507d011391446ba9a631ecbf2c4c4895.ppt
- Количество слайдов: 48

 Fragmenting genomic DNA for cloning – Random methods are best • Mechanical shearing: sonication, nebulizer • Nuclease treatment (usually restriction digest): 4 base cutters, partial digest – Large fragments better than small, fewer clones to get coverage of large genome
Fragmenting genomic DNA for cloning – Random methods are best • Mechanical shearing: sonication, nebulizer • Nuclease treatment (usually restriction digest): 4 base cutters, partial digest – Large fragments better than small, fewer clones to get coverage of large genome
 Random fragmentation of genomic DNA: Hydrodynamic shear (physical breakage) -- sonication (vibrating metal probe) -- nebulization (like asthma inhalers) -- passage through small needle orifice DNA must be repaired with DNA polymerase after these treatments Enzymatic breakage -- Restriction enzyme (4 cutter, partial digest) Cvi. J (py. GCpy and pu. GCpu) -- DNAse I (semi-random cleavage)
Random fragmentation of genomic DNA: Hydrodynamic shear (physical breakage) -- sonication (vibrating metal probe) -- nebulization (like asthma inhalers) -- passage through small needle orifice DNA must be repaired with DNA polymerase after these treatments Enzymatic breakage -- Restriction enzyme (4 cutter, partial digest) Cvi. J (py. GCpy and pu. GCpu) -- DNAse I (semi-random cleavage)
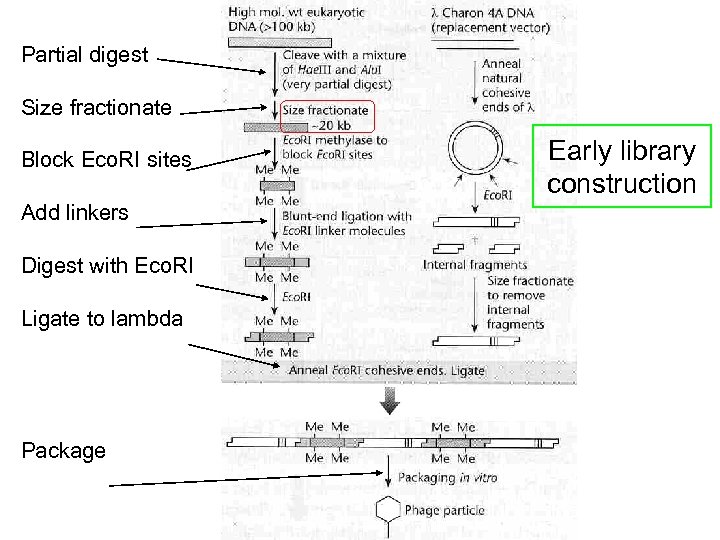 Partial digest Size fractionate Block Eco. RI sites Add linkers Digest with Eco. RI Ligate to lambda Package Early library construction
Partial digest Size fractionate Block Eco. RI sites Add linkers Digest with Eco. RI Ligate to lambda Package Early library construction
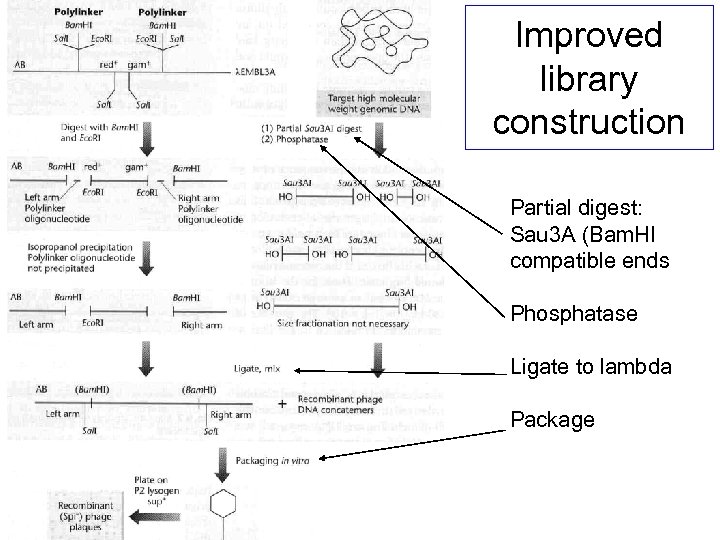 Improved library construction Partial digest: Sau 3 A (Bam. HI compatible ends Phosphatase Ligate to lambda Package
Improved library construction Partial digest: Sau 3 A (Bam. HI compatible ends Phosphatase Ligate to lambda Package
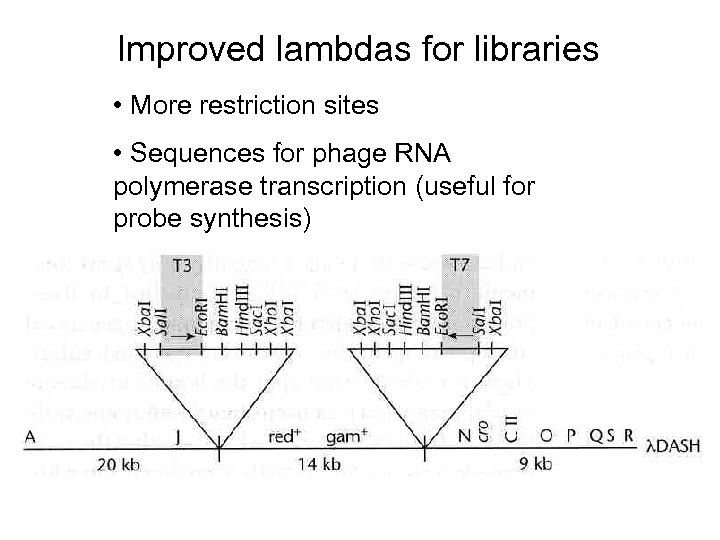 Improved lambdas for libraries • More restriction sites • Sequences for phage RNA polymerase transcription (useful for probe synthesis)
Improved lambdas for libraries • More restriction sites • Sequences for phage RNA polymerase transcription (useful for probe synthesis)
 But…. • • Cosmids BACs PACs YACs …can be used for cloning larger DNAs using similar methods… Why use lambda libraries?
But…. • • Cosmids BACs PACs YACs …can be used for cloning larger DNAs using similar methods… Why use lambda libraries?
