3e3d353363a31905901c9f2ddee45fc9.ppt
- Количество слайдов: 39
Formal Cell Biology in Biocham François Fages Constraint Programming Group http: //contraintes. inria. fr/ INRIA Paris-Rocquencourt France To tackle the complexity of biological systems, investigate: • Programming Theory Concepts • Formal Methods of Circuit and Program Verification • Constraint Modeling and Automated Reasoning Tools Implementation in the Biochemical Abstract Machine BIOCHAM v 3. 3 Shonan village 14/11/11 François Fages

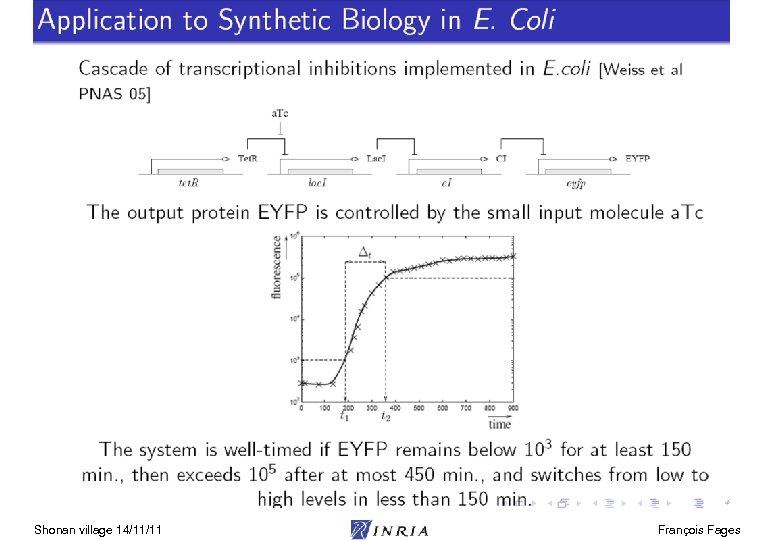
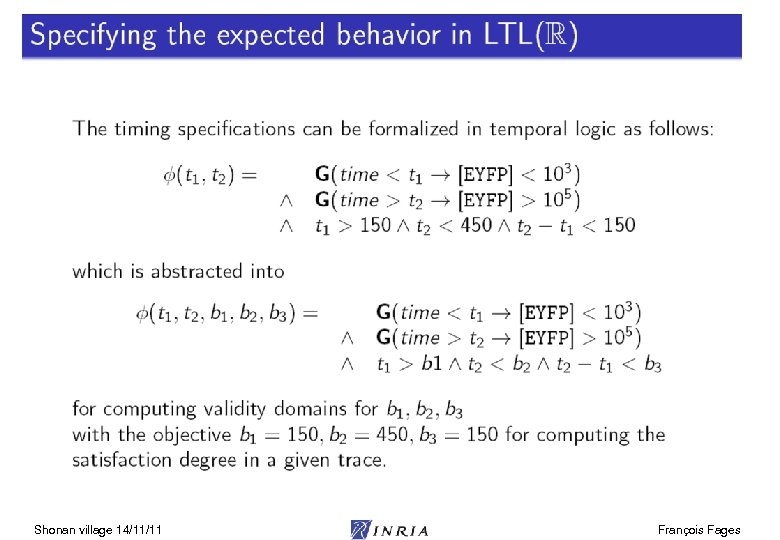
Systems Biology Challenge Aims at system-level understanding of multiscale biological processes in terms of their elementary interactions at the molecular level. Mitosis movie [ Lodish et al. 03] Systems Biology Markup Language (SBML): model exchange format Model repositories: e. g. biomodels. net 1000 models of cell processes Modeling environments (Cell designer, Cytoscape, Copasi, Biocham, …) The limits on what is observable and uncertainty of data make reasoning with partial information structures necessary symbolic, logic, constraints Shonan village 14/11/11 François Fages
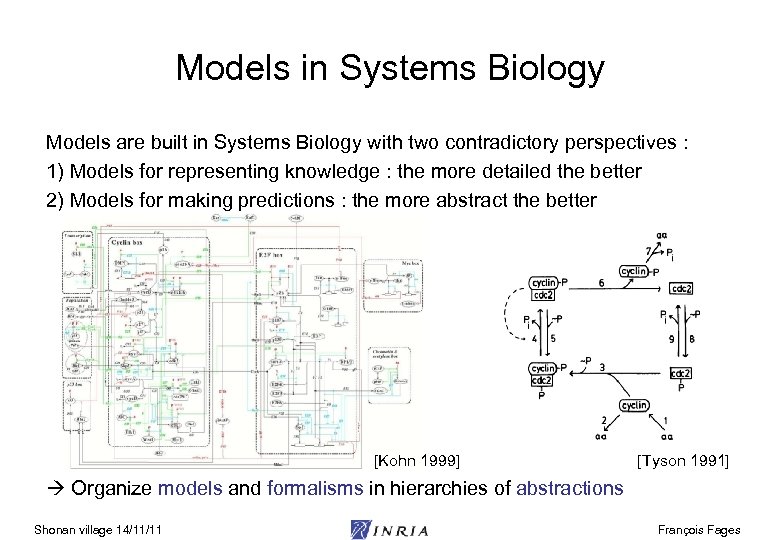
Models in Systems Biology Models are built in Systems Biology with two contradictory perspectives : 1) Models for representing knowledge : the more detailed the better 2) Models for making predictions : the more abstract the better [Kohn 1999] [Tyson 1991] Organize models and formalisms in hierarchies of abstractions Shonan village 14/11/11 François Fages
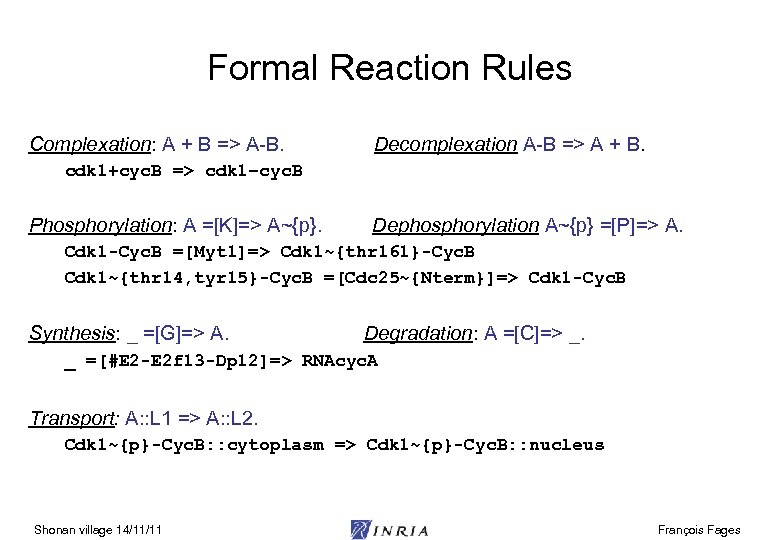
