aac5893d64327a70857809ef3fd3a91f.ppt
- Количество слайдов: 48
Expression & Purification of Recombinant Proteins August 23, 2009 Biochemistry 201 D. Worthylake, 7152 MEB, x 5176
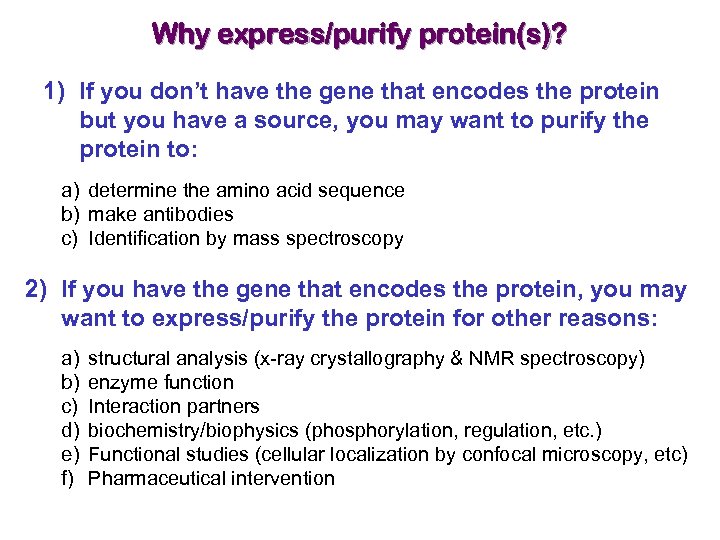
Why express/purify protein(s)? 1) If you don’t have the gene that encodes the protein but you have a source, you may want to purify the protein to: a) determine the amino acid sequence b) make antibodies c) Identification by mass spectroscopy 2) If you have the gene that encodes the protein, you may want to express/purify the protein for other reasons: a) b) c) d) e) f) structural analysis (x-ray crystallography & NMR spectroscopy) enzyme function Interaction partners biochemistry/biophysics (phosphorylation, regulation, etc. ) Functional studies (cellular localization by confocal microscopy, etc) Pharmaceutical intervention

TOP 10 Things to consider 1) Which protein construct to express 2) Expression host (bacterial, insect cell, yeast, or mammalian) 3) Cell line for expression 4) Promoter for induction of protein production 5) Codon optimization (for mammalian proteins expressed in bacteria) 6) Cloning method 7) May require expression as a fusion protein 8) May require co-expression with molecular chaperones 9) Affinity tag(s) for purification & protease cleavage site to remove the tag 10) Purification protocol & buffer to keep protein happy and active
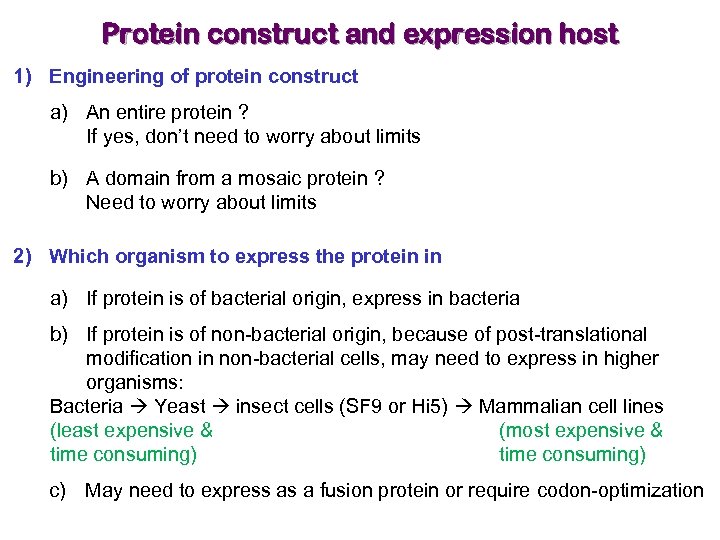
Protein construct and expression host 1) Engineering of protein construct a) An entire protein ? If yes, don’t need to worry about limits b) A domain from a mosaic protein ? Need to worry about limits 2) Which organism to express the protein in a) If protein is of bacterial origin, express in bacteria b) If protein is of non-bacterial origin, because of post-translational modification in non-bacterial cells, may need to express in higher organisms: Bacteria Yeast insect cells (SF 9 or Hi 5) Mammalian cell lines (least expensive & (most expensive & time consuming) c) May need to express as a fusion protein or require codon-optimization
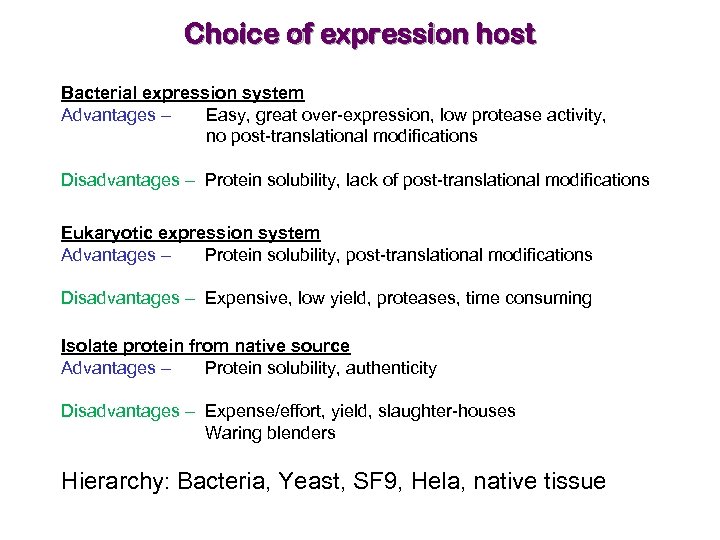