 • Cosmids replicate as high copy number plasmids--tend to be unstable, deleting insert DNA (to reduce drag on cells) • BAC and YAC libraries difficult to prepare • larger-sized DNA more difficult to work with
• Cosmids replicate as high copy number plasmids--tend to be unstable, deleting insert DNA (to reduce drag on cells) • BAC and YAC libraries difficult to prepare • larger-sized DNA more difficult to work with
 Cloning c. DNAs • Prepared by reverse transcription of m. RNA • Eukaryotic m. RNAs--lack introns, often show variable splicing, c. DNAs of these RNAs indicate how genes are actually expressed • Individual m. RNA abundance varies widely: to isolate low abundance m. RNAs by c. DNA cloning, need to make libraries
Cloning c. DNAs • Prepared by reverse transcription of m. RNA • Eukaryotic m. RNAs--lack introns, often show variable splicing, c. DNAs of these RNAs indicate how genes are actually expressed • Individual m. RNA abundance varies widely: to isolate low abundance m. RNAs by c. DNA cloning, need to make libraries
 Key points of c. DNA cloning 1) m. RNA source (tissue type) matters a lot 2) m. RNA must be of high quality (no Rnases…. ) 3) Rare m. RNAs can be enriched e. g. “Subtractive cloning” hybridize sample c. DNA against immobilized RNA/c. DNA from a “driver”, clone only those m. RNAs that are not bound by the driver This relies on differential m. RNA expression between sample and “driver” m. RNA populations
Key points of c. DNA cloning 1) m. RNA source (tissue type) matters a lot 2) m. RNA must be of high quality (no Rnases…. ) 3) Rare m. RNAs can be enriched e. g. “Subtractive cloning” hybridize sample c. DNA against immobilized RNA/c. DNA from a “driver”, clone only those m. RNAs that are not bound by the driver This relies on differential m. RNA expression between sample and “driver” m. RNA populations
 Gubler/Hoffman method (MC Chapter 11) 1) Synthesize first strand c. DNA 2) Second strand c. DNA 3) Methylate c. DNA 4) Attach linkers or adaptors for cloning 5) Fractionate c. DNA by size (select 2 -8 kb) 6) Ligate c. DNA into bacteriophage arms
Gubler/Hoffman method (MC Chapter 11) 1) Synthesize first strand c. DNA 2) Second strand c. DNA 3) Methylate c. DNA 4) Attach linkers or adaptors for cloning 5) Fractionate c. DNA by size (select 2 -8 kb) 6) Ligate c. DNA into bacteriophage arms
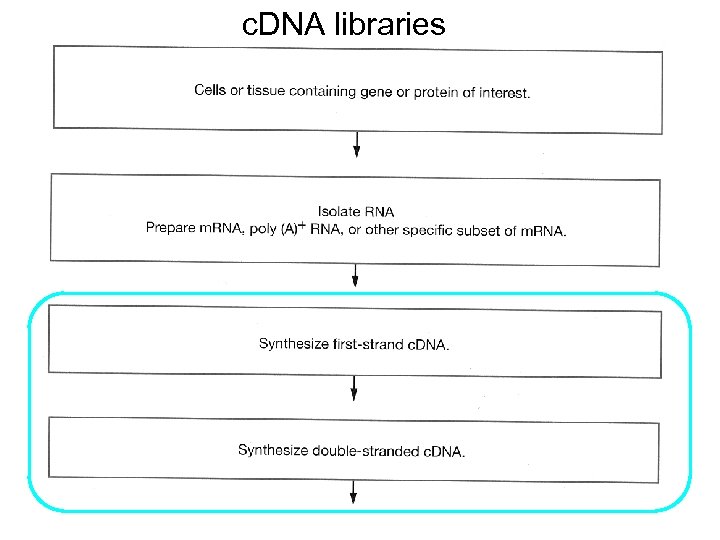 c. DNA libraries
