efe71fdb9f05f0bec7582dde33a75fee.ppt
- Количество слайдов: 52

“First generation" sequencing technologies and genome assembly Roger Bumgarner Associate Professor, Microbiology, UW Rogerb@u. washington. edu

Overview • How to sequence any DNA • How to sequence a lot of DNA • What have we learned from 20 years of the genome project? • What’s next?

Intended outcomes • An understanding of: – The process of DNA sequencing – the types/rates of errors in DNA sequence data • A historical perspective of genome sequencing • An understanding of the outcomes of the genome project and the post-genome challenges • A introduction to some of the related ethical issues

Automated DNA Sequencing

Goal - To Read the Sequence of the Basepairs in a region of DNA

DNA Structure

DNA Sequencing: Process Overview • • Generation of a nested set of fragments Separation of the fragments Detection Analysis or base calling

Maxim-Gilbert Sequencing
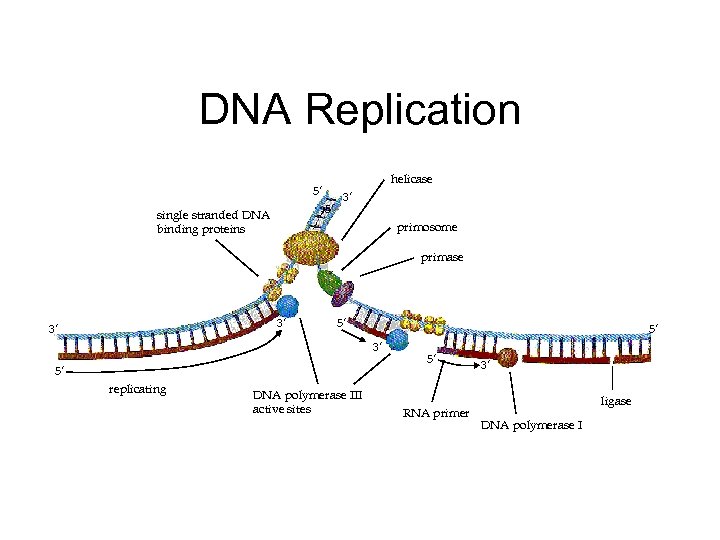
DNA Replication helicase 5’ 5’ single stranded DNA binding proteins 3’ primosome primase 3’ 3’ 5’ 5’ 3’ 5’ replicating DNA polymerase III active sites 5’ RNA primer 3’ ligase DNA polymerase I

The 3’ hydroxyl group is the point of attachment of the next base What happens if the 3’ OH is not there? X

Sanger Sequencing

An “Auto. Rad” of a Sequencing Gel ACGT

With 4 -colors, all reaction can be run in one lane C A C G A C C A A A T C G A A C T T C A A T G T C A Label each with a different color C G A C C A A A T C G A A C T T C A A T G T Mix all reactions prior to loading C G A C C A A A T C G A A C T T C A A T

The Principle of 4 -color Fluorescent DNA Sequencing

The Perkin Elmer/ABI 373 Fluorescence Based DNA Sequencer

A Sequencing Gel Image

Automated DNA Sequencing ACGTT…. - A AC ACGTT +

Gel Analysis Process • Lane Finding - Look for local correlations in the vertical dimension • Lane extraction - sum up pixel across the lanes, straighten if necessary • Transform from wavelength domain to concentration domain • Apply mobility and spacing correction • Filter noise from data - low and high pass filters • Find and identify peaks - numerical derivative • Output called data

Raw Sequencing Data

Idealized Dye Spectra

Actual Dye Spectra

“Chromaticity” Transformation • Measured - Signal in four filters (channels) • Want - Signal in four concentrations [dye]=[fragments] basepairs 4 equations, 4 unknowns I 1 = a 1[A] + c 1[C] + g 1[G] + t 1[T] I 2 = a 2[A] + c 2[C] + g 2[G] + t 2[T] I 3 = a 3[A] + c 3[C] + g 3[G] + t 3[T] I 4 = a 4[A] + c 4[C] + g 4[G] + t 4[T] Matrix formulation I = {x} Conc = {x}-1 I

Gel Analysis Process • Lane Finding - Look for local correlations in the vertical dimension • Lane extraction - sum up pixel across the lanes, straighten if necessary • Transform from wavelength domain to concentration domain • Apply mobility and spacing correction • Filter noise from data - low and high pass filters • Find and identify peaks - numerical derivative • Output called data

Processed Electropherogram

Higher Voltages Produce Faster rates of Electrophoresis • Speed is proportional to Voltage (V) • Current (I) is depends on the resistance of the gel I=V/R • Energy in Watts is W = V*I • Thinner gels give higher R. • Hence, thin or otherwise small gels must be used for higher voltages.