Choice of expression host Bacterial expression system Advantages – Easy, great over-expression, low protease activity, no post-translational modifications Disadvantages – Protein solubility, lack of post-translational modifications Eukaryotic expression system Advantages – Protein solubility, post-translational modifications Disadvantages – Expensive, low yield, proteases, time consuming Isolate protein from native source Advantages – Protein solubility, authenticity Disadvantages – Expense/effort, yield, slaughter-houses Waring blenders Hierarchy: Bacteria, Yeast, SF 9, Hela, native tissue
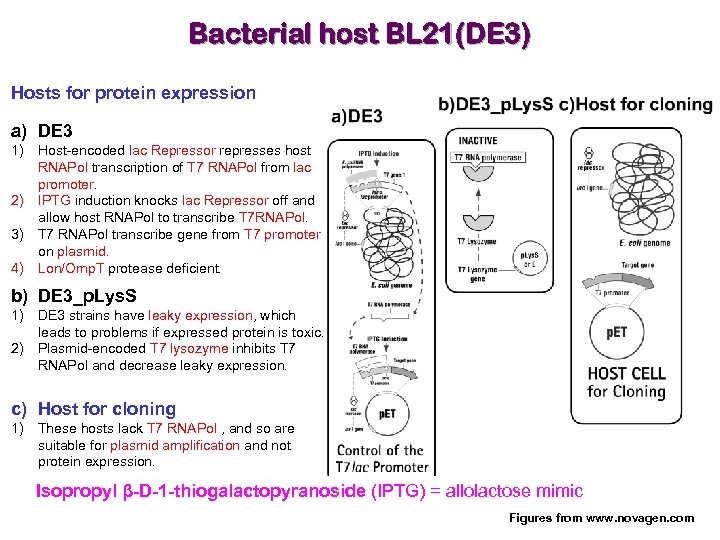
Bacterial host BL 21(DE 3) Hosts for protein expression a) DE 3 1) Host-encoded lac Repressor represses host RNAPol transcription of T 7 RNAPol from lac promoter. 2) IPTG induction knocks lac Repressor off and allow host RNAPol to transcribe T 7 RNAPol. 3) T 7 RNAPol transcribe gene from T 7 promoter on plasmid. 4) Lon/Omp. T protease deficient. b) DE 3_p. Lys. S 1) DE 3 strains have leaky expression, which leads to problems if expressed protein is toxic. 2) Plasmid-encoded T 7 lysozyme inhibits T 7 RNAPol and decrease leaky expression. c) Host for cloning 1) These hosts lack T 7 RNAPol , and so are suitable for plasmid amplification and not protein expression. Isopropyl β-D-1 -thiogalactopyranoside (IPTG) = allolactose mimic Figures from www. novagen. com
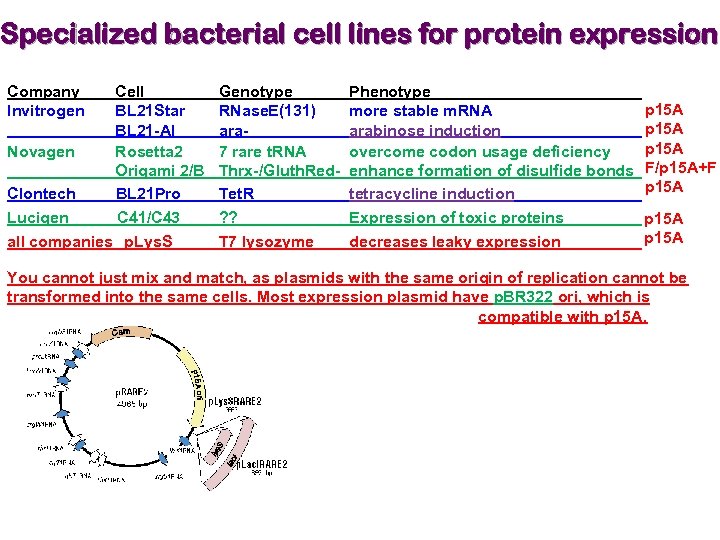
Specialized bacterial cell lines for protein expression Company Invitrogen Cell BL 21 Star BL 21 -AI Novagen Rosetta 2 Origami 2/B Clontech BL 21 Pro Lucigen C 41/C 43 all companies p. Lys. S Genotype RNase. E(131) ara 7 rare t. RNA Thrx-/Gluth. Red. Tet. R ? ? T 7 lysozyme Phenotype more stable m. RNA arabinose induction overcome codon usage deficiency enhance formation of disulfide bonds tetracycline induction Expression of toxic proteins decreases leaky expression p 15 A F/p 15 A+F p 15 A You cannot just mix and match, as plasmids with the same origin of replication cannot be transformed into the same cells. Most expression plasmid have p. BR 322 ori, which is compatible with p 15 A.