c. DNA libraries

 c. DNA synthesis • Make the first DNA strand from the m. RNA template using reverse transcriptase • Remove the RNA • Make the second DNA strand from the first DNA strand
c. DNA synthesis • Make the first DNA strand from the m. RNA template using reverse transcriptase • Remove the RNA • Make the second DNA strand from the first DNA strand
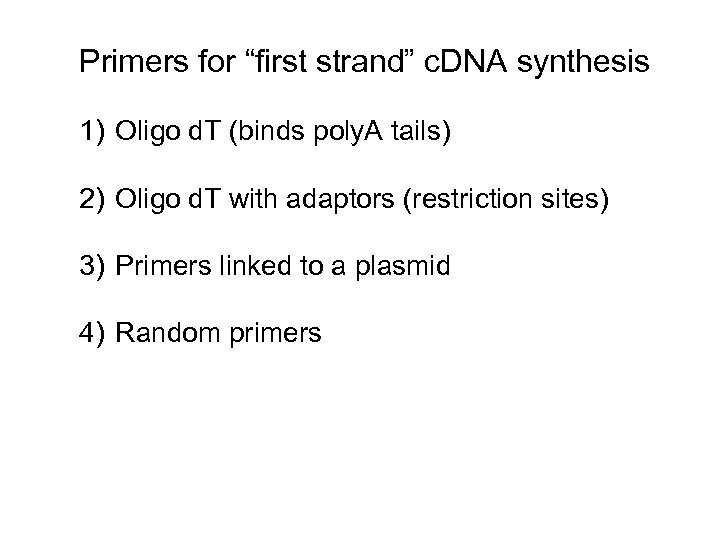 Primers for “first strand” c. DNA synthesis 1) Oligo d. T (binds poly. A tails) 2) Oligo d. T with adaptors (restriction sites) 3) Primers linked to a plasmid 4) Random primers
Primers for “first strand” c. DNA synthesis 1) Oligo d. T (binds poly. A tails) 2) Oligo d. T with adaptors (restriction sites) 3) Primers linked to a plasmid 4) Random primers
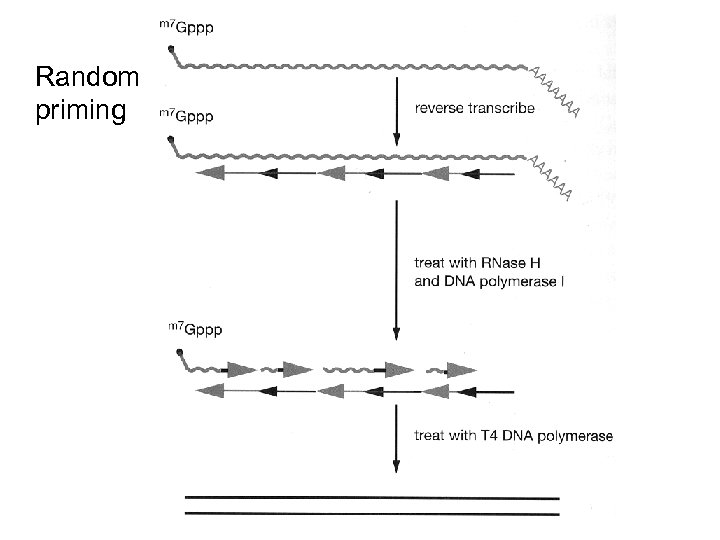 Random priming
Random priming
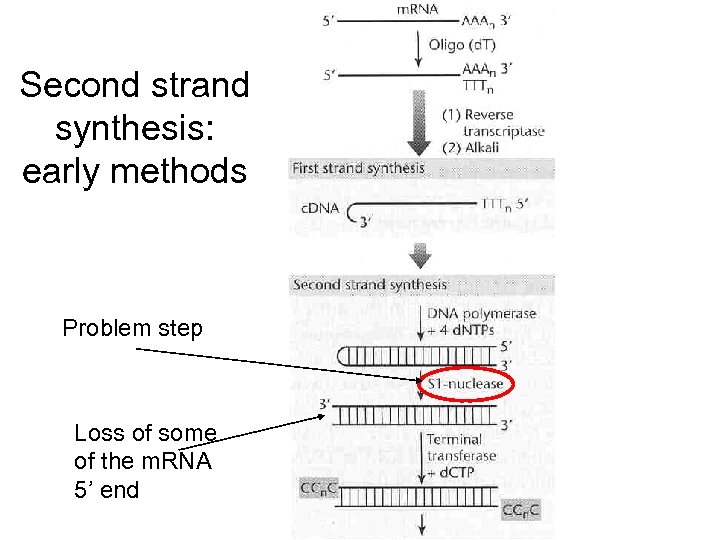 Second strand synthesis: early methods Problem step Loss of some of the m. RNA 5’ end
Second strand synthesis: early methods Problem step Loss of some of the m. RNA 5’ end
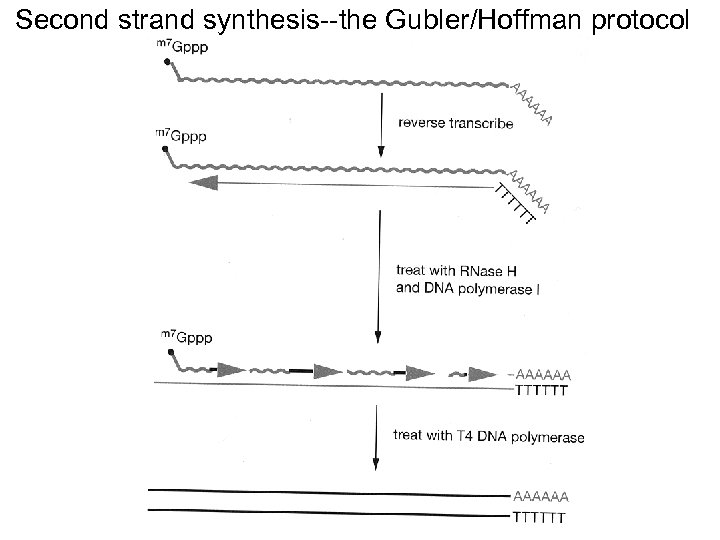 Second strand synthesis--the Gubler/Hoffman protocol
Second strand synthesis--the Gubler/Hoffman protocol
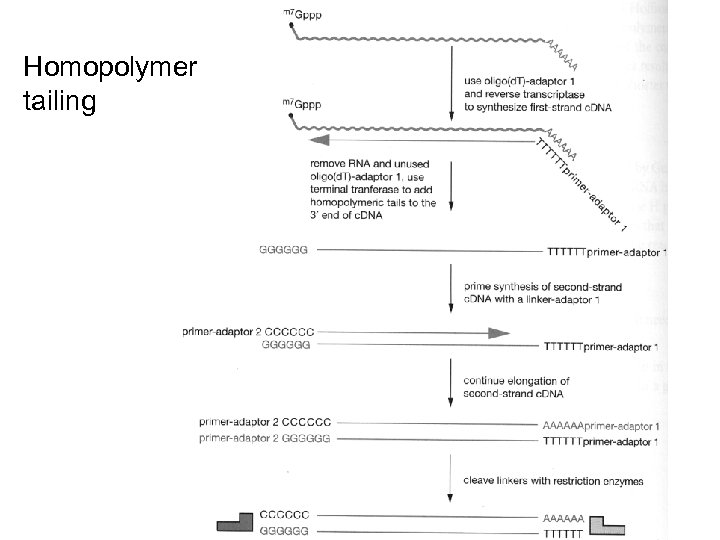 Homopolymer tailing
Homopolymer tailing
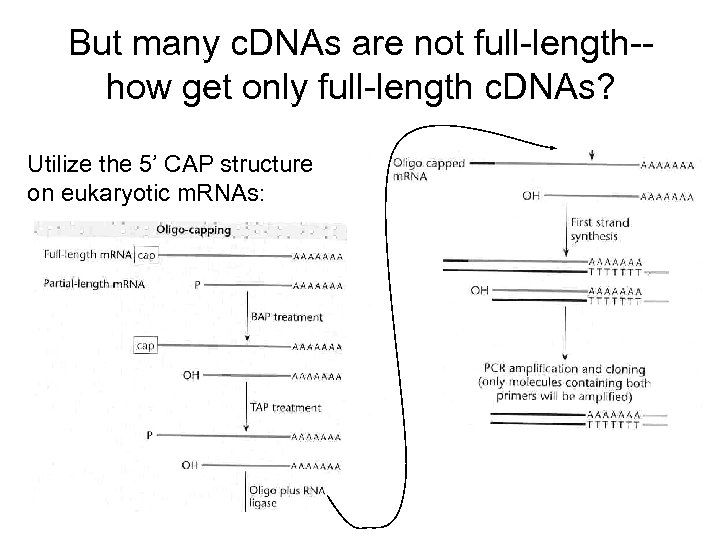 But many c. DNAs are not full-length-how get only full-length c. DNAs? Utilize the 5’ CAP structure on eukaryotic m. RNAs:
But many c. DNAs are not full-length-how get only full-length c. DNAs? Utilize the 5’ CAP structure on eukaryotic m. RNAs:
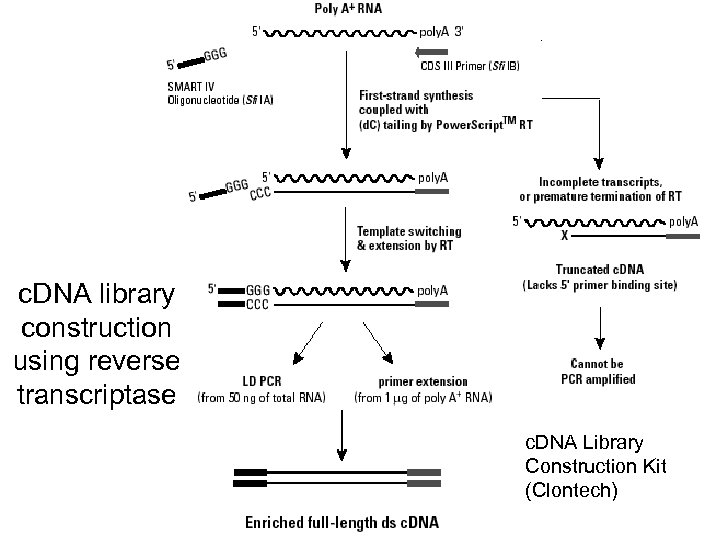 c. DNA library construction using reverse transcriptase c. DNA Library Construction Kit (Clontech)
c. DNA library construction using reverse transcriptase c. DNA Library Construction Kit (Clontech)
 ESTs: Expressed Sequence Tags • Full length c. DNAs hard to get, difficult to scale up • But short c. DNA sequences are often useful – ID and map specific genes – “High throughput” allows very fast generation of 200 -300 bp sequences, or ESTs • Millions of ESTs in database • Useful in designing “microarrays” (later)
ESTs: Expressed Sequence Tags • Full length c. DNAs hard to get, difficult to scale up • But short c. DNA sequences are often useful – ID and map specific genes – “High throughput” allows very fast generation of 200 -300 bp sequences, or ESTs • Millions of ESTs in database • Useful in designing “microarrays” (later)
