357b5a01f4cb1246eaf5a844674a72db.ppt
- Количество слайдов: 17
 Faculty of Computer Science Towards Applying Text Mining and Natural Language Processing for Biomedical Ontology Acquisition Inniss T. , Light M. , Thomas G. , Lee J. , Grassi M. , Williams A. TMBIO(2006) Amit Satsangi amit@cs. ualberta. ca CMPUT 603 © 2006
Faculty of Computer Science Towards Applying Text Mining and Natural Language Processing for Biomedical Ontology Acquisition Inniss T. , Light M. , Thomas G. , Lee J. , Grassi M. , Williams A. TMBIO(2006) Amit Satsangi amit@cs. ualberta. ca CMPUT 603 © 2006
 Department of Computing Science Focus Ontology for describing age-related macular degeneration (AMD) Comparison of the accuracy of three methods for Ontology – Natural Language Processing (NLP) – Text Mining (SAS Text Miner) – Human Expert Manual and adhoc knowledge acquisition IDOCS (Intelligent Distributed Ontology Consensus System) CMPUT 605 © 2006
Department of Computing Science Focus Ontology for describing age-related macular degeneration (AMD) Comparison of the accuracy of three methods for Ontology – Natural Language Processing (NLP) – Text Mining (SAS Text Miner) – Human Expert Manual and adhoc knowledge acquisition IDOCS (Intelligent Distributed Ontology Consensus System) CMPUT 605 © 2006
 Department of Computing Science Introduction No existing common and standardized vocabulary for classification of disease types for certain eyediseases Clinicians, dispersed geographically, may use different terms to describe the same condition Research aimed at extracting the feature and attribute descriptions for the vocabulary of AMD, and build an Ontology from that. CMPUT 605 © 2006
Department of Computing Science Introduction No existing common and standardized vocabulary for classification of disease types for certain eyediseases Clinicians, dispersed geographically, may use different terms to describe the same condition Research aimed at extracting the feature and attribute descriptions for the vocabulary of AMD, and build an Ontology from that. CMPUT 605 © 2006