Formal Reaction Rules Complexation: A + B => A-B. Decomplexation A-B => A + B. cdk 1+cyc. B => cdk 1–cyc. B Phosphorylation: A =[K]=> A~{p}. Dephosphorylation A~{p} =[P]=> A. Cdk 1 -Cyc. B =[Myt 1]=> Cdk 1~{thr 161}-Cyc. B Cdk 1~{thr 14, tyr 15}-Cyc. B =[Cdc 25~{Nterm}]=> Cdk 1 -Cyc. B Synthesis: _ =[G]=> A. Degradation: A =[C]=> _. _ =[#E 2 -E 2 f 13 -Dp 12]=> RNAcyc. A Transport: A: : L 1 => A: : L 2. Cdk 1~{p}-Cyc. B: : cytoplasm => Cdk 1~{p}-Cyc. B: : nucleus Shonan village 14/11/11 François Fages
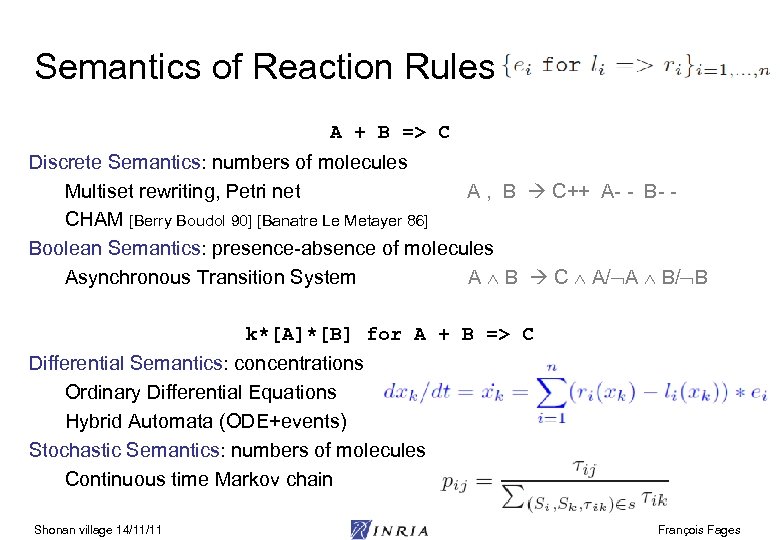
Semantics of Reaction Rules A + B => C Discrete Semantics: numbers of molecules Multiset rewriting, Petri net A , B C++ A- - B- CHAM [Berry Boudol 90] [Banatre Le Metayer 86] Boolean Semantics: presence-absence of molecules Asynchronous Transition System A B C A/ A B/ B k*[A]*[B] for A + B => C Differential Semantics: concentrations Ordinary Differential Equations Hybrid Automata (ODE+events) Stochastic Semantics: numbers of molecules Continuous time Markov chain Shonan village 14/11/11 François Fages
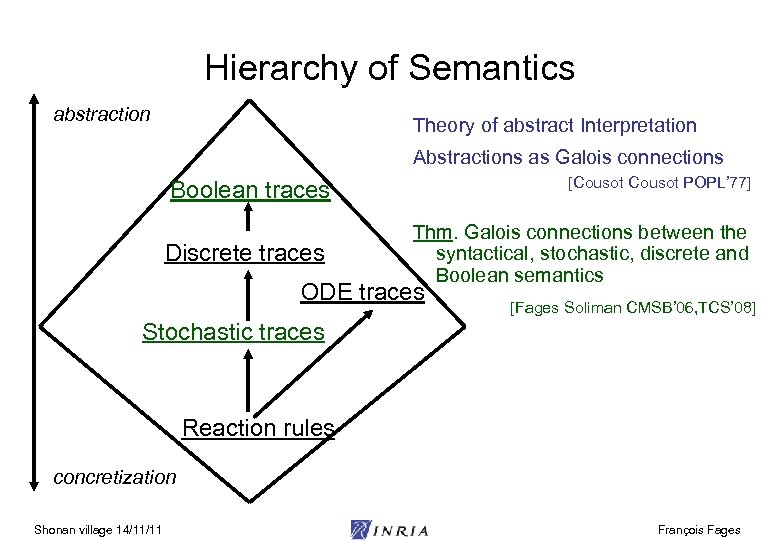
Hierarchy of Semantics abstraction Theory of abstract Interpretation Abstractions as Galois connections Boolean traces Discrete traces [Cousot POPL’ 77] Thm. Galois connections between the syntactical, stochastic, discrete and Boolean semantics ODE traces [Fages Soliman CMSB’ 06, TCS’ 08] Stochastic traces Reaction rules concretization Shonan village 14/11/11 François Fages
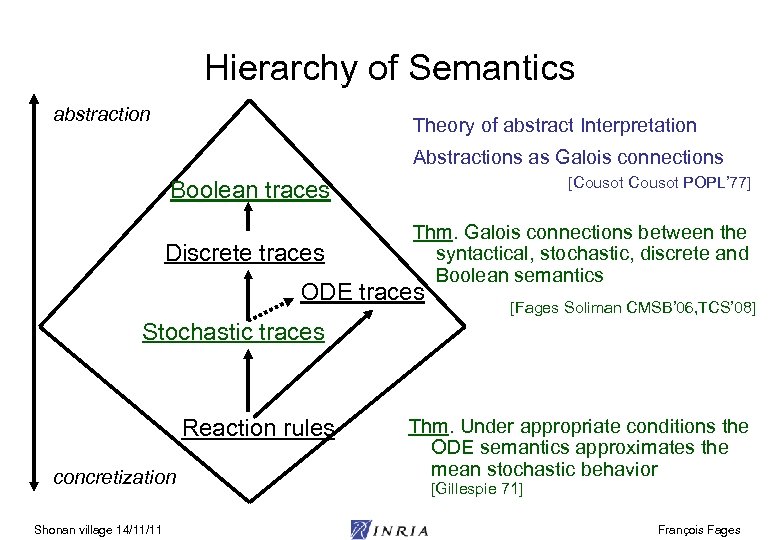
Hierarchy of Semantics abstraction Theory of abstract Interpretation Abstractions as Galois connections Boolean traces Discrete traces [Cousot POPL’ 77] Thm. Galois connections between the syntactical, stochastic, discrete and Boolean semantics ODE traces [Fages Soliman CMSB’ 06, TCS’ 08] Stochastic traces Reaction rules concretization Shonan village 14/11/11 Thm. Under appropriate conditions the ODE semantics approximates the mean stochastic behavior [Gillespie 71] François Fages