 c. DNA libraries: the easy way out Pre-made c. DNA libraries (organisms, tissues, variable conditions Custom made c. DNA libraries (you supply the m. RNA) “kits” for making your own c. DNA library (See Table 11 -6 of Molecular Cloning for a directory)
c. DNA libraries: the easy way out Pre-made c. DNA libraries (organisms, tissues, variable conditions Custom made c. DNA libraries (you supply the m. RNA) “kits” for making your own c. DNA library (See Table 11 -6 of Molecular Cloning for a directory)
 Library construction 1) DNA (entire genome…) a) Fragment the DNA b) Clone in lambda phage vector 2) m. RNA (only the expressed genes) a) First strand c. DNA b) Second strand c. DNA c) Expressed sequence tags (ESTs)
Library construction 1) DNA (entire genome…) a) Fragment the DNA b) Clone in lambda phage vector 2) m. RNA (only the expressed genes) a) First strand c. DNA b) Second strand c. DNA c) Expressed sequence tags (ESTs)
 Screening libraries for specific genes (finding the needle in the haystack) I. II. Isolating individual clones Screening by sequence A. B. III. IV. Hybridization PCR Screening by protein structure/biological function Gene identification--diseases Course reading #29
Screening libraries for specific genes (finding the needle in the haystack) I. II. Isolating individual clones Screening by sequence A. B. III. IV. Hybridization PCR Screening by protein structure/biological function Gene identification--diseases Course reading #29
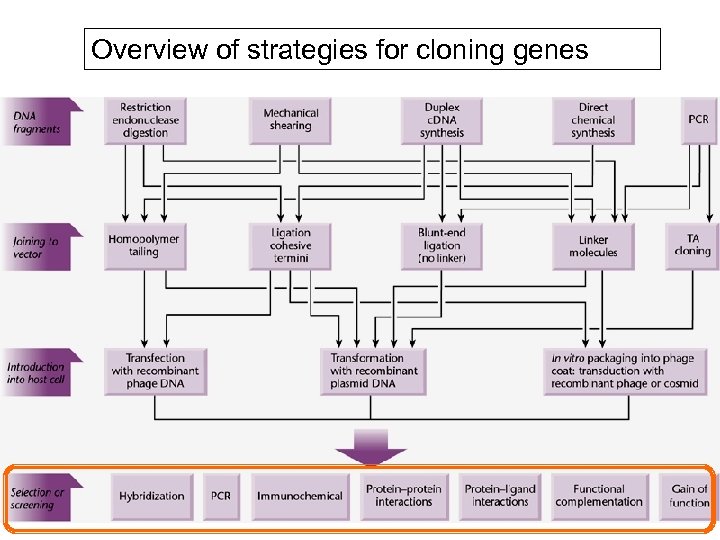 Overview of strategies for cloning genes
Overview of strategies for cloning genes
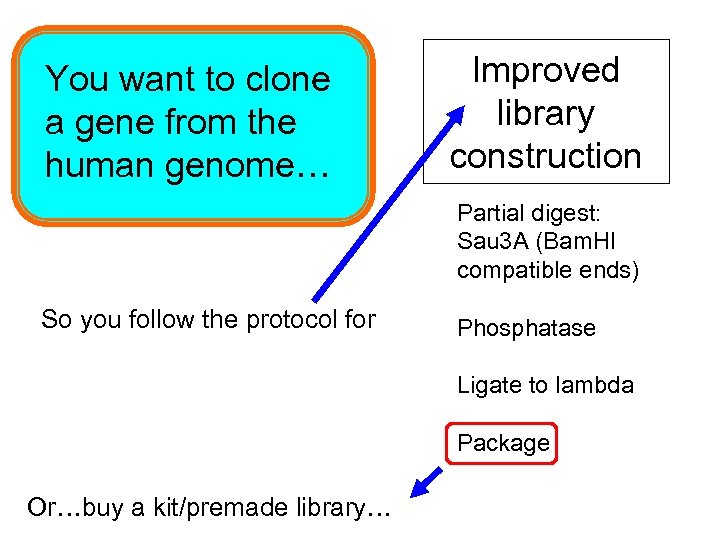 You want to clone a gene from the human genome… Improved library construction Partial digest: Sau 3 A (Bam. HI compatible ends) So you follow the protocol for Phosphatase Ligate to lambda Package Or…buy a kit/premade library…
You want to clone a gene from the human genome… Improved library construction Partial digest: Sau 3 A (Bam. HI compatible ends) So you follow the protocol for Phosphatase Ligate to lambda Package Or…buy a kit/premade library…
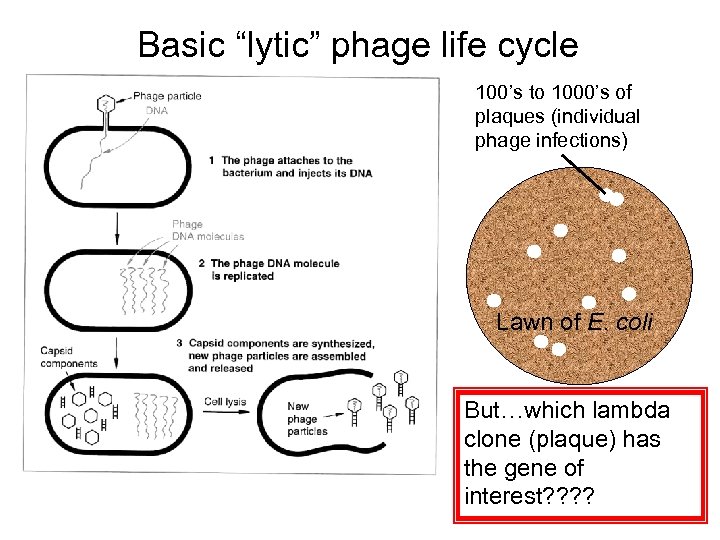 Basic “lytic” phage life cycle 100’s to 1000’s of plaques (individual phage infections) Lawn of E. coli But…which lambda clone (plaque) has the gene of interest? ?