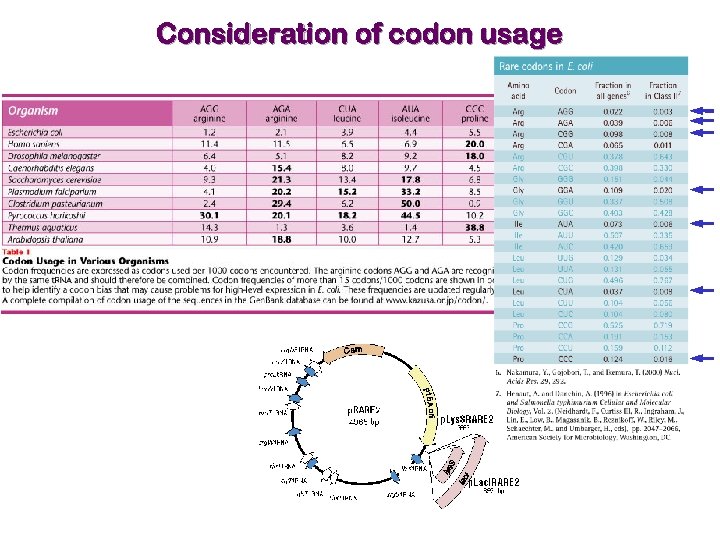
Consideration of codon usage
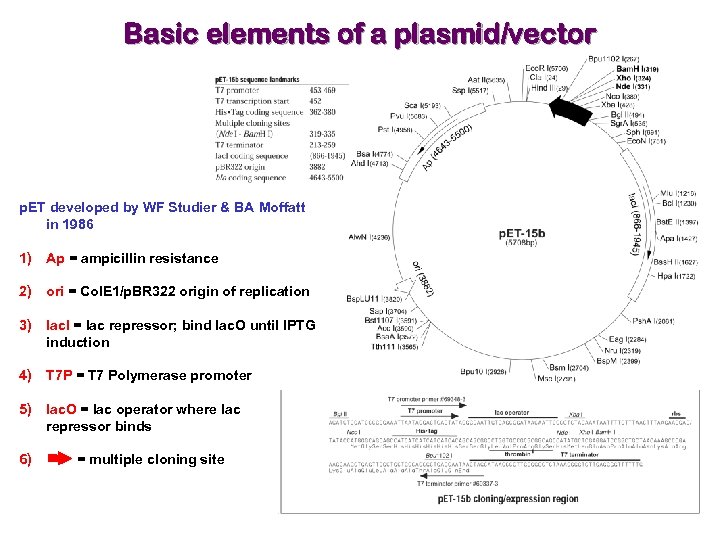
Basic elements of a plasmid/vector p. ET developed by WF Studier & BA Moffatt in 1986 1) Ap = ampicillin resistance 2) ori = Col. E 1/p. BR 322 origin of replication 3) lac. I = lac repressor; bind lac. O until IPTG induction 4) T 7 P = T 7 Polymerase promoter 5) lac. O = lac operator where lac repressor binds 6) = multiple cloning site
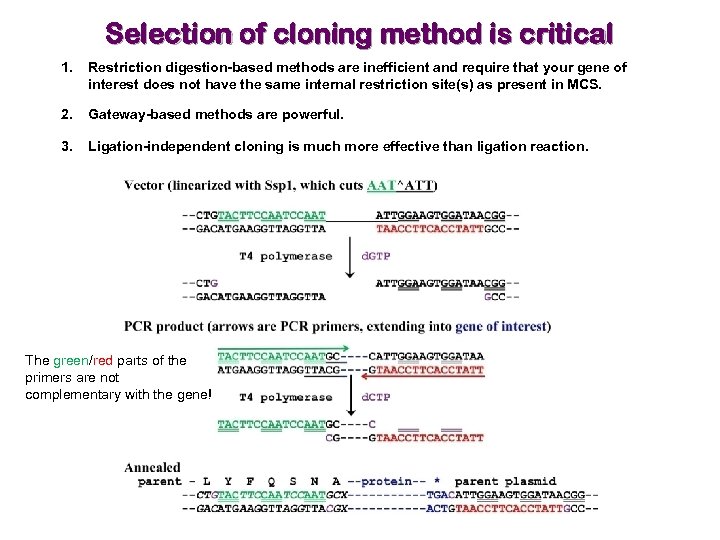
Selection of cloning method is critical 1. Restriction digestion-based methods are inefficient and require that your gene of interest does not have the same internal restriction site(s) as present in MCS. 2. Gateway-based methods are powerful. 3. Ligation-independent cloning is much more effective than ligation reaction. The green/red parts of the primers are not complementary with the gene!
