761634a0aa3bde33a8dbf64ca47c103a.ppt
- Количество слайдов: 20
 Fluxion: The Compara. GRID Data Integration Architecture Matthew Pocock, Tony Burdett, Rob Davey, Andrew Gibson, Trevor Paterson Bio. Models 2007
Fluxion: The Compara. GRID Data Integration Architecture Matthew Pocock, Tony Burdett, Rob Davey, Andrew Gibson, Trevor Paterson Bio. Models 2007
 The Collaboration “Developing a GRID-based system for integrating and exploring data from comparative genomics, to discover biological knowledge that can not be discovered from any one source” ● Collaborative BBSRC project – ● 5 sites across the UK 3/19/2018 http: //3/19/2018 www. comparagrid. org 3/1 9/2018 Bio. Models 2007
The Collaboration “Developing a GRID-based system for integrating and exploring data from comparative genomics, to discover biological knowledge that can not be discovered from any one source” ● Collaborative BBSRC project – ● 5 sites across the UK 3/19/2018 http: //3/19/2018 www. comparagrid. org 3/1 9/2018 Bio. Models 2007
 Fluxion ● ● ● Our data-integration platform Must support comparative genomics We would like it to be broadly re-useable Bio. Models 2007
Fluxion ● ● ● Our data-integration platform Must support comparative genomics We would like it to be broadly re-useable Bio. Models 2007
 Motivations ● Data and “knowledge” about genomics is in many databases – – ● ● ● General Species-specific Process-specific … Then there’s all the things we use to interpret genomics No unified schema / format / formalisation No common location But ● We are pretty sure that useful stuff is waiting to be uncovered by joining these together Bio. Models 2007
Motivations ● Data and “knowledge” about genomics is in many databases – – ● ● ● General Species-specific Process-specific … Then there’s all the things we use to interpret genomics No unified schema / format / formalisation No common location But ● We are pretty sure that useful stuff is waiting to be uncovered by joining these together Bio. Models 2007
 Same problem everywhere ● These issues are common among all of us – – ● Genomics Pathways/modelling Buying a house … Want to minimise how much of the process is performed by people So ● Need to describe what we want formally ● Tools to work with these descriptions Bio. Models 2007
Same problem everywhere ● These issues are common among all of us – – ● Genomics Pathways/modelling Buying a house … Want to minimise how much of the process is performed by people So ● Need to describe what we want formally ● Tools to work with these descriptions Bio. Models 2007
 Target users? ● ● ● Want wide adoption Provide maximal choice about real-life deployment Hide all details from end-user Standard deployable stack with minimal effort Allow 3 rd parties to publish different views of raw data sources Different views for different communities Bio. Models 2007