Basic “lytic” phage life cycle 100’s to 1000’s of plaques (individual phage infections) Lawn of E. coli But…which lambda clone (plaque) has the gene of interest? ?
 How many recombinant DNA molecules are required in a library to get complete coverage of a genome? N= ln(1 -p) ln(1 -f) p = probability of getting a specific piece of DNA f = fractional size of clone DNA relative to genome N = number of clones needed
How many recombinant DNA molecules are required in a library to get complete coverage of a genome? N= ln(1 -p) ln(1 -f) p = probability of getting a specific piece of DNA f = fractional size of clone DNA relative to genome N = number of clones needed
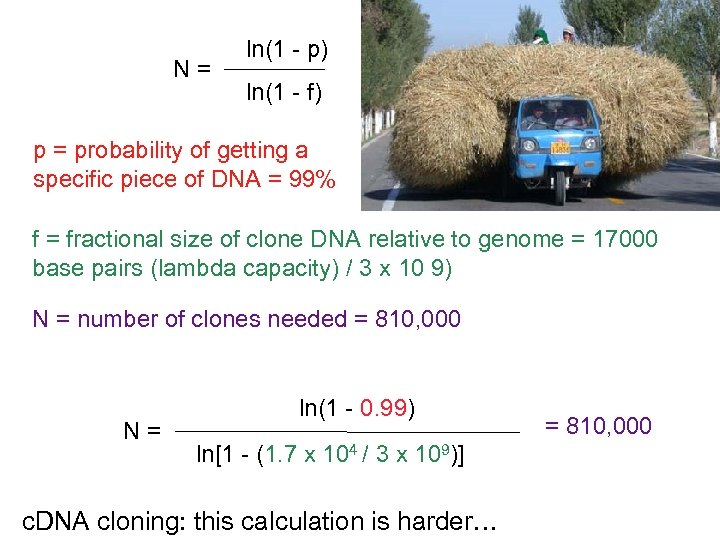 N= ln(1 - p) ln(1 - f) p = probability of getting a specific piece of DNA = 99% f = fractional size of clone DNA relative to genome = 17000 base pairs (lambda capacity) / 3 x 10 9) N = number of clones needed = 810, 000 N= ln(1 - 0. 99) ln[1 - (1. 7 x 104 / 3 x 109)] c. DNA cloning: this calculation is harder… = 810, 000
N= ln(1 - p) ln(1 - f) p = probability of getting a specific piece of DNA = 99% f = fractional size of clone DNA relative to genome = 17000 base pairs (lambda capacity) / 3 x 10 9) N = number of clones needed = 810, 000 N= ln(1 - 0. 99) ln[1 - (1. 7 x 104 / 3 x 109)] c. DNA cloning: this calculation is harder… = 810, 000
 Screen by hybridization § Very fast § Applicable to a large number of clones § Can identify clones that are not full length § But you need to know at least some of the sequence of the gene you are after (more on this later)
Screen by hybridization § Very fast § Applicable to a large number of clones § Can identify clones that are not full length § But you need to know at least some of the sequence of the gene you are after (more on this later)
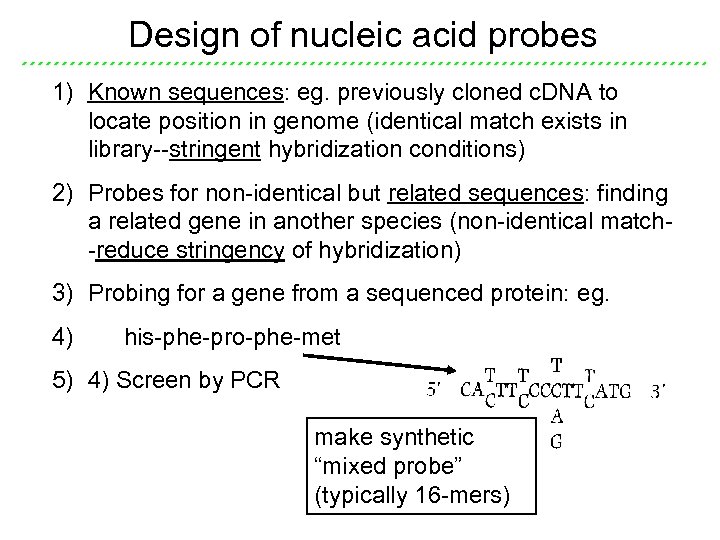 Design of nucleic acid probes 1) Known sequences: eg. previously cloned c. DNA to locate position in genome (identical match exists in library--stringent hybridization conditions) 2) Probes for non-identical but related sequences: finding a related gene in another species (non-identical match-reduce stringency of hybridization) 3) Probing for a gene from a sequenced protein: eg. 4) his-phe-pro-phe-met 5) 4) Screen by PCR make synthetic “mixed probe” (typically 16 -mers)