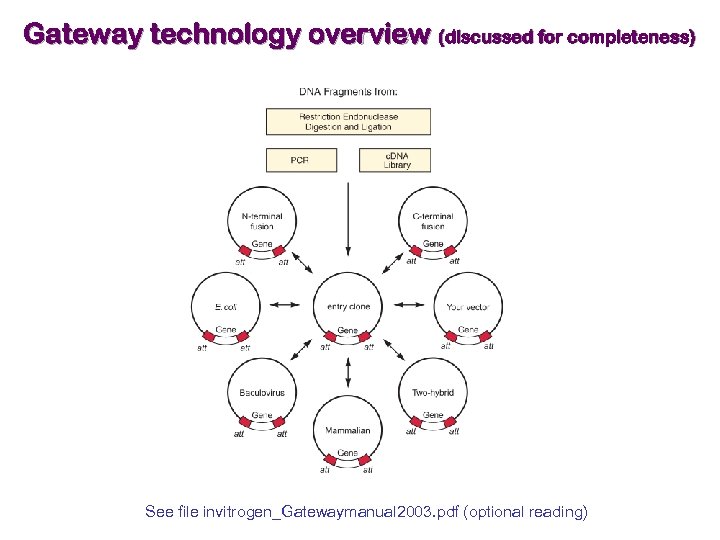
Gateway technology overview (discussed for completeness) See file invitrogen_Gatewaymanual 2003. pdf (optional reading)
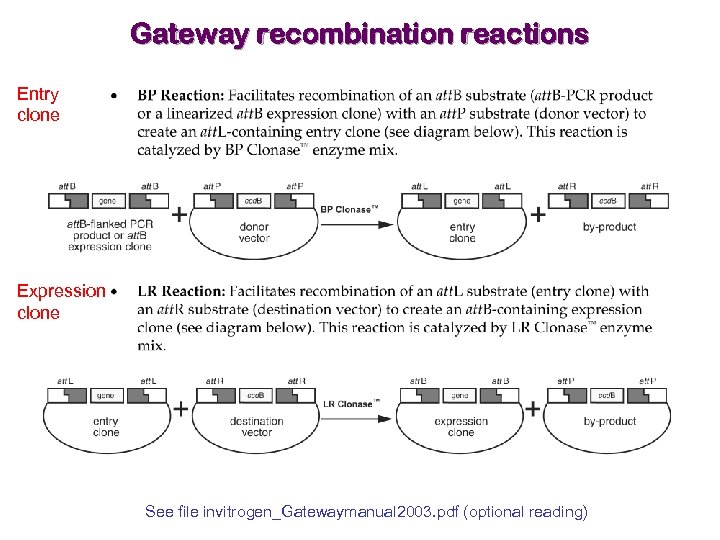
Gateway recombination reactions Entry clone Expression clone See file invitrogen_Gatewaymanual 2003. pdf (optional reading)
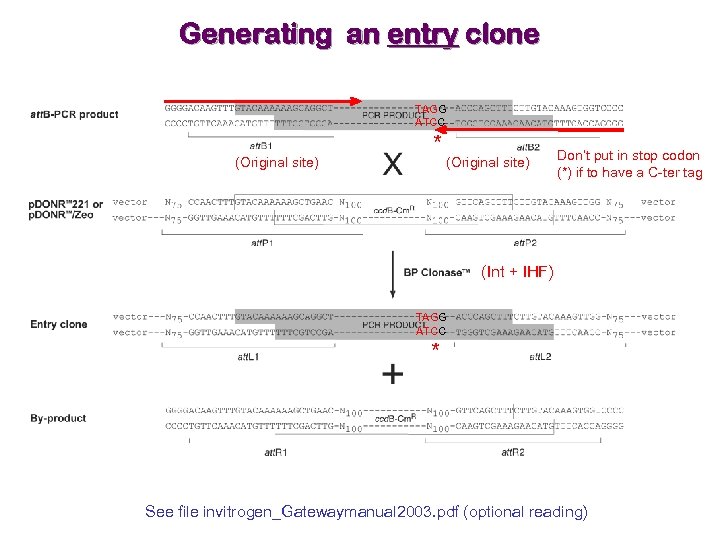
Generating an entry clone TAGG ATCC (Original site) * (Original site) Don’t put in stop codon (*) if to have a C-ter tag (Int + IHF) TAGG ATCC * See file invitrogen_Gatewaymanual 2003. pdf (optional reading)
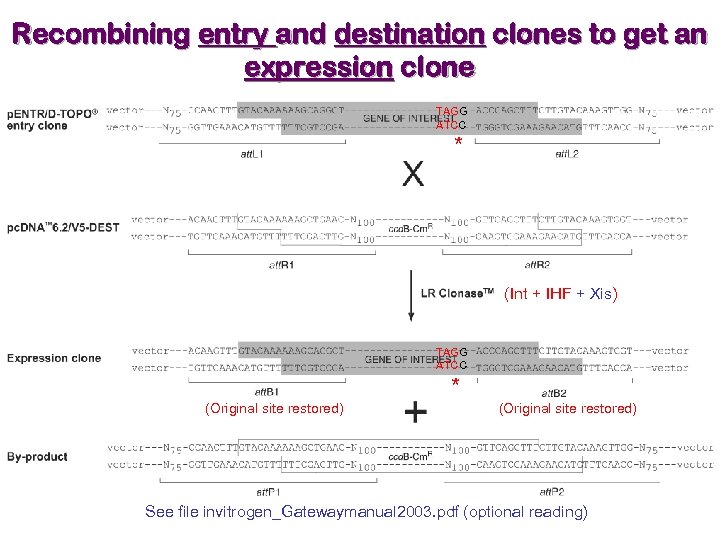
Recombining entry and destination clones to get an expression clone TAGG ATCC * (Int + IHF + Xis) TAGG ATCC * (Original site restored) See file invitrogen_Gatewaymanual 2003. pdf (optional reading)