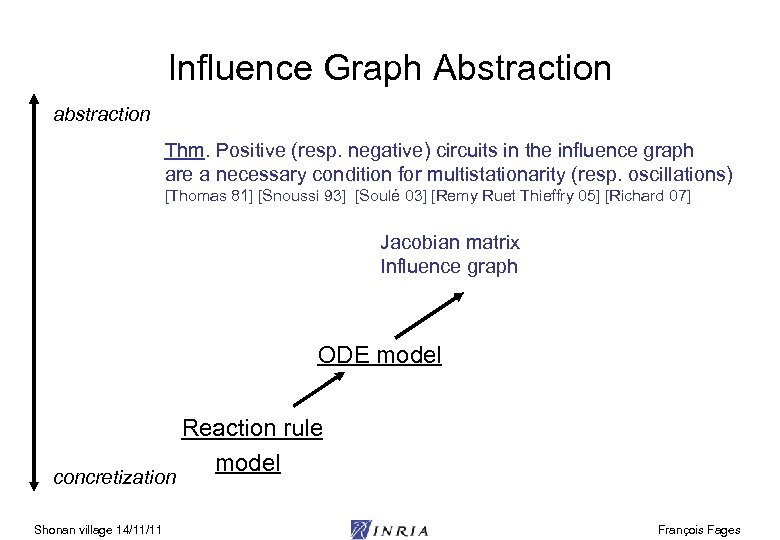
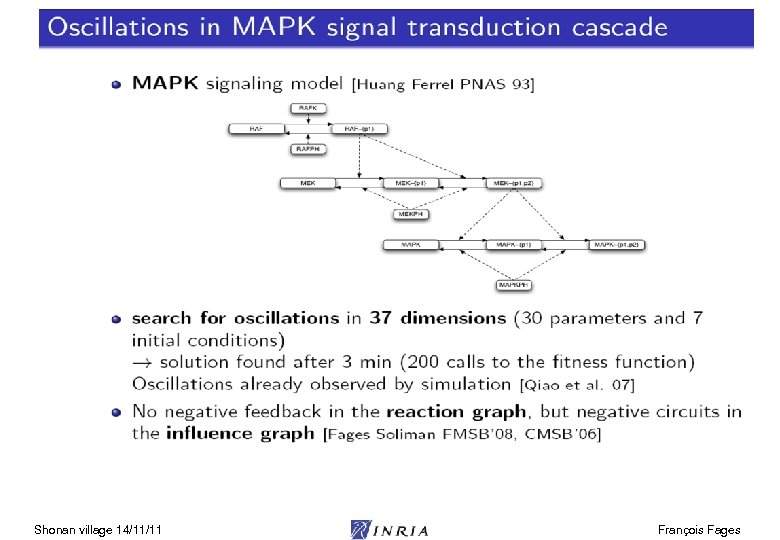
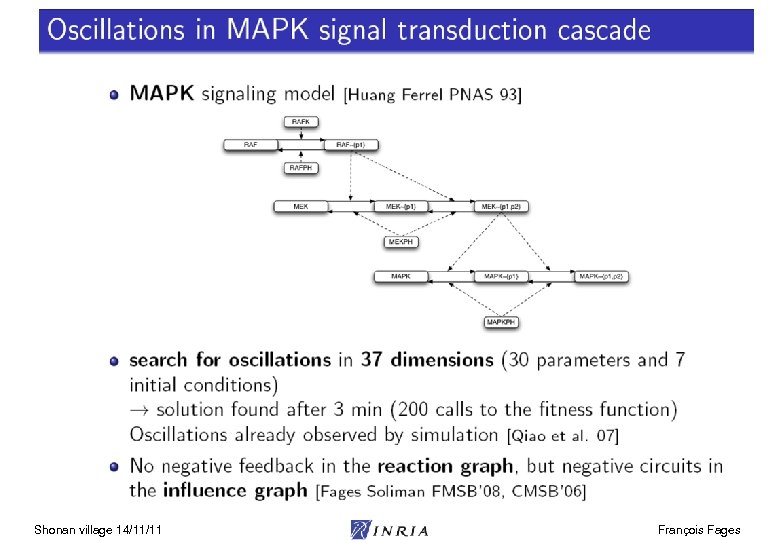
Influence Graph Abstraction abstraction Thm. Positive (resp. negative) circuits in the influence graph are a necessary condition for multistationarity (resp. oscillations) [Thomas 81] [Snoussi 93] [Soulé 03] [Remy Ruet Thieffry 05] [Richard 07] Jacobian matrix Influence graph ODE model concretization Shonan village 14/11/11 Reaction rule model François Fages
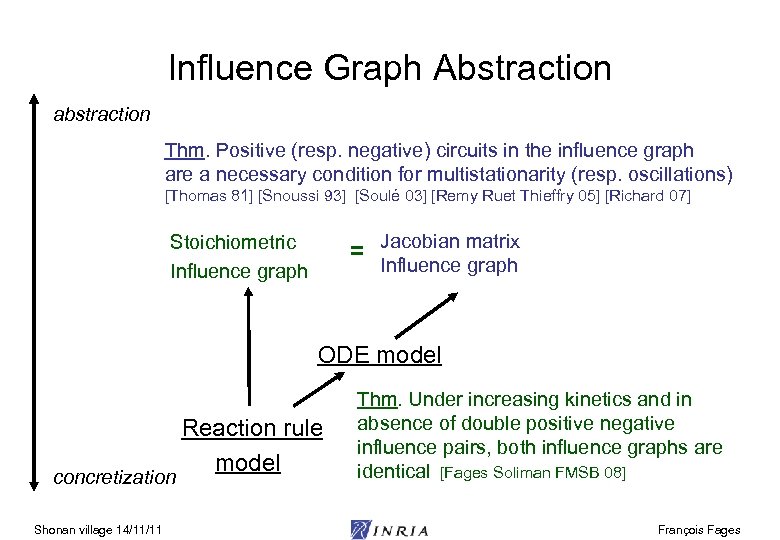
Influence Graph Abstraction abstraction Thm. Positive (resp. negative) circuits in the influence graph are a necessary condition for multistationarity (resp. oscillations) [Thomas 81] [Snoussi 93] [Soulé 03] [Remy Ruet Thieffry 05] [Richard 07] = Jacobian matrix Stoichiometric Influence graph ODE model concretization Shonan village 14/11/11 Reaction rule model Thm. Under increasing kinetics and in absence of double positive negative influence pairs, both influence graphs are identical [Fages Soliman FMSB 08] François Fages
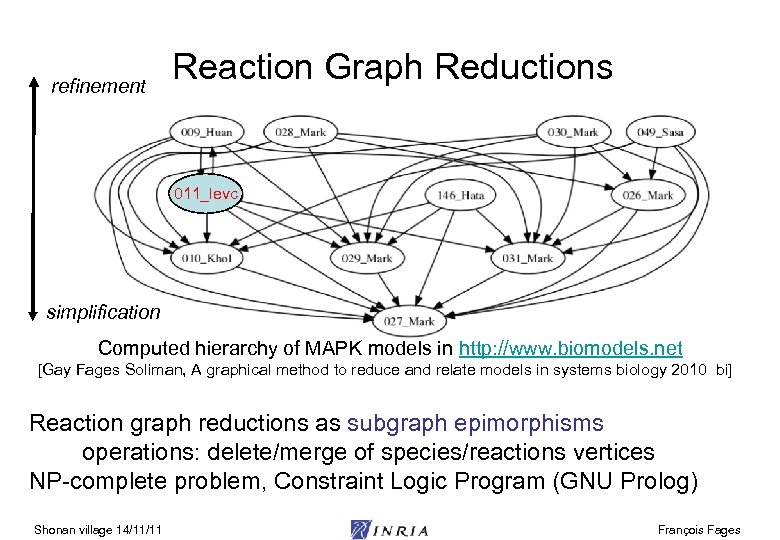
refinement Reaction Graph Reductions 011_levc simplification Computed hierarchy of MAPK models in http: //www. biomodels. net [Gay Fages Soliman, A graphical method to reduce and relate models in systems biology 2010 bi] Reaction graph reductions as subgraph epimorphisms operations: delete/merge of species/reactions vertices NP-complete problem, Constraint Logic Program (GNU Prolog) Shonan village 14/11/11 François Fages
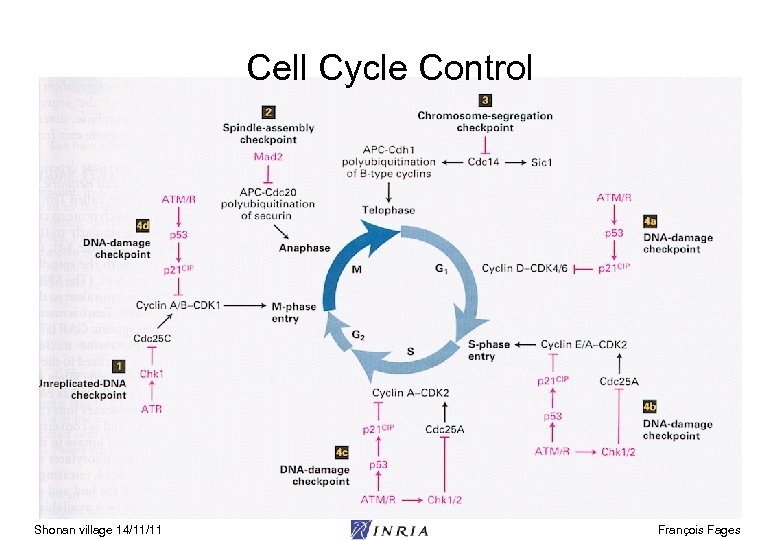
Cell Cycle Control Shonan village 14/11/11 François Fages