Design of nucleic acid probes 1) Known sequences: eg. previously cloned c. DNA to locate position in genome (identical match exists in library--stringent hybridization conditions) 2) Probes for non-identical but related sequences: finding a related gene in another species (non-identical match-reduce stringency of hybridization) 3) Probing for a gene from a sequenced protein: eg. 4) his-phe-pro-phe-met 5) 4) Screen by PCR make synthetic “mixed probe” (typically 16 -mers)
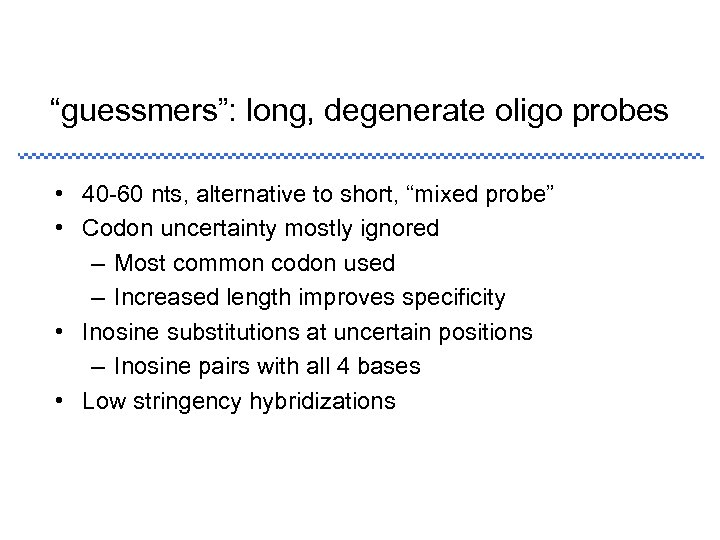 “guessmers”: long, degenerate oligo probes • 40 -60 nts, alternative to short, “mixed probe” • Codon uncertainty mostly ignored – Most common codon used – Increased length improves specificity • Inosine substitutions at uncertain positions – Inosine pairs with all 4 bases • Low stringency hybridizations
“guessmers”: long, degenerate oligo probes • 40 -60 nts, alternative to short, “mixed probe” • Codon uncertainty mostly ignored – Most common codon used – Increased length improves specificity • Inosine substitutions at uncertain positions – Inosine pairs with all 4 bases • Low stringency hybridizations
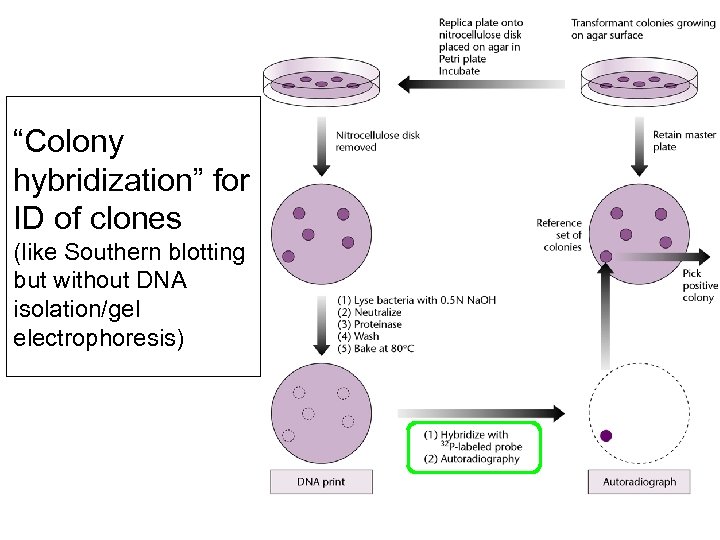 “Colony hybridization” for ID of clones (like Southern blotting but without DNA isolation/gel electrophoresis)
“Colony hybridization” for ID of clones (like Southern blotting but without DNA isolation/gel electrophoresis)
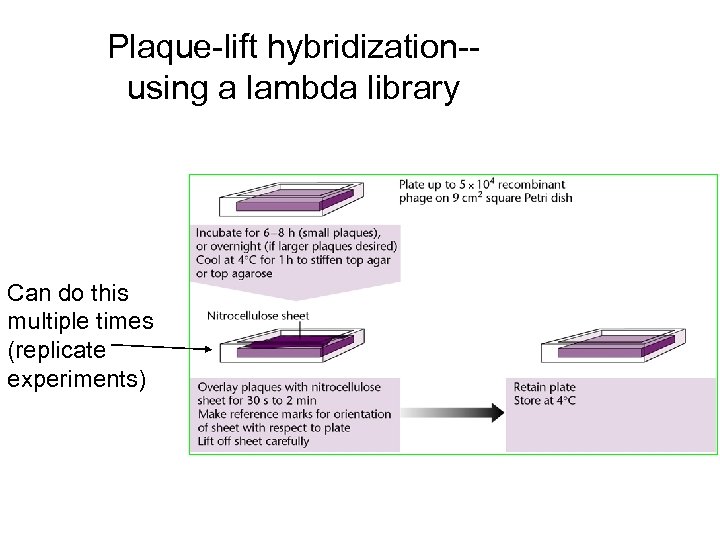 Plaque-lift hybridization-using a lambda library Can do this multiple times (replicate experiments)
Plaque-lift hybridization-using a lambda library Can do this multiple times (replicate experiments)

 Alternative to plating: arrayed libraries • Individual clones of library spotted onto membranes in high density arrays (tens of thousands of genes) • Membranes probed as described (a la microarrays) • Standardizable, centralizable
Alternative to plating: arrayed libraries • Individual clones of library spotted onto membranes in high density arrays (tens of thousands of genes) • Membranes probed as described (a la microarrays) • Standardizable, centralizable
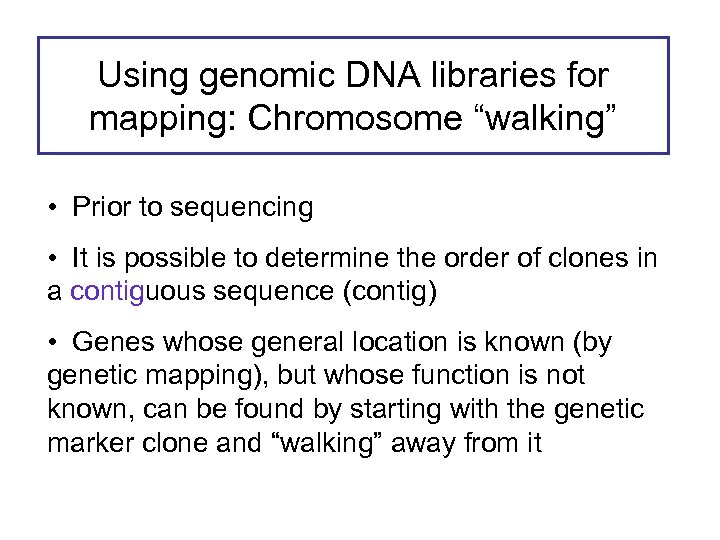 Using genomic DNA libraries for mapping: Chromosome “walking” • Prior to sequencing • It is possible to determine the order of clones in a contiguous sequence (contig) • Genes whose general location is known (by genetic mapping), but whose function is not known, can be found by starting with the genetic marker clone and “walking” away from it