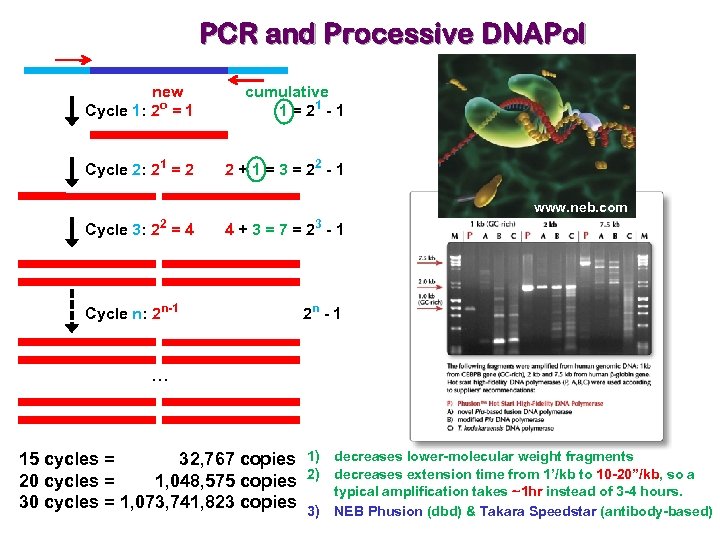
PCR and Processive DNAPol new Cycle 1: 2 o = 1 cumulative 1 = 21 - 1 Cycle 2: 21 = 2 2 + 1 = 3 = 22 - 1 www. neb. com 2 Cycle 3: 2 = 4 Cycle n: 2 n-1 3 4+3=7=2 -1 2 n - 1 … 15 cycles = 32, 767 copies 1) 20 cycles = 1, 048, 575 copies 2) 30 cycles = 1, 073, 741, 823 copies 3) decreases lower-molecular weight fragments decreases extension time from 1’/kb to 10 -20”/kb, so a typical amplification takes ~1 hr instead of 3 -4 hours. NEB Phusion (dbd) & Takara Speedstar (antibody-based)
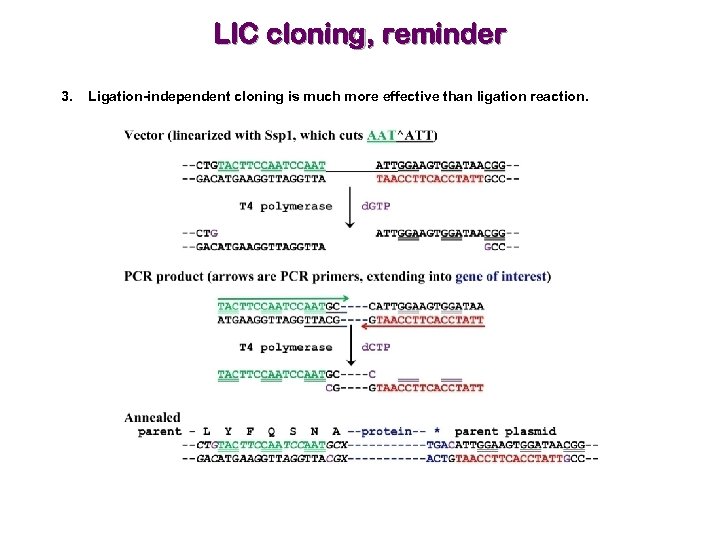
LIC cloning, reminder 3. Ligation-independent cloning is much more effective than ligation reaction.
