with extreme and variable physico-chemical conditions including very high temperatures (from 20 to over 80°C), anoxic conditions, low p. H, high concentration of heavy metals and sulfide…This environment makes A. pompejana an ideal model for studies aimed at deciphering adaptation in general as well as a unique source of thermostable proteins of eukaryotic origin for structural studies. For these reasons, the Alvinella consortium initiated a massive c. DNA sequencing project. To exploit the first 70, 000 reads, we have designed a semi automated protocol starting from Alvinella c. DNA collection up to annotated proteins. This protocol includes chromatograms base calling, raw sequences cleaning and assembling as well as original strategies for protein creation and annotation. gills dorsal face with epibiotic bacteria Whole animals as well as dissected tissues were been collected during the oceanographic Biospeedo cruise on the Pacific Ridge in 2004. The sequencing of the 5’ ends is ongoing at Genoscope on a ABI 3730 sequencer using dyeterminator fluorescent DNA sequencing technology. A total of 200, 000 reads will be achieved. We will select about 10, 000 full-length c. DNA using the sequence data and the entire sequence of the selected clones will be determined. Semi automated c. DNA sequence analysis protocol Cleaning and assembling process Protein creation and integrative annotation with MACSIMS Contigs and singlets are annotated by the software platform, GScope, developed at the LBGI (R. Ripp, manuscript in preparation). GScope manages, integrates, validates, analyses and visualizes high-throughput information (genome & proteic sequences, transcriptomics…). Classical tools for similarity search, gene prediction, codon usage determination are implemented as well as in-house programs for specialised analysis (start codon validation, frameshift detection, oligonucleotide design, target analysis, phylogenetic distribution…). chromatograms PHRED: low-quality region trimming Protein sequence prediction PHRED: sequence and quality extraction Blast. X-based protein sequence prediction. The significant assembled sequence Blast. X HSPs are reported on the corresponding c. DNA segment to be translated in correct reading frame. Unmatched c. DNA segments and covering HSPs segments are padded with ‘X’ characters. Finally protein is extended in both directions until stop codon or c. DNA extremities. Cross-match: vector masking ad hoc script: poly. A masking We developed an original Blast. X-based approach to detect and translate Alvinella CDS segments complementary to hidden Markov Model CDS prediction program ESTscan 2 (Lottaz et al. ). Due to the limited number of Alvinella c. DNA coding versus non-coding sequences, robust HMM model could not be constructed leading to the use of the bundled human model that proved to be efficient. This result is linked to the close relationships existing between A. pompejana and vertebrates (Alvinella consortium, manuscript in preparation). MACS creation All the annotation process programs rely on high quality clustered multiple alignments generated by the Pipe. Align (http: //bips. u-strasbg. fr/Pipe. Align/) protein analysis toolkit. This allows the reliable characterization of a target protein sequence in its evolutionary context. ad hoc scripts: sequence trimming and parsing Propagation of functional and structural information using MACSIMS Annotation File synchronization eliminated sequences (<100 bp, chimera) For the 70, 000 available reads, base-calling and low-quality (Q≤ 13) region trimming were performed using the Phred program. Vector sequences and other contaminants were masked using Cross-match. Poly(A/T) regions as well as repetitive sequences were masked using ad hoc scripts. After sequence trimming and masking, sequences with fewer than 100 unmasked bases were excluded from further processing. Cleaned sequences of each library were assembled separately using Cap 3, leading to a total of 13, 000 contigs and singlets. Mean contig length is > 900 bp and the library redundancy ranges from 53 to 79%. Multiple Alignment of Complete Sequences (MACS) creation using Pipe. Align. We used MACSIMS (http: //bips. u-strasbg. fr/MACSIMS) to propagate to Alvinella sequences structural and functional information mined from the public databases. In addition, the Go. Anno program (http: //bips. u-strasbg. fr/GOAnno/) annotates