Target users? ● ● ● Want wide adoption Provide maximal choice about real-life deployment Hide all details from end-user Standard deployable stack with minimal effort Allow 3 rd parties to publish different views of raw data sources Different views for different communities Bio. Models 2007
 Tech Choices ● ● ● Java 5 Haskell Web services – – ● SOAP / WSDL XFire – – Protégé 4 Wonderweb OWL API OWL-DL Bio. Models 2007
Tech Choices ● ● ● Java 5 Haskell Web services – – ● SOAP / WSDL XFire – – Protégé 4 Wonderweb OWL API OWL-DL Bio. Models 2007
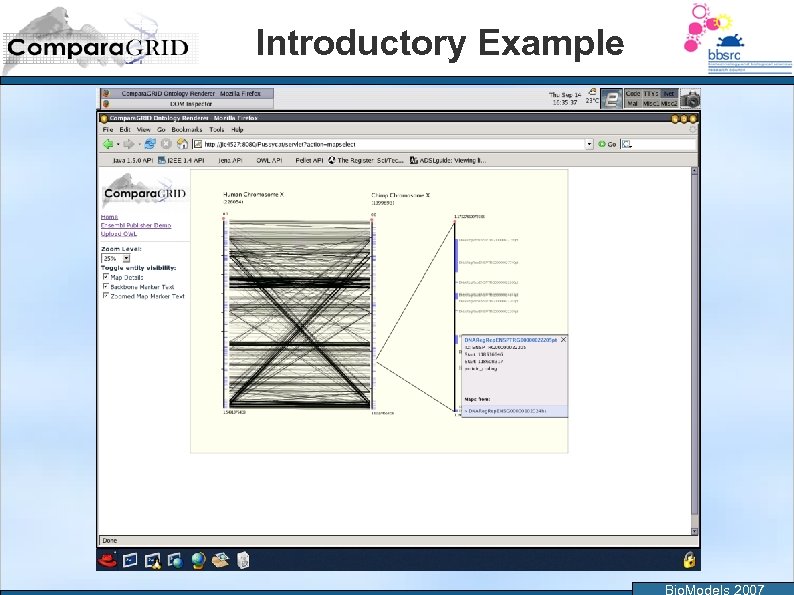 Introductory Example Bio. Models 2007
Introductory Example Bio. Models 2007
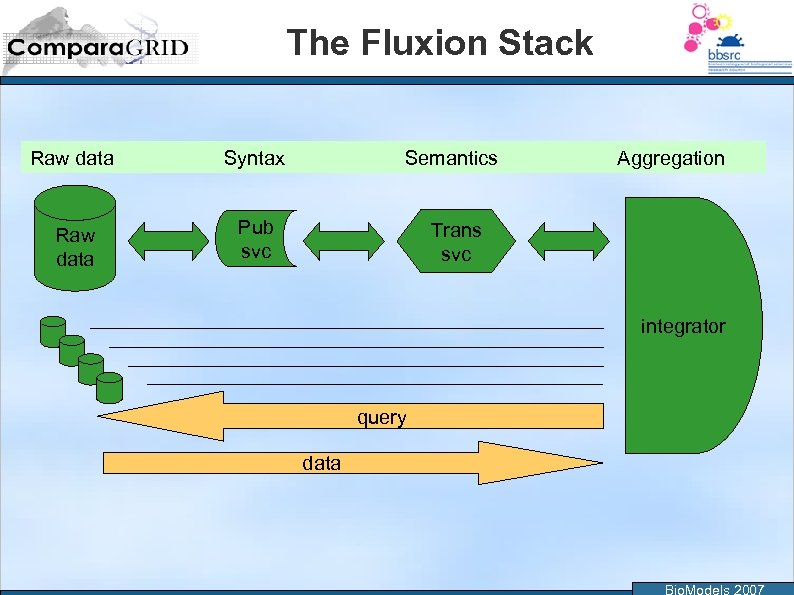 The Fluxion Stack Raw data Syntax Semantics Raw data Pub svc Aggregation Trans svc integrator query data Bio. Models 2007
The Fluxion Stack Raw data Syntax Semantics Raw data Pub svc Aggregation Trans svc integrator query data Bio. Models 2007
 Query Semantics ● Query by providing an OWL class – ● Against knowledge-base exposed by that data-source, not The World Result is a knowledge-base fragment – – – ● All entailed by queried KB (it’s a subset) Can be assertions from the KB, or any entalements Allow a reasoner to classify all the individuals who match the query correctly Preferably using properties, not asserted types (a-box preferred over t-box) Contains at least the statements needed to – ● An application should always run the result + query through an OWL reasoner Bio. Models 2007
Query Semantics ● Query by providing an OWL class – ● Against knowledge-base exposed by that data-source, not The World Result is a knowledge-base fragment – – – ● All entailed by queried KB (it’s a subset) Can be assertions from the KB, or any entalements Allow a reasoner to classify all the individuals who match the query correctly Preferably using properties, not asserted types (a-box preferred over t-box) Contains at least the statements needed to – ● An application should always run the result + query through an OWL reasoner Bio. Models 2007
 Rationale ● ● Low barrier-to-entry for implementers Support a range of implementations – – – ● ● Speed for accuracy Implementation complexity for data-volume Return all instances of known classes e. g. db table with minimal filtering – if in doubt, return it Simplistic implementations Complex implementations – Can compute exactly the minimal amount of data that needs to be returned, but potentially requires a full OWL -DL reasoning cycle for each piece of data Bio. Models 2007
Rationale ● ● Low barrier-to-entry for implementers Support a range of implementations – – – ● ● Speed for accuracy Implementation complexity for data-volume Return all instances of known classes e. g. db table with minimal filtering – if in doubt, return it Simplistic implementations Complex implementations – Can compute exactly the minimal amount of data that needs to be returned, but potentially requires a full OWL -DL reasoning cycle for each piece of data Bio. Models 2007