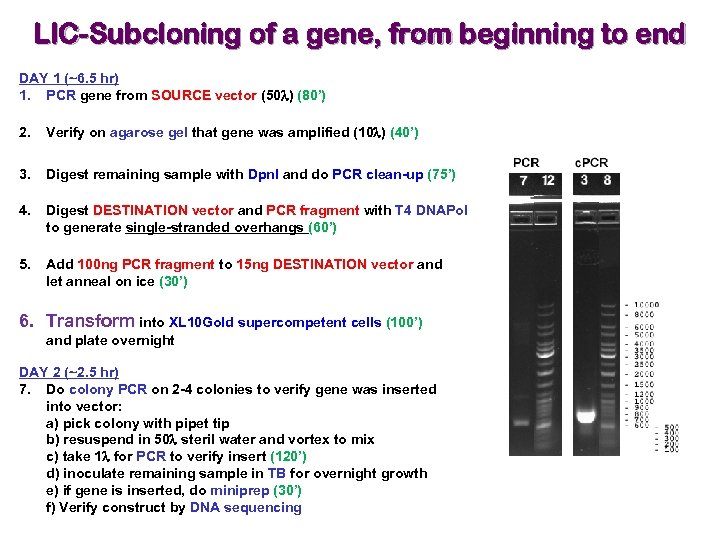
LIC-Subcloning of a gene, from beginning to end DAY 1 (~6. 5 hr) 1. PCR gene from SOURCE vector (50 l) (80’) 2. Verify on agarose gel that gene was amplified (10 l) (40’) 3. Digest remaining sample with Dpn. I and do PCR clean-up (75’) 4. Digest DESTINATION vector and PCR fragment with T 4 DNAPol to generate single-stranded overhangs (60’) 5. Add 100 ng PCR fragment to 15 ng DESTINATION vector and let anneal on ice (30’) 6. Transform into XL 10 Gold supercompetent cells (100’) and plate overnight DAY 2 (~2. 5 hr) 7. Do colony PCR on 2 -4 colonies to verify gene was inserted into vector: a) pick colony with pipet tip b) resuspend in 50 l steril water and vortex to mix c) take 1 l for PCR to verify insert (120’) d) inoculate remaining sample in TB for overnight growth e) if gene is inserted, do miniprep (30’) f) Verify construct by DNA sequencing
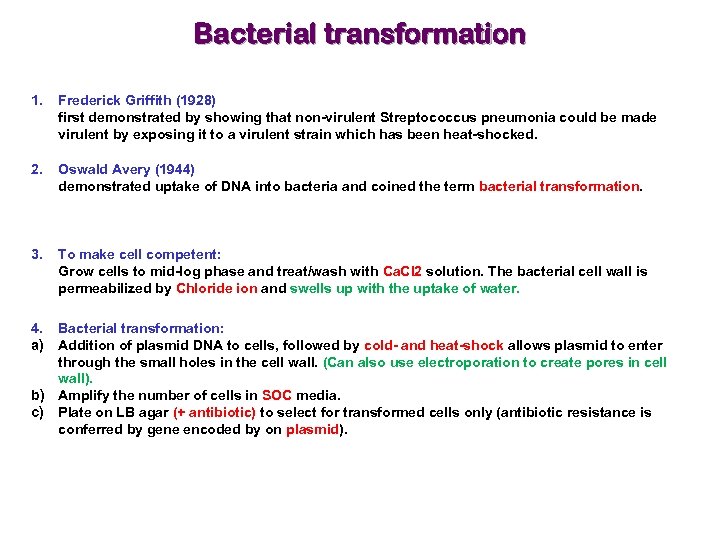
Bacterial transformation 1. Frederick Griffith (1928) first demonstrated by showing that non-virulent Streptococcus pneumonia could be made virulent by exposing it to a virulent strain which has been heat-shocked. 2. Oswald Avery (1944) demonstrated uptake of DNA into bacteria and coined the term bacterial transformation. 3. To make cell competent: Grow cells to mid-log phase and treat/wash with Ca. Cl 2 solution. The bacterial cell wall is permeabilized by Chloride ion and swells up with the uptake of water. 4. Bacterial transformation: a) Addition of plasmid DNA to cells, followed by cold- and heat-shock allows plasmid to enter through the small holes in the cell wall. (Can also use electroporation to create pores in cell wall). b) Amplify the number of cells in SOC media. c) Plate on LB agar (+ antibiotic) to select for transformed cells only (antibiotic resistance is conferred by gene encoded by on plasmid).