 Department of Computing Science Related Work Lot of research done, since 1990’s, for applying NLP techniques in medicine, bio-medicine etc. NLP & Text Data Mining have been recognized to play an important role in this endeavor Research focused on online repositories such as Medline & Pub. Med NLP systems developed: Med. Lee, UMLS, GENIES etc. CMPUT 605 © 2006
Department of Computing Science Related Work Lot of research done, since 1990’s, for applying NLP techniques in medicine, bio-medicine etc. NLP & Text Data Mining have been recognized to play an important role in this endeavor Research focused on online repositories such as Medline & Pub. Med NLP systems developed: Med. Lee, UMLS, GENIES etc. CMPUT 605 © 2006
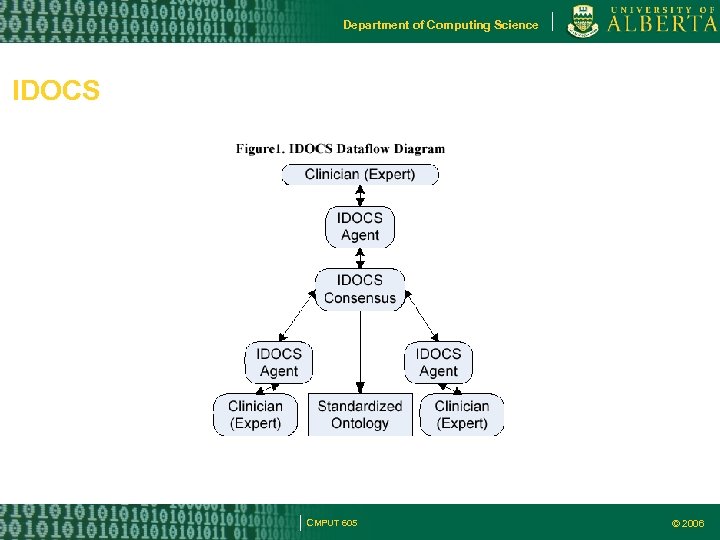 Department of Computing Science IDOCS CMPUT 605 © 2006
Department of Computing Science IDOCS CMPUT 605 © 2006
 Department of Computing Science Methodology Four clinical experts in retinal diseases enlisted to view 100 eye sample images of AMD Experts in different geographic locations Described the observations using digital voice recorders – no artificially imposed vocabulary constraints Another retinal expert for manual parsing of the transcribed text – extracting key words, organization of key-words into categories etc. CMPUT 605 © 2006
Department of Computing Science Methodology Four clinical experts in retinal diseases enlisted to view 100 eye sample images of AMD Experts in different geographic locations Described the observations using digital voice recorders – no artificially imposed vocabulary constraints Another retinal expert for manual parsing of the transcribed text – extracting key words, organization of key-words into categories etc. CMPUT 605 © 2006
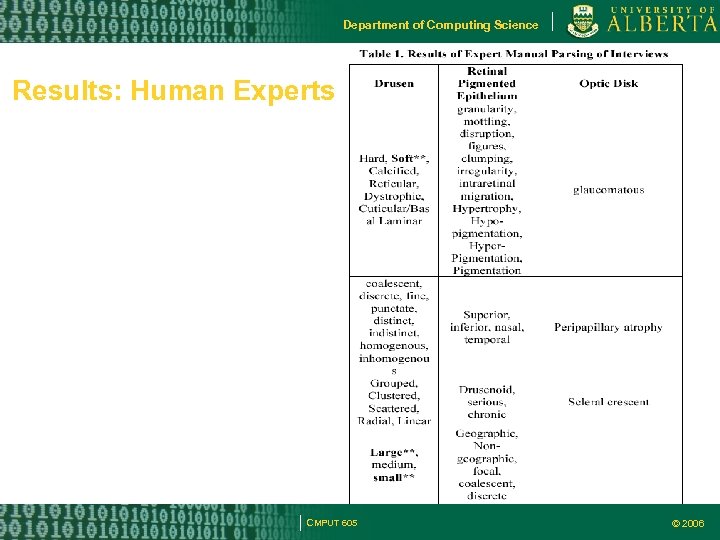 Department of Computing Science Results: Human Experts CMPUT 605 © 2006
Department of Computing Science Results: Human Experts CMPUT 605 © 2006