 Role of Ontology in Fluxion ● ● ● A domain ontology defines what Fluxion integrates Must be endorsed by the target community Needs to capture both the structure and the meaning of the domain – – ● Sbml provides some structure Sbo provides some terminology But – 142 ‘extra’ validation rules Would need to encode all important bits of this in the ontology Developing a ‘good’ domain ontology is – – Hard work Poorly scoped No widely-validated methodology Biologist Modeller so language gap Bio. Models 2007
Role of Ontology in Fluxion ● ● ● A domain ontology defines what Fluxion integrates Must be endorsed by the target community Needs to capture both the structure and the meaning of the domain – – ● Sbml provides some structure Sbo provides some terminology But – 142 ‘extra’ validation rules Would need to encode all important bits of this in the ontology Developing a ‘good’ domain ontology is – – Hard work Poorly scoped No widely-validated methodology Biologist Modeller so language gap Bio. Models 2007
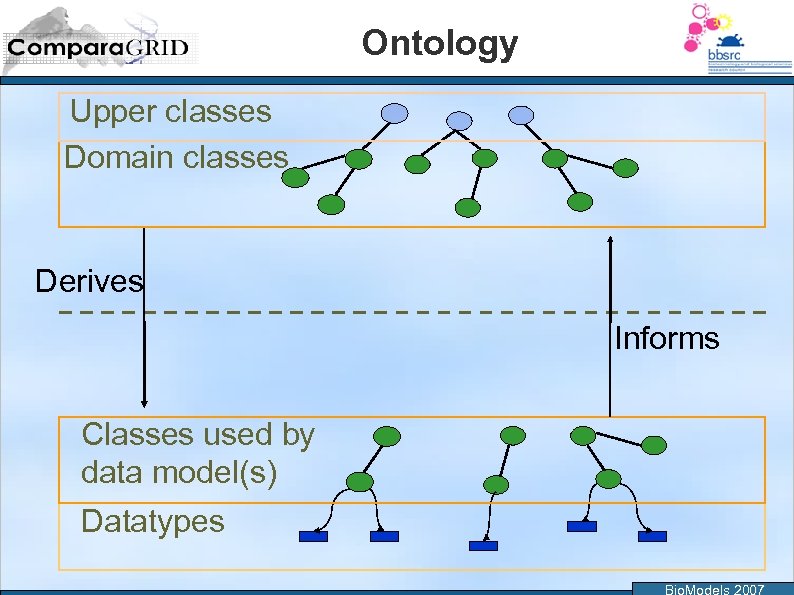 Ontology Upper classes Domain classes Derives Informs Classes used by data model(s) Datatypes Bio. Models 2007
Ontology Upper classes Domain classes Derives Informs Classes used by data model(s) Datatypes Bio. Models 2007
 Publishing Data ● Vast amounts of data in ‘legacy’ databases – – – SQL Text/flat-file Custom/proprietary formats ● Implicit and under-defined semantics ● Data Publisher Role – – ● Schema as OWL concepts Queries populate OWL instances Supported formats automated – ‘mix-in’ knowledge Bio. Models 2007
Publishing Data ● Vast amounts of data in ‘legacy’ databases – – – SQL Text/flat-file Custom/proprietary formats ● Implicit and under-defined semantics ● Data Publisher Role – – ● Schema as OWL concepts Queries populate OWL instances Supported formats automated – ‘mix-in’ knowledge Bio. Models 2007
 Runcible Rules ● Source databases have different models – – ● ● ● Application-specific Mutually incompatible Ontology could become ‘universal union’ Subsumption not the solution Expert knowledge required to map from source schema to domain ontology – – Do not want this ‘fossilized’ in application code Map a source schema to multiple domains Bio. Models 2007
Runcible Rules ● Source databases have different models – – ● ● ● Application-specific Mutually incompatible Ontology could become ‘universal union’ Subsumption not the solution Expert knowledge required to map from source schema to domain ontology – – Do not want this ‘fossilized’ in application code Map a source schema to multiple domains Bio. Models 2007