![Mammalian Cell Cycle Control Map [Kohn 99] Shonan village 14/11/11 François Fages Mammalian Cell Cycle Control Map [Kohn 99] Shonan village 14/11/11 François Fages](https://present5.com/presentation/3e3d353363a31905901c9f2ddee45fc9/image-12.jpg)
Mammalian Cell Cycle Control Map [Kohn 99] Shonan village 14/11/11 François Fages
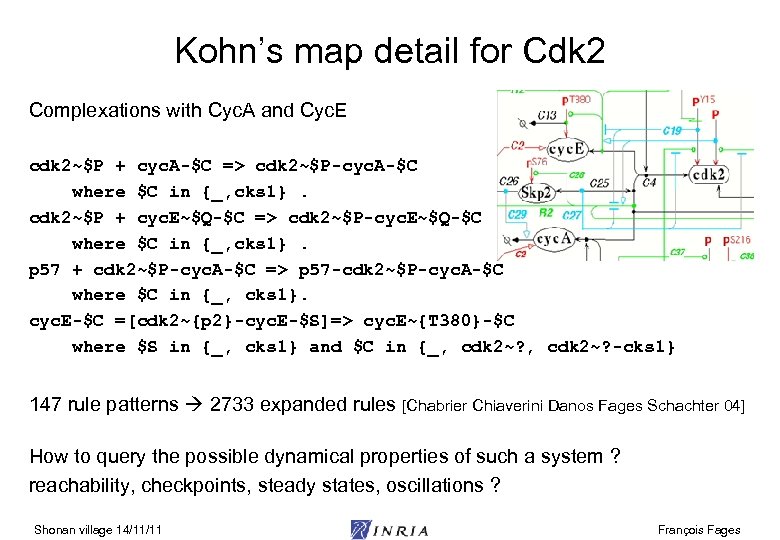
Kohn’s map detail for Cdk 2 Complexations with Cyc. A and Cyc. E cdk 2~$P + cyc. A-$C => cdk 2~$P-cyc. A-$C where $C in {_, cks 1}. cdk 2~$P + cyc. E~$Q-$C => cdk 2~$P-cyc. E~$Q-$C where $C in {_, cks 1}. p 57 + cdk 2~$P-cyc. A-$C => p 57 -cdk 2~$P-cyc. A-$C where $C in {_, cks 1}. cyc. E-$C =[cdk 2~{p 2}-cyc. E-$S]=> cyc. E~{T 380}-$C where $S in {_, cks 1} and $C in {_, cdk 2~? -cks 1} 147 rule patterns 2733 expanded rules [Chabrier Chiaverini Danos Fages Schachter 04] How to query the possible dynamical properties of such a system ? reachability, checkpoints, steady states, oscillations ? Shonan village 14/11/11 François Fages
![Temporal Logic Queries Temporal logics introduced for program verification by [Pnueli 77] Computation Tree Temporal Logic Queries Temporal logics introduced for program verification by [Pnueli 77] Computation Tree](https://present5.com/presentation/3e3d353363a31905901c9f2ddee45fc9/image-14.jpg)
Temporal Logic Queries Temporal logics introduced for program verification by [Pnueli 77] Computation Tree Logic CTL [Emerson Clarke 80] Non-det. Time E exists A always X next time EX( ) AX( ) F finally EF( ) AG( ) AF( ) liveness G globally EG( ) AF( ) AG( ) safety U until E ( 1 U 2) A ( 1 U 2) Shonan village 14/11/11 Non-determinism E, A AG F, G, U EF Time François Fages
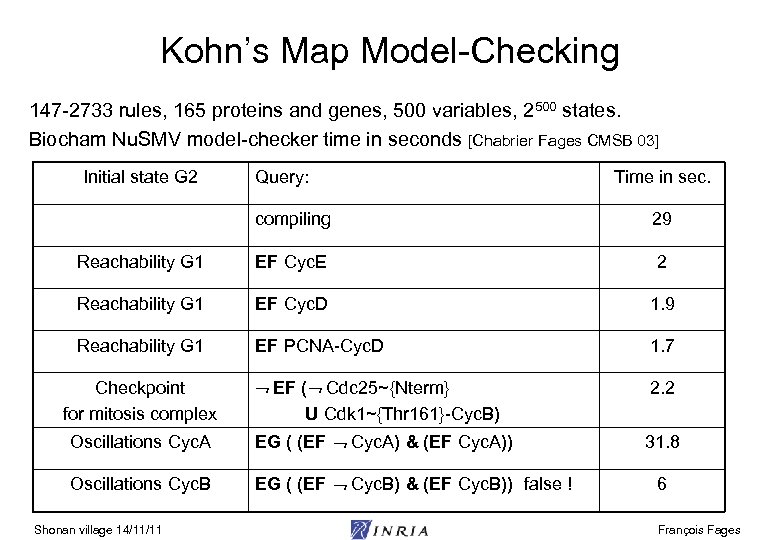