proteins according to the Gene Ontology and a data mining programs generates a consensus functional definition and a consensus EC number from close homologs. Throughout the whole analysis protocol, fine grained information about c. DNAs (tissular origin, cloning errors, sequence quality, …) are maintained in a relational database to facilitate tissue libraries comparison, variant comparison and efficient exploitation of A. pompejana c. DNAs. PFAM-A annotation display using Jal. View (www. jalview. org). Propagated features appear in a lighter color than database mined features. Conclusion and perspectives Annotation results summary No annotation 5% 70, 000 c. DNAs Cleaning protocol Ongoing developments To facilitate and speed up oligo design for future protein expression tests, we have developed a new program called Oli. DA (Oligo Design Automatization) to automatically determine optimized c. DNAs and protein boundaries through MACSIMS results analysis. Boundary determination combines PFAM-A domains or PDB structure boundaries with phylogenetic distribution and conservation patterns. This program is integrated into the GScope platform upstream to oligo ordering for PCR and will be available as a web application. Annotation protocol 50, 000 cleaned c. DNAs Per library assembly Annotated 95% 4, 000 contigs 9, 000 singlets Proposed boundary Propagated helix yes Select full sequence Select complete domains yes Generate 5’ & 3’ oligos no Generate 3’ oligo Generate 5’ oligo No Gene Ontology 14% No definition 13% No EC number 26% Order oligos 2 and more 8% 1 Pfam-A 60% Insert long enough to include C terminal end ? Use run-off oligo 6, 600 proteins No Pfam-A 32% no Correct the region by comparing to aligned PDBs Propagated strand Blast. X based protein creation Protein complete ? With Gene Ontology 86% With definition 87% With EC number 74% • About 30% of initial c. DNA sequences have been discarded from the assembly by the cleaning process. Although some short sequences of good quality were removed, the vast majority of these sequences were empty vector sequences and chimeric inserts. • From the 13, 000 assembled sequences, only half of them have significant Blast. X homologs for protein creation and annotation. ESTscan 2 prediction using human model on the sequences without homologs showed many long open reading frames with biased composition. • Almost all the proteins have been annotated with either PFAM-A domains, Gene Ontology, functional definition or EC number. Annotation verification is in progress, nevertheless we will also implement a scoring function that will help to semi automatically check the consistency of the annotation for each sequence. Beta version of Oli. DA Web 2. 0 results page. The red lines indicate the proposed boundaries. User can correct cloning boundaries by clicking on the alignment. Overview of the Oli. DA decision tree. Since sequenced 3’ c. DNA extremities are often unusable , when the C terminus extremity of the protein is expected to be in the 1, 200 mean base pairs of the insert, the program will use vector specific hand designed oligos called ‘run-off oligos’. These oligos match the vector downstream to the insert and then the endogenous protein stop codon should be used. References • Chalmel F, Lardenois A, Thompson JD, Muller J, Sahel JA, Leveillard T, Poch. O. GOAnno: GO annotation based on multiple alignment. Bioinformatics. 2005 • Clamp, M. , Cuff, J. , Searle, SM, Barton, GJ. The Jalview Java Alignment Editor. Bioinformatics. 2004 • Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. Genome Res. 1998 • Huang X, Madan A. CAP 3: A DNA sequence assembly program. Genome Res. 1999 • Lecompte O, Thompson JD, Plewniak F, Thierry J, Poch O. Multiple alignment of complete sequences (MACS) in the post-genomic era. Gene. 2001 • Lottaz C, Iseli C, Jongeneel CV, Bucher P. Modeling sequencing errors by combining Hidden Markov models. Bioinformatics. 2003 • Plewniak F, Bianchetti L, Brelivet Y, Carles A, Chalmel F, Lecompte O, Mochel T, Moulinier L, Muller A, Muller J, Prigent V, Ripp R, Thierry JC, Thompson JD, Wicker N, Poch O. Pipe. Align: A new toolkit for protein family analysis. Nucleic Acids Res. 2003 • Thompson JD, Muller A, Waterhouse A, Procter J, Barton GJ, Plewniak F, Poch O. MACSIMS: multiple alignment of complete sequences information management system. BMC Bioinformatics. 2006