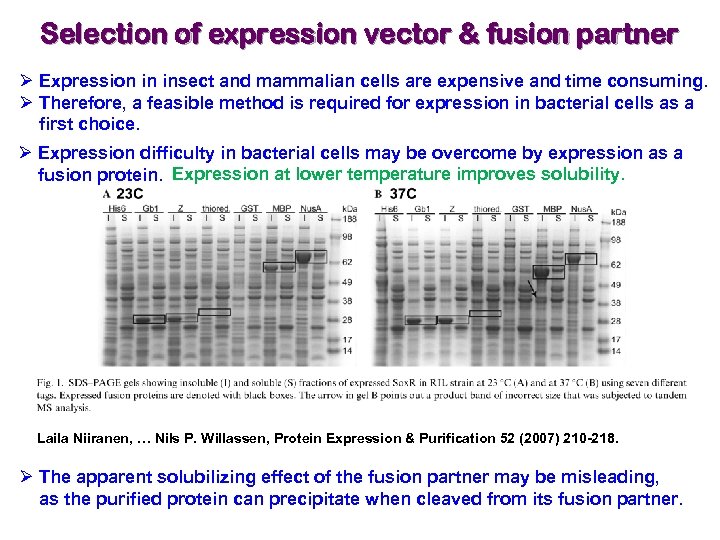
Selection of expression vector & fusion partner Ø Expression in insect and mammalian cells are expensive and time consuming. Ø Therefore, a feasible method is required for expression in bacterial cells as a first choice. Ø Expression difficulty in bacterial cells may be overcome by expression as a fusion protein. Expression at lower temperature improves solubility. Laila Niiranen, … Nils P. Willassen, Protein Expression & Purification 52 (2007) 210 -218. Ø The apparent solubilizing effect of the fusion partner may be misleading, as the purified protein can precipitate when cleaved from its fusion partner.
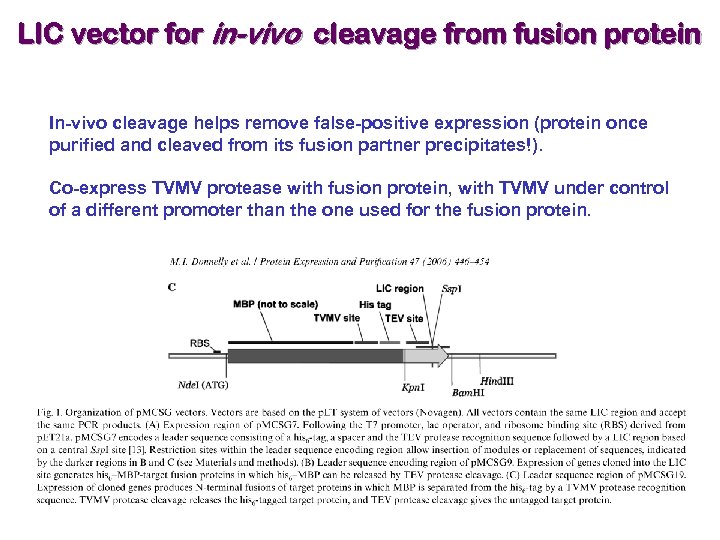
LIC vector for in-vivo cleavage from fusion protein In-vivo cleavage helps remove false-positive expression (protein once purified and cleaved from its fusion partner precipitates!). Co-express TVMV protease with fusion protein, with TVMV under control of a different promoter than the one used for the fusion protein.