 Runcible Rules ● Declarative – ● Patterns – – – ● OWL class expressions with ‘holes’ Match against source database Bind variables Generate domain/application OWL – ● Like xpath/xquery, xslt Fill in ‘template’ OWL statements using bound variables Rule application semantics are reversible – – Given source->domain rules, domain->source rules can be machine-generated Supports a wide range of optimization strategies Bio. Models 2007
Runcible Rules ● Declarative – ● Patterns – – – ● OWL class expressions with ‘holes’ Match against source database Bind variables Generate domain/application OWL – ● Like xpath/xquery, xslt Fill in ‘template’ OWL statements using bound variables Rule application semantics are reversible – – Given source->domain rules, domain->source rules can be machine-generated Supports a wide range of optimization strategies Bio. Models 2007
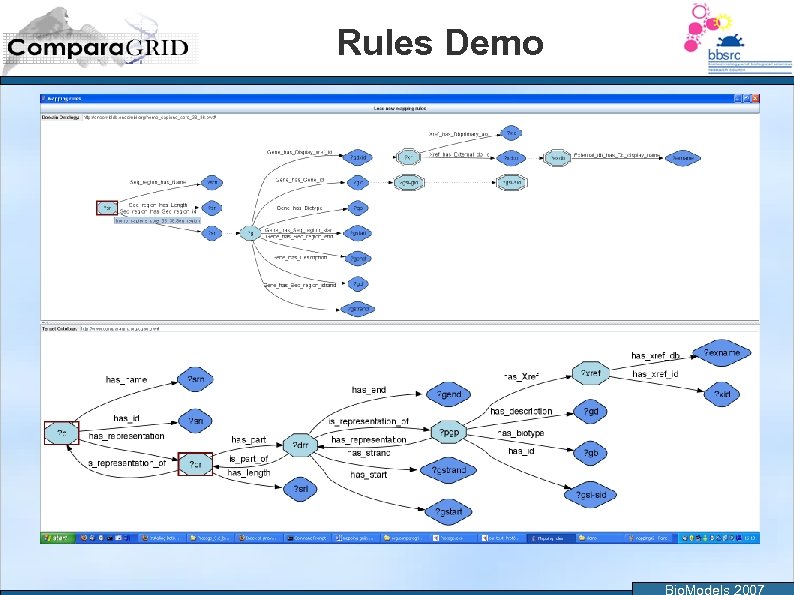 Rules Demo Bio. Models 2007
Rules Demo Bio. Models 2007
 Where Are We? ● Did first live demos in Oct, Nov – – ● ● ● Held together by string Got 1 week to make them work for everyone Web services work as of Xmas Automated publishing of SQL -> OWL works now Data protection rules work for expert user Browser has been working since ISMB, but constantly improves Protégé plugins need re-writing for Protégé 4 Will be available to download (alpha users) once our ISMB paper is in Bio. Models 2007
Where Are We? ● Did first live demos in Oct, Nov – – ● ● ● Held together by string Got 1 week to make them work for everyone Web services work as of Xmas Automated publishing of SQL -> OWL works now Data protection rules work for expert user Browser has been working since ISMB, but constantly improves Protégé plugins need re-writing for Protégé 4 Will be available to download (alpha users) once our ISMB paper is in Bio. Models 2007
 Acknowledgements • Newcastle – – – – Anil Wipat Darren Wilkinson Richard Boys Matthew Pocock Madhu Bhattacharjee Dan Swan Phil Lord • EBI – Peter Rice – Tony Burdett • Manchester – Robert Stevens – Andrew Gibson • Roslin – Andy Law – Trevor Patterson • John Innes Centre – Jo Dicks – Rob Davey 3/19/2018 http: //deanmoor. ncl. ac. uk/blogs 3/19/2018 http: //deanmoor. ncl. ac. uk/websvn 3/19/2018 http: //www. comparagrid. org 3/19/2018 mailto: comparagrid@lists. bbsrc. ac. uk Bio. Models 2007
Acknowledgements • Newcastle – – – – Anil Wipat Darren Wilkinson Richard Boys Matthew Pocock Madhu Bhattacharjee Dan Swan Phil Lord • EBI – Peter Rice – Tony Burdett • Manchester – Robert Stevens – Andrew Gibson • Roslin – Andy Law – Trevor Patterson • John Innes Centre – Jo Dicks – Rob Davey 3/19/2018 http: //deanmoor. ncl. ac. uk/blogs 3/19/2018 http: //deanmoor. ncl. ac. uk/websvn 3/19/2018 http: //www. comparagrid. org 3/19/2018 mailto: comparagrid@lists. bbsrc. ac. uk Bio. Models 2007