Using genomic DNA libraries for mapping: Chromosome “walking” • Prior to sequencing • It is possible to determine the order of clones in a contiguous sequence (contig) • Genes whose general location is known (by genetic mapping), but whose function is not known, can be found by starting with the genetic marker clone and “walking” away from it
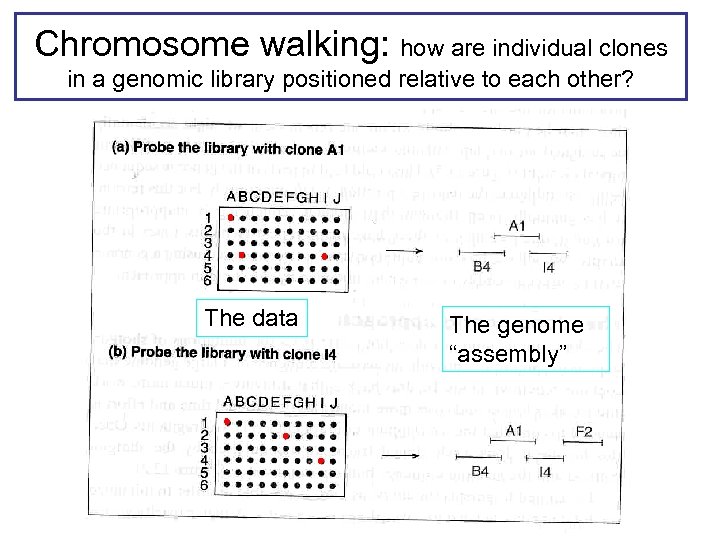 Chromosome walking: how are individual clones in a genomic library positioned relative to each other? The data The genome “assembly”
Chromosome walking: how are individual clones in a genomic library positioned relative to each other? The data The genome “assembly”
 Chromosome walking • Probing can be restricted to one direction with RNA probes generated from clone ends • Beware of “warping” to another chromosome because of repetitive sequence probes • Use YAC and BAC libraries to take larger steps
Chromosome walking • Probing can be restricted to one direction with RNA probes generated from clone ends • Beware of “warping” to another chromosome because of repetitive sequence probes • Use YAC and BAC libraries to take larger steps
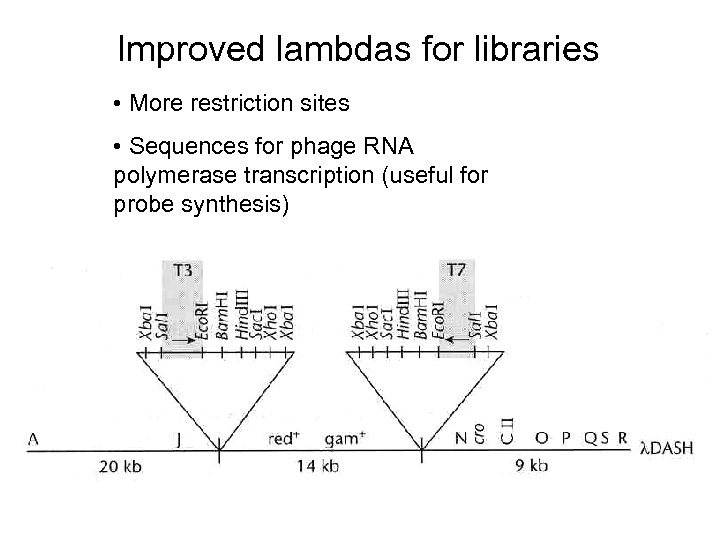 Improved lambdas for libraries • More restriction sites • Sequences for phage RNA polymerase transcription (useful for probe synthesis)
Improved lambdas for libraries • More restriction sites • Sequences for phage RNA polymerase transcription (useful for probe synthesis)
 Expression libraries-alternative to hybridization • Gene product (protein) is made (by E. coli) and detected by variety of methods • Eukaryotic genes: c. DNA library is essential (no introns, gene size small) Screening: • Immunological • Functional
Expression libraries-alternative to hybridization • Gene product (protein) is made (by E. coli) and detected by variety of methods • Eukaryotic genes: c. DNA library is essential (no introns, gene size small) Screening: • Immunological • Functional
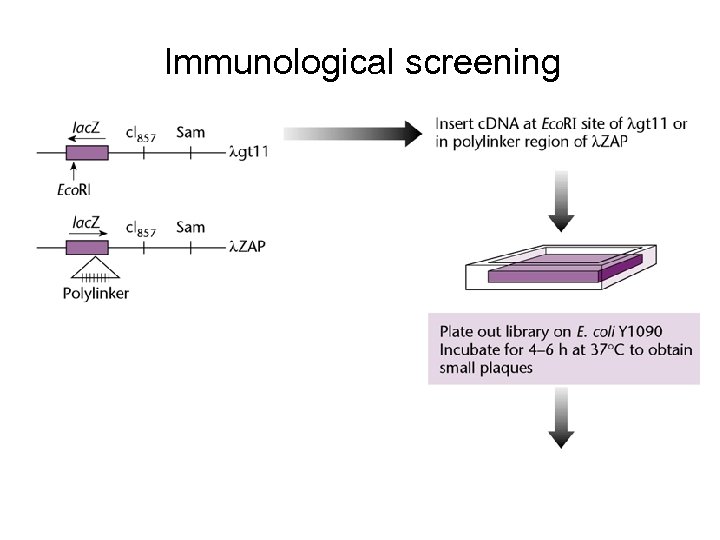 Immunological screening
Immunological screening
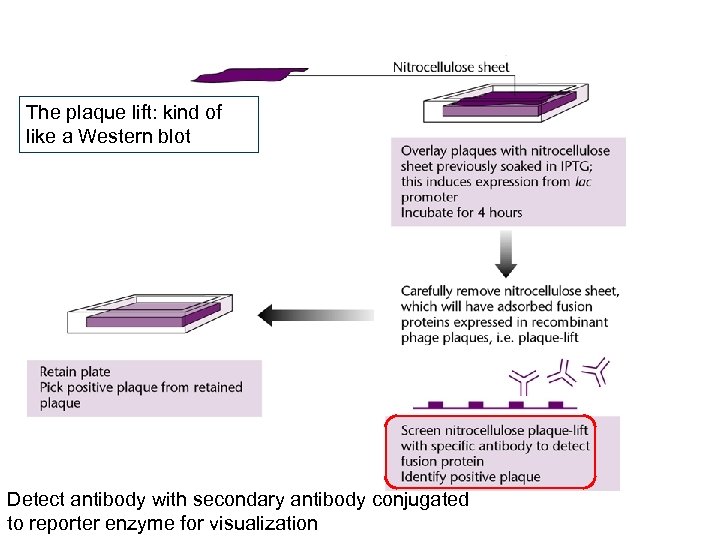 The plaque lift: kind of like a Western blot Detect antibody with secondary antibody conjugated to reporter enzyme for visualization
The plaque lift: kind of like a Western blot Detect antibody with secondary antibody conjugated to reporter enzyme for visualization