Kohn’s Map Model-Checking 147 -2733 rules, 165 proteins and genes, 500 variables, 2500 states. Biocham Nu. SMV model-checker time in seconds [Chabrier Fages CMSB 03] Initial state G 2 Query: Time in sec. compiling 29 Reachability G 1 EF Cyc. E 2 Reachability G 1 EF Cyc. D 1. 9 Reachability G 1 EF PCNA-Cyc. D 1. 7 EF ( Cdc 25~{Nterm} 2. 2 Checkpoint for mitosis complex U Cdk 1~{Thr 161}-Cyc. B) Oscillations Cyc. A EG ( (EF Cyc. A) & (EF Cyc. A)) Oscillations Cyc. B EG ( (EF Cyc. B) & (EF Cyc. B)) false ! Shonan village 14/11/11 31. 8 6 François Fages
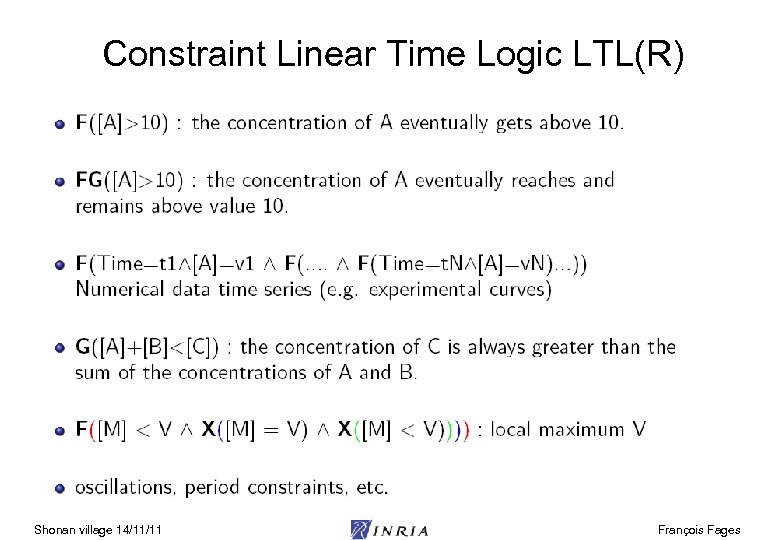
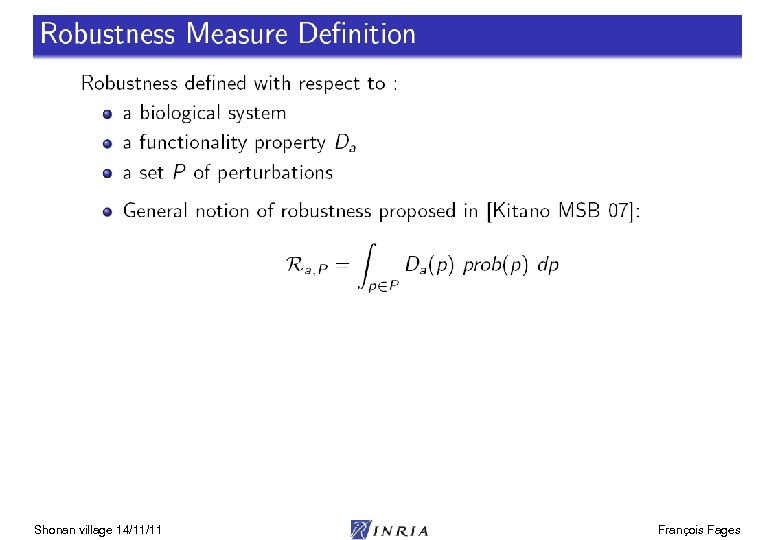
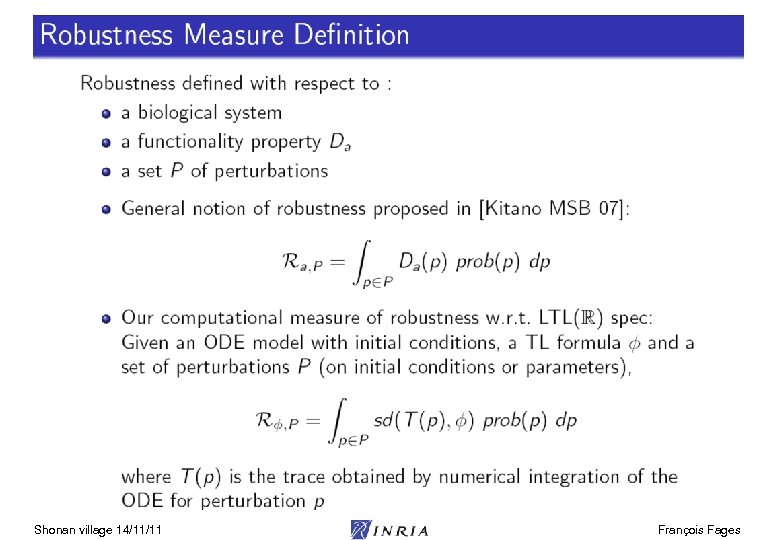
Constraint Linear Time Logic LTL(R) Shonan village 14/11/11 François Fages
![Quantitative Model of Cell Cycle Control [Tyson 91] k 1 for _ => Cyclin. Quantitative Model of Cell Cycle Control [Tyson 91] k 1 for _ => Cyclin.](https://present5.com/presentation/3e3d353363a31905901c9f2ddee45fc9/image-17.jpg)
Quantitative Model of Cell Cycle Control [Tyson 91] k 1 for _ => Cyclin. k 3*[Cyclin]*[Cdc 2~{p 1}] for Cyclin + Cdc 2~{p 1} => Cdc 2~{p 1}-Cyclin~{p 1}. k 6*[Cdc 2 -Cyclin~{p 1}] for Cdc 2 -Cyclin~{p 1} => Cdc 2 + Cyclin~{p 1}. k 7*[Cyclin~{p 1}] for Cyclin~{p 1} => _. k 8*[Cdc 2] for Cdc 2 => Cdc 2~{p 1}. k 9*[Cdc 2~{p 1}] for Cdc 2~{p 1} => Cdc 2. k 4 p*[Cdc 2~{p 1}-Cyclin~{p 1}] for Cdc 2~{p 1}-Cyclin~{p 1} => Cdc 2 -Cyclin~{p 1}. k 4*[Cdc 2 -Cyclin~{p 1}]^2*[Cdc 2~{p 1}-Cyclin~{p 1}] for Cdc 2~{p 1}-Cyclin~{p 1} =[Cdc 2 -Cyclin~{p 1}]=> Cdc 2 -Cyclin~{p 1}. Shonan village 14/11/11 François Fages
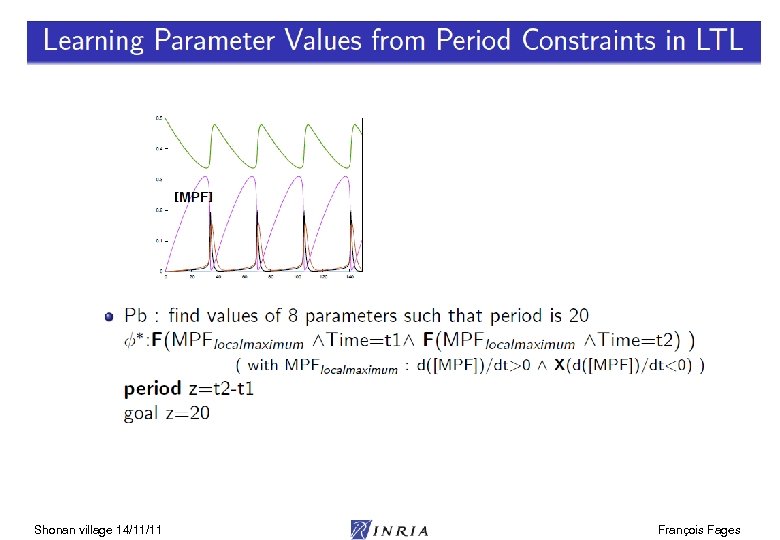