8 Capillary Array

Beckman CEQ 8000 DNA Sequencer • • 8 Capillary Array Linear polyacrylamide separation matrix 4 color terminator sequencing chemistry Windows NT based operating system

Beckman CEQ 8000

Different Labeling Chemistries can be used • Dye Primer - dye is attached to the 5’ end of the sequencing primer. • Dye Terminator - dye is attached to the dd. NTP - allows all 4 reactions to be run in same tube. • Internal Labeling - dye is attached to a d. NTP - signal/molecule increases with length

Large Scale Sequencing

The (Human) Genome Project. The ultimate goal of the Human Genome Project is to decode, letter by letter, the exact sequence of all 3 billion nucleotide bases that make up the human genome. Just a single misplaced letter is sufficient to cause disease. GCTTACTGAGTACATGTGCTAATCGT 3, 400, 000 letters total

The (Human) Genome Project. • Begun in 1990 with a 15 year budget of $3. 0 B overall. • Goals: – To obtain the sequences of human and model Organisms - E-Coli, Drosophila (fruit fly), C-Elegans (a worm), Yeast, Mouse – Develop the necessary technologies to obtain the above.

Sizes and status of a sampling of Genomes

Overview of the goal

How do we begin to analyze a genome? • We want DNA sequence for the entire genome (3. 5 Bbp for human, 4 Mbp for a bacterium). • Sequencing allows one to read about 750 base pairs/sample. • We need a method to sequence bigger pieces.

Primer Walking Vector Primer Clone to sequence Sequence New Primer Sequence Repeat

“Shotgun” sequencing Copy Clone to sequence Sequence and “assemble” …. GTCTACCTGTACTGATCTAGC. . . …. CCTGTACTGATCTAGCATTA. . . …. GTACTGATCTAGCATTACG. . . Subclone

Shotgun vs. walking

Methods for very large scale sequencing • A hierarchical approach – Map on a large scale (physical mapping), sequence specific clones whose position in the genome is known • Shot gun sequencing – “Tear up” the genome and sequence random fragments until it is done • Sequence tagged connectors (STC) – Sequence the ends of many clones and use this info to pick overlapping clones

Making a genomic “library” Isolate DNA Fragment DNA Cells “Library” Clone {

Library Types Ø Chromosome specific libraries Chromosomes can be sorted from one another based on size and GC content. Ø Genomic Libraries - made from the entire genome. Ø Large insert/small insert : combination of vector choice (YAC, BAC, plasmid, m 13), fragmentation method (enzymatic, shearing, sonication), and size selection (by gel or other method).

Another view of a library Multiple copies of the genome (streched out) Randomly fragment and clone Can we order these fragments relative to one another?

Restriction Enzymes - 1970 Copyright 1998 Access Excellence www. gene. com

Physical Mapping : Digest and look for common features in clones A B

Repeat a “minimal tiling path” a many times to construct Pick physical map Sequence these mapped clones (typically by the shotgun method).

Path that was used for genome sequencing YACs BACs or Cosmids m 13, plasmid map (MBP) map (200 k. BP) sequence (kbp)

“Shotgun” the genome Genome to sequence Subclone Sequence and “assemble” …. GTCTACCTGTACTGATCTAGC. . . …. CCTGTACTGATCTAGCATTA. . . …. GTACTGATCTAGCATTACG. . .

Sequence tagged connectors (STC) Genome to sequence Subclone Sequence the ends and store in a d. B Sequence a clone, look for overlaps in the d. B

Which method? • Whole genome shot-gun – Very successful for bacteria – Celera’s approach to the human genome, but what about repeats? • Physical mapping – Traditional method • STC – Hybrid method, not a difficult as physical mapping, can resolve some issue with repeats.

Issues with genome sequencing • • • Whose genome? Quality Contiguity Publication Patenting

What are the fruits of genome sequencing? • A nearly complete list of genes • A reference against which to compare other sequences – Identify polymorphisms in the population – Comparative genomics • Identify highly conserved regions • Evolutionary inferences • A tremendously enabling reagent resource – PCR primers – Microarrays for expression – SNP’s for genetic mapping
efe71fdb9f05f0bec7582dde33a75fee.ppt