 Department of Computing Science Methodology: NLP: Used for information extraction and automatic summarization. Identify short sequences of words having meaning over and above a meaning composed directly from their parts – “extreme programming” Ngram Statistics Package (NSP) used for collocation discovery in case of bi-grams Word-pair associations measured by PMI CMPUT 605 © 2006
Department of Computing Science Methodology: NLP: Used for information extraction and automatic summarization. Identify short sequences of words having meaning over and above a meaning composed directly from their parts – “extreme programming” Ngram Statistics Package (NSP) used for collocation discovery in case of bi-grams Word-pair associations measured by PMI CMPUT 605 © 2006
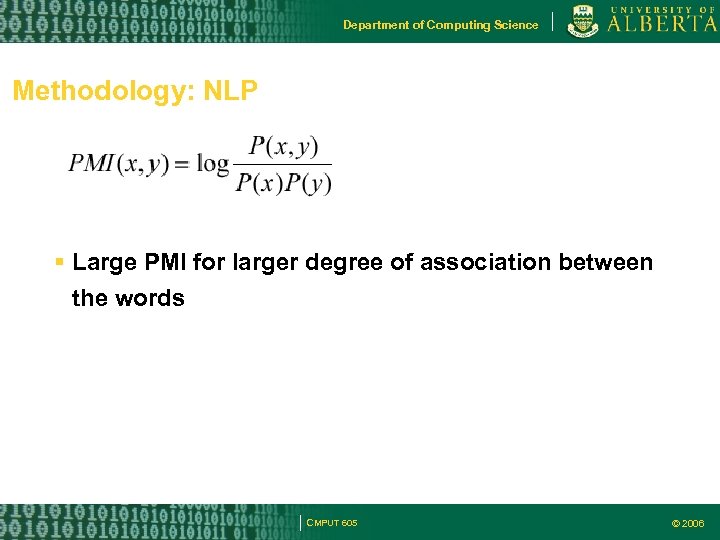 Department of Computing Science Methodology: NLP Large PMI for larger degree of association between the words CMPUT 605 © 2006
Department of Computing Science Methodology: NLP Large PMI for larger degree of association between the words CMPUT 605 © 2006
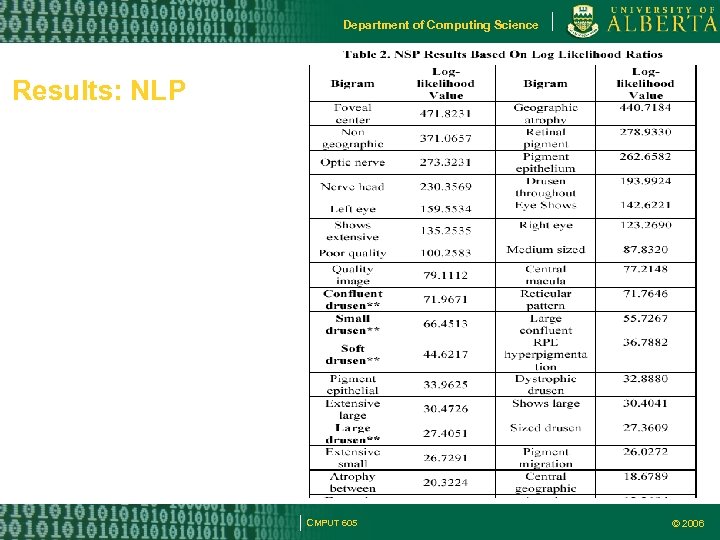 Department of Computing Science Results: NLP CMPUT 605 © 2006
Department of Computing Science Results: NLP CMPUT 605 © 2006
 Department of Computing Science Methodology: Text Mining (SAS Text Miner) Collection of documents (corpus) used as input to any text mining algorithm Corpus broken into tokens or terms (tokens in a particular language) Term weighting Measures: Entropy, Inverse Document Frequency (IDF), Global Frequency (GF) IDF, None (Global weight of 1) & Normal term wt. CMPUT 605 © 2006
Department of Computing Science Methodology: Text Mining (SAS Text Miner) Collection of documents (corpus) used as input to any text mining algorithm Corpus broken into tokens or terms (tokens in a particular language) Term weighting Measures: Entropy, Inverse Document Frequency (IDF), Global Frequency (GF) IDF, None (Global weight of 1) & Normal term wt. CMPUT 605 © 2006
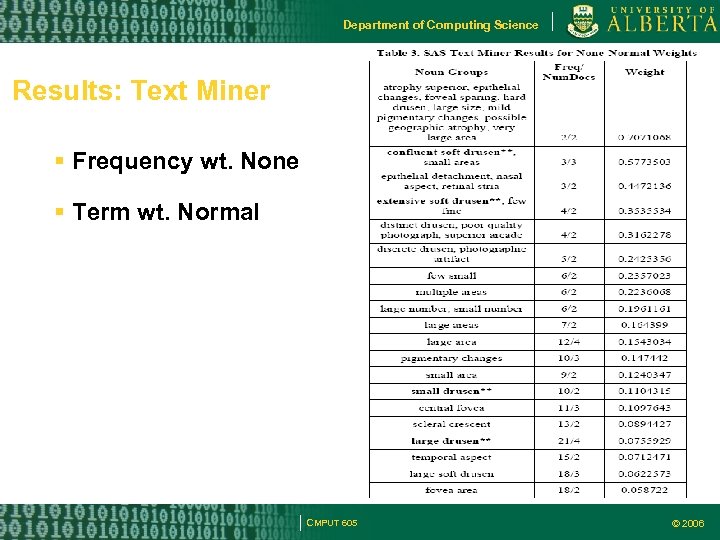 Department of Computing Science Results: Text Miner Frequency wt. None Term wt. Normal CMPUT 605 © 2006