 Functional cloning • Genetic complementation: – Cloned DNA sequence corrects defect in host strain • Gain of function – Cloned DNA confers new function to host Both of these require cloned DNA to be transcribed, translated into functional protein in host (eukaryotic protein in E. coli could cause problems) And you need a good assay for expression!
Functional cloning • Genetic complementation: – Cloned DNA sequence corrects defect in host strain • Gain of function – Cloned DNA confers new function to host Both of these require cloned DNA to be transcribed, translated into functional protein in host (eukaryotic protein in E. coli could cause problems) And you need a good assay for expression!
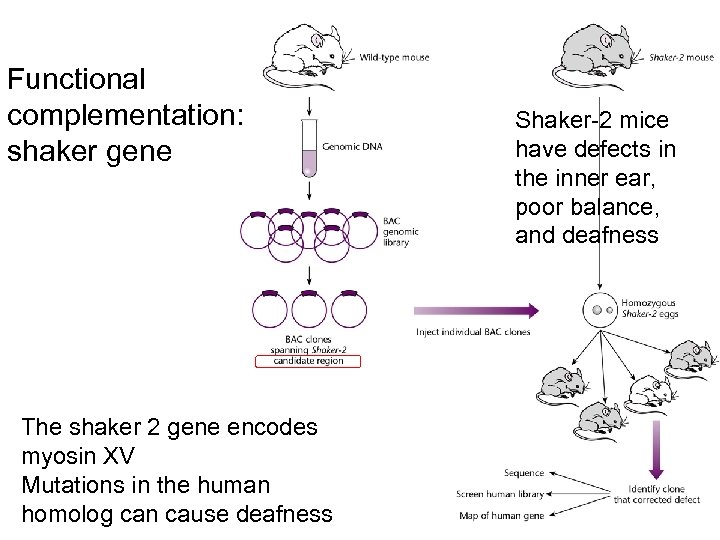 Functional complementation: shaker gene The shaker 2 gene encodes myosin XV Mutations in the human homolog can cause deafness Shaker-2 mice have defects in the inner ear, poor balance, and deafness
Functional complementation: shaker gene The shaker 2 gene encodes myosin XV Mutations in the human homolog can cause deafness Shaker-2 mice have defects in the inner ear, poor balance, and deafness
 Subtractive cloning – Remove c. DNAs that are common to two sources – Useful for isolation and detections of differentially expressed rare c. DNAs – Example: differential expression from physiological change – “driver” DNA - immobilized – “test” c. DNA (single stranded): labelled and then annealed to driver DNA – Remaining DNA has no counterpart in the driver cells--probe library to locate genes – Or use the remaining DNA to probe a microarray
Subtractive cloning – Remove c. DNAs that are common to two sources – Useful for isolation and detections of differentially expressed rare c. DNAs – Example: differential expression from physiological change – “driver” DNA - immobilized – “test” c. DNA (single stranded): labelled and then annealed to driver DNA – Remaining DNA has no counterpart in the driver cells--probe library to locate genes – Or use the remaining DNA to probe a microarray
 Screening libraries for specific genes (finding the needle in the haystack) I. II. Isolating individual clones Screening by sequence A. B. III. IV. Hybridization PCR Screening by protein structure/biological function Gene identification--diseases
Screening libraries for specific genes (finding the needle in the haystack) I. II. Isolating individual clones Screening by sequence A. B. III. IV. Hybridization PCR Screening by protein structure/biological function Gene identification--diseases