![Parameter Search from LTL(R) Properties biocham: learn_parameter([k 3, k 4], [(0, 200), (0, 200)], Parameter Search from LTL(R) Properties biocham: learn_parameter([k 3, k 4], [(0, 200), (0, 200)],](https://present5.com/presentation/3e3d353363a31905901c9f2ddee45fc9/image-18.jpg)
Parameter Search from LTL(R) Properties biocham: learn_parameter([k 3, k 4], [(0, 200), (0, 200)], 20, oscil(Cdc 2 -Cyclin~{p 1}, 3), 150). Shonan village 14/11/11 François Fages
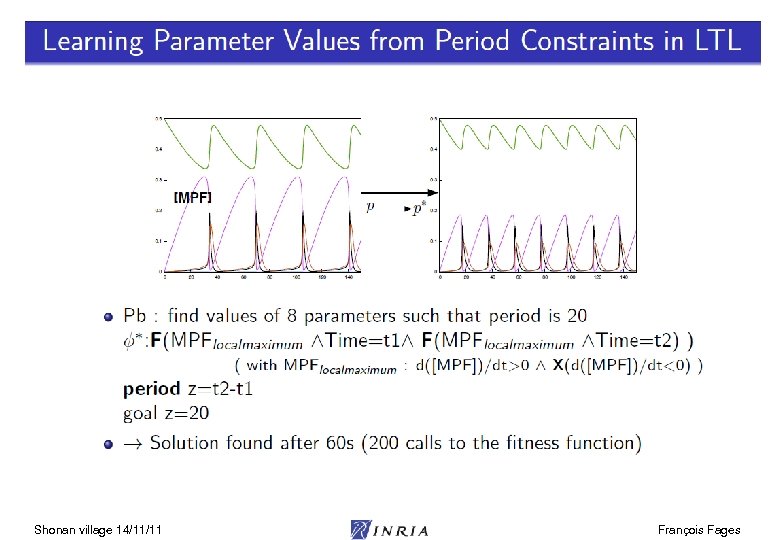
![Parameter Search from LTL(R) Properties biocham: learn_parameter([k 3, k 4], [(0, 200), (0, 200)], Parameter Search from LTL(R) Properties biocham: learn_parameter([k 3, k 4], [(0, 200), (0, 200)],](https://present5.com/presentation/3e3d353363a31905901c9f2ddee45fc9/image-19.jpg)
Parameter Search from LTL(R) Properties biocham: learn_parameter([k 3, k 4], [(0, 200), (0, 200)], 20, oscil(Cdc 2 -Cyclin~{p 1}, 3), 150). First values found : parameter(k 3, 10). parameter(k 4, 70). Shonan village 14/11/11 François Fages
![Parameter Search from LTL(R) Properties biocham: learn_parameter([k 3, k 4], [(0, 200), (0, 200)], Parameter Search from LTL(R) Properties biocham: learn_parameter([k 3, k 4], [(0, 200), (0, 200)],](https://present5.com/presentation/3e3d353363a31905901c9f2ddee45fc9/image-20.jpg)
Parameter Search from LTL(R) Properties biocham: learn_parameter([k 3, k 4], [(0, 200), (0, 200)], 20, oscil(Cdc 2 -Cyclin~{p 1}, 3) & F([Cdc 2 -Cyclin~{p 1}]>0. 15), 150). First values found : parameter(k 3, 10). parameter(k 4, 120). Shonan village 14/11/11 François Fages
![Parameter Search from LTL(R) Properties biocham: learn_parameter([k 3, k 4], [(0, 200), (0, 200)], Parameter Search from LTL(R) Properties biocham: learn_parameter([k 3, k 4], [(0, 200), (0, 200)],](https://present5.com/presentation/3e3d353363a31905901c9f2ddee45fc9/image-21.jpg)