Department of Computing Science Results: Text Miner Frequency wt. None Term wt. Normal CMPUT 605 © 2006
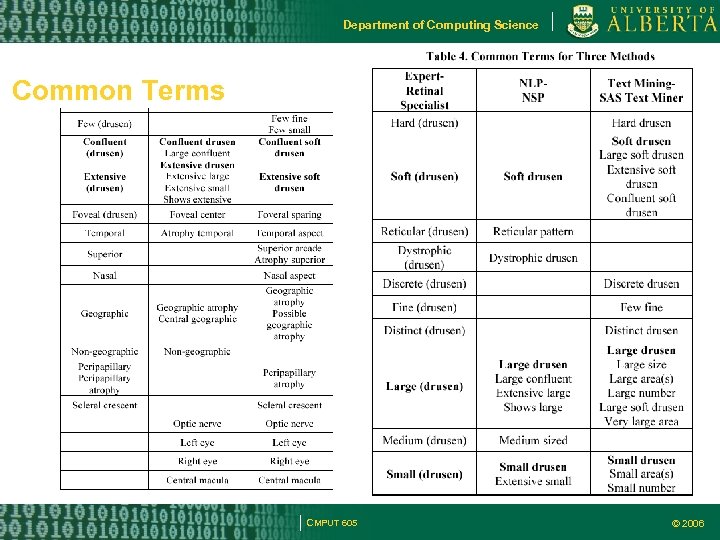 Department of Computing Science Common Terms sss CMPUT 605 © 2006
Department of Computing Science Common Terms sss CMPUT 605 © 2006
 Department of Computing Science Comparison Thus text mining is a viable and effective method for determining vocabulary to describe a particular disease Text Mining found a lot of terms that NLP found Human Expert is the best Ground Truth CMPUT 605 © 2006
Department of Computing Science Comparison Thus text mining is a viable and effective method for determining vocabulary to describe a particular disease Text Mining found a lot of terms that NLP found Human Expert is the best Ground Truth CMPUT 605 © 2006
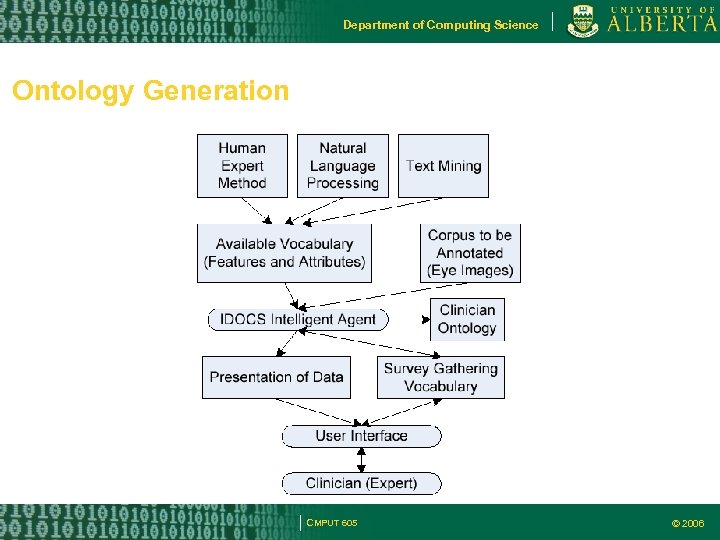 Department of Computing Science Ontology Generation CMPUT 605 © 2006
Department of Computing Science Ontology Generation CMPUT 605 © 2006
 Department of Computing Science Conclusion and Future Work Human experts are the best, but they did miss some key descriptors Text Mining and NLP can enhance the generation of feature generations, by preventing the above case As a consequence more robust vocabulary can be generated Extension – evaluate the effectiveness of the automated tools, text mining & NLP Different weighting schemes to be tried in the future CMPUT 605 © 2006
Department of Computing Science Conclusion and Future Work Human experts are the best, but they did miss some key descriptors Text Mining and NLP can enhance the generation of feature generations, by preventing the above case As a consequence more robust vocabulary can be generated Extension – evaluate the effectiveness of the automated tools, text mining & NLP Different weighting schemes to be tried in the future CMPUT 605 © 2006
 Department of Computing Science Thank You For Your Attention! CMPUT 605 © 2006
Department of Computing Science Thank You For Your Attention! CMPUT 605 © 2006