Parameter Search from LTL(R) Properties biocham: learn_parameter([k 3, k 4], [(0, 200), (0, 200)], 20, period(Cdc 2 -Cyclin~{p 1}, 35), 150). First values found: parameter(k 3, 10). parameter(k 4, 280). Shonan village 14/11/11 François Fages
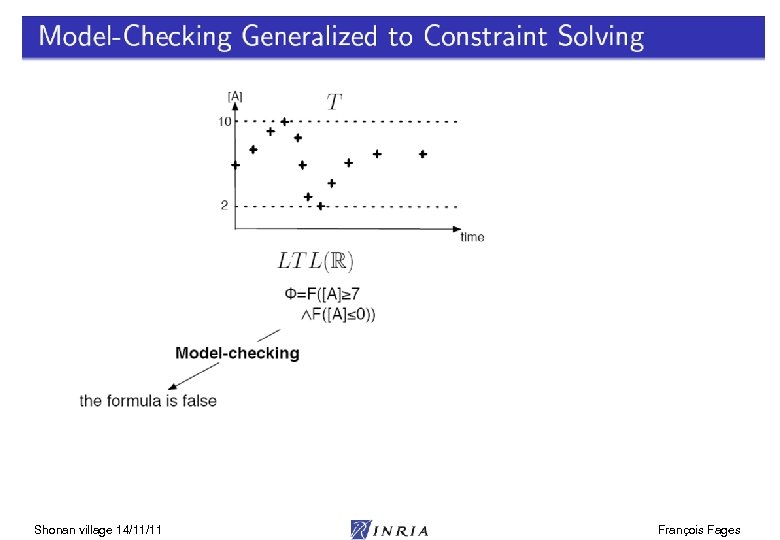
Shonan village 14/11/11 François Fages
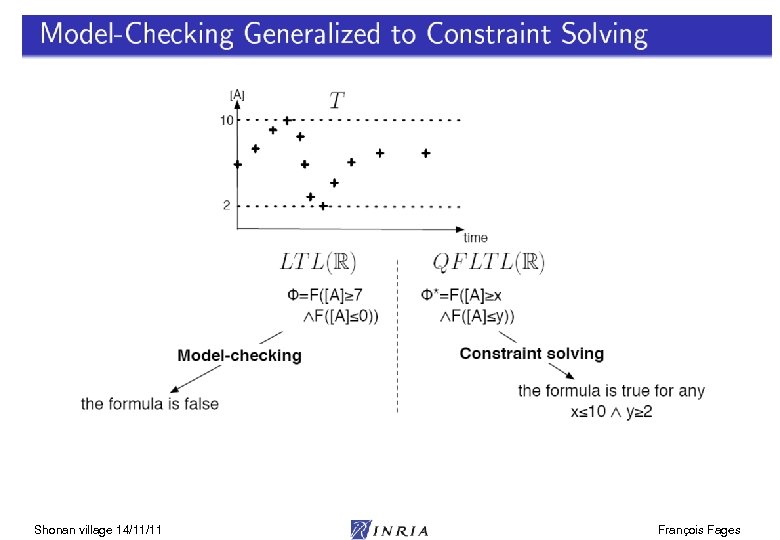
Shonan village 14/11/11 François Fages
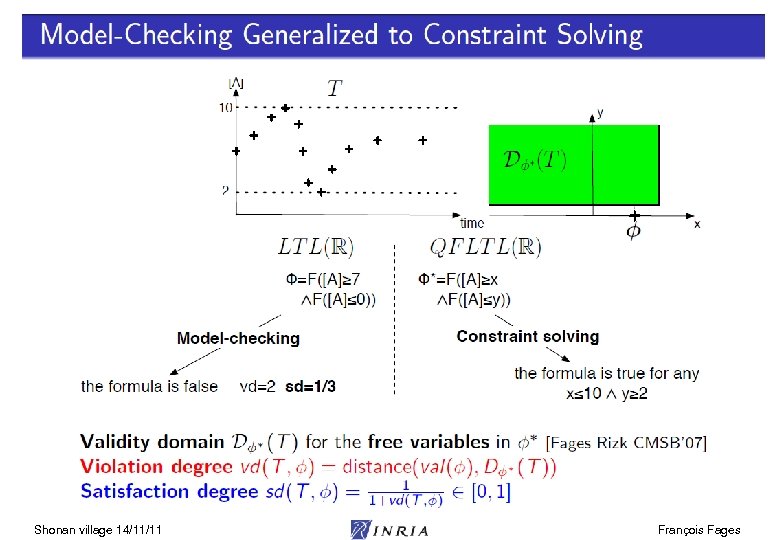
Shonan village 14/11/11 François Fages
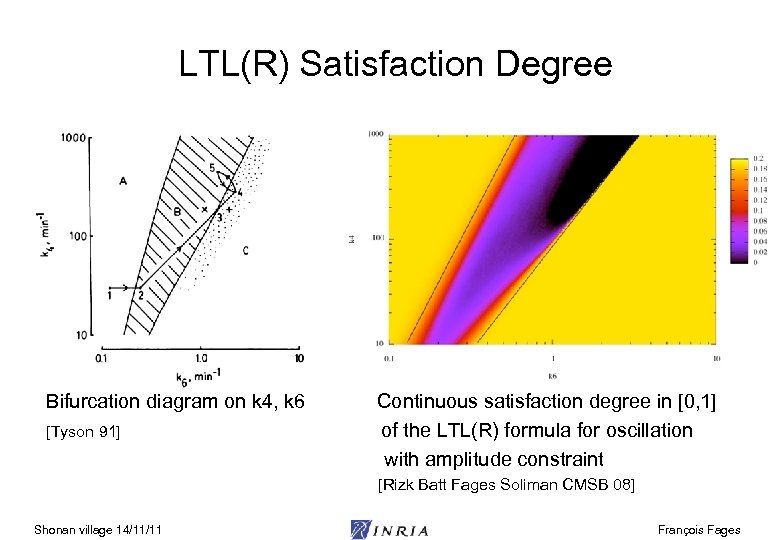
LTL(R) Satisfaction Degree Bifurcation diagram on k 4, k 6 Continuous satisfaction degree in [0, 1] [Tyson 91] of the LTL(R) formula for oscillation with amplitude constraint [Rizk Batt Fages Soliman CMSB 08] Shonan village 14/11/11 François Fages
Shonan village 14/11/11 François Fages
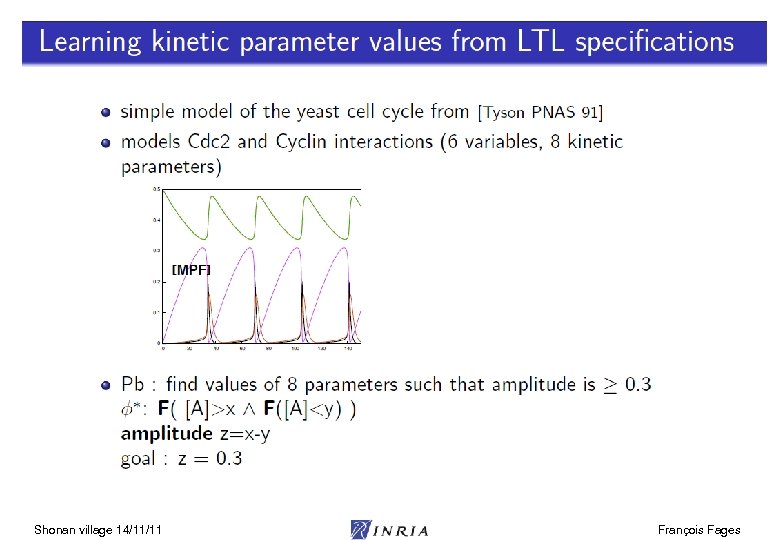
Shonan village 14/11/11 François Fages
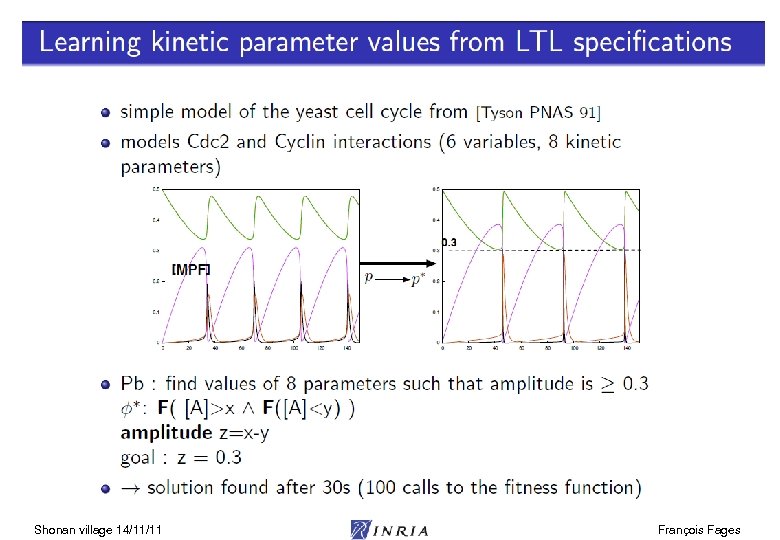
Shonan village 14/11/11 François Fages
Shonan village 14/11/11 François Fages
Shonan village 14/11/11 François Fages
Shonan village 14/11/11 François Fages
Shonan village 14/11/11 François Fages
Shonan village 14/11/11 François Fages
Shonan village 14/11/11 François Fages
Shonan village 14/11/11 François Fages
Shonan village 14/11/11 François Fages
![[Rizk Batt Fages Soliman ISMB’ 09 Bioinformatics] Shonan village 14/11/11 François Fages [Rizk Batt Fages Soliman ISMB’ 09 Bioinformatics] Shonan village 14/11/11 François Fages](https://present5.com/presentation/3e3d353363a31905901c9f2ddee45fc9/image-37.jpg)
[Rizk Batt Fages Soliman ISMB’ 09 Bioinformatics] Shonan village 14/11/11 François Fages
Conclusion • New focus in Systems Biology: formal methods from Computer Science – Beyond diagrammatic notations: formal semantics, abstract interpretation – Beyond simulation: symbolic execution – Beyond curve fitting: high-level specifications in temporal logic – Automatic model-checking. Parameter optimization. Model reduction • New focus in Programming: numerical methods – Beyond discrete machines: stochastic or continuous or hybrid dynamics – Quantitative transition systems, hybrid systems – Continuous satisfaction degree of Temporal Logic specifications, optimization – From model checking (back) to model synthesis [Pnueli 77] Shonan village 14/11/11 François Fages
Acknowledgments • • Contraintes group at INRIA Paris-Rocquencourt on this topic: Grégory Batt, Sylvain Soliman, Elisabetta De Maria, Aurélien Rizk, Steven Gay, Dragana Jovanovska, Faten Nabli, Xavier Duportet, Janis Ulhendorf Era. Net Sys. Bio C 5 Sys (follow up of FP 6 Tempo) on cancer chronotherapies, coord. Francis Lévi, INSERM; Jean Clairambault INRIA; … Coupled models of cell and circadian cycles, p 53/mdm 2, cytotoxic drugs. INRIA/INRA project Regate coord. F. Clément INRIA; E. Reiter, D. Heitzler Modeling of GPCR Angiotensine and FSH signaling networks ANR project Calamar, coord. C. Chaouiya, D. Thieffry , ENS, L. Calzone, Curie. . . Modularity and Compositionality in regulatory networks. OSEO Biointelligence, coord. Dassault-Systèmes ANR Iceberg, coord Grégory Batt, with P. Hersen, Physics, O Gandrillon CNRS, … Model-based control of gene expression in microfluidic ANR Syne 2 arti, coord. Grégory Batt, INRIA with D. Drasdo INRIA, O. Maler, CNRS Verimag, R. Weiss MIT Model-based control of tissue growth in synthetic biology Shonan village 14/11/11 François Fages
3e3d353363a31905901c9f2ddee45fc9.ppt