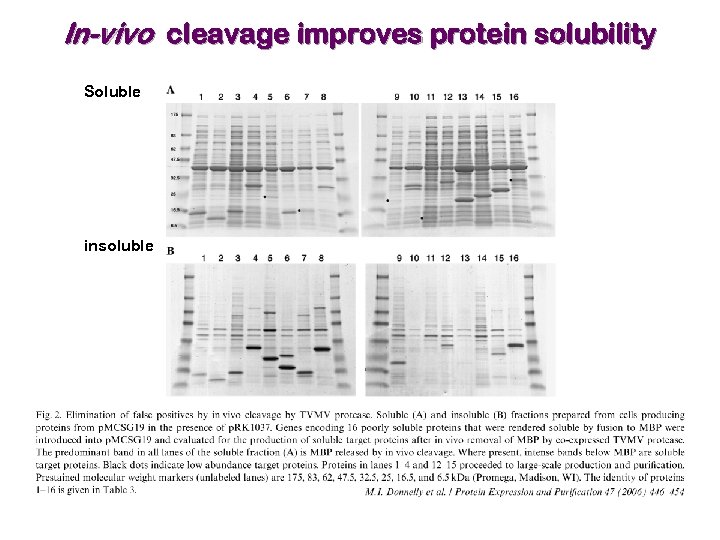
In-vivo cleavage improves protein solubility Soluble insoluble
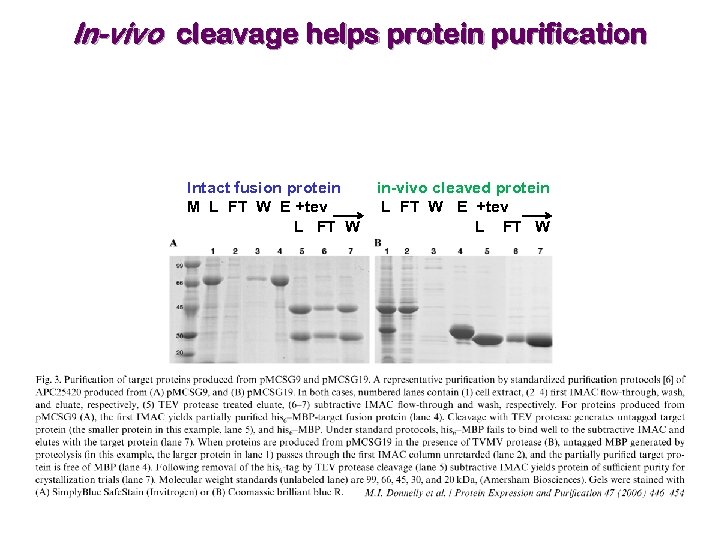
In-vivo cleavage helps protein purification Intact fusion protein M L FT W E +tev L FT W in-vivo cleaved protein L FT W E +tev L FT W
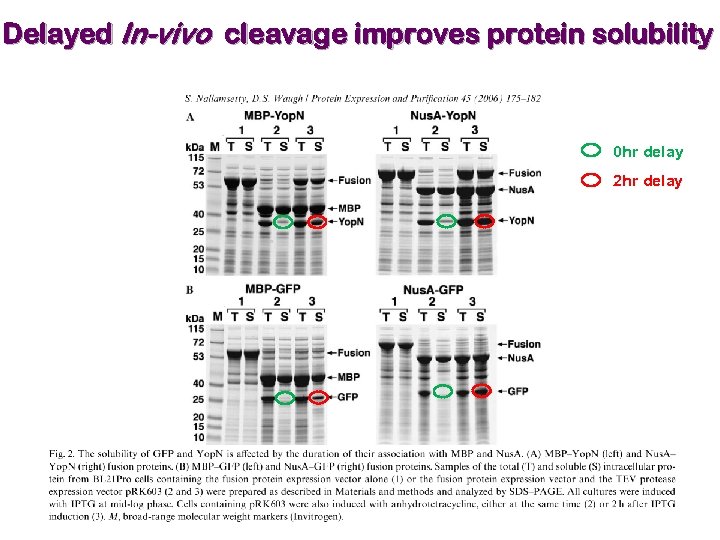
Delayed In-vivo cleavage improves protein solubility 0 hr delay 2 hr delay
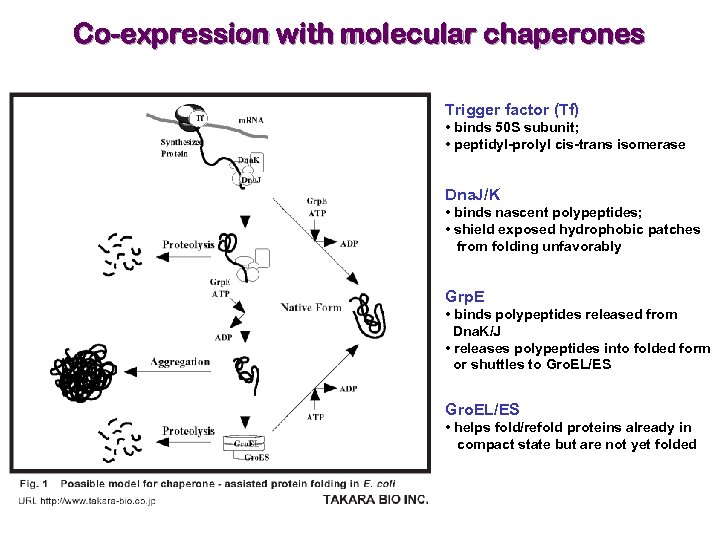
Co-expression with molecular chaperones Trigger factor (Tf) • binds 50 S subunit; • peptidyl-prolyl cis-trans isomerase Dna. J/K • binds nascent polypeptides; • shield exposed hydrophobic patches from folding unfavorably Grp. E • binds polypeptides released from Dna. K/J • releases polypeptides into folded form or shuttles to Gro. EL/ES • helps fold/refold proteins already in compact state but are not yet folded
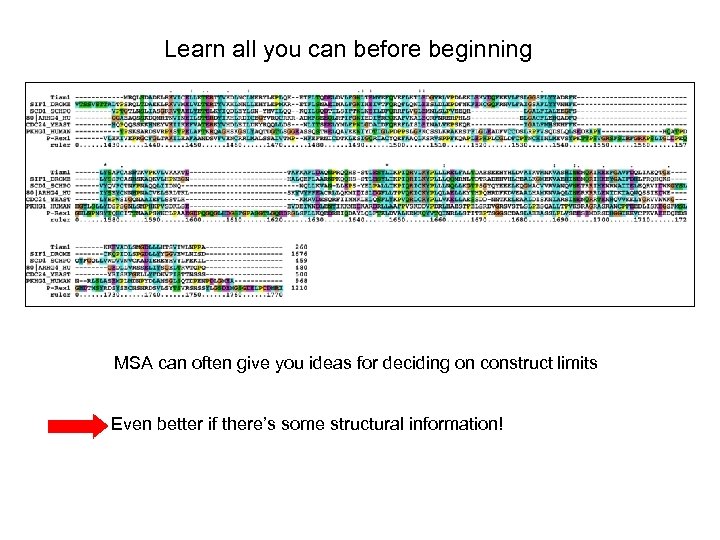
Learn all you can before beginning MSA can often give you ideas for deciding on construct limits Even better if there’s some structural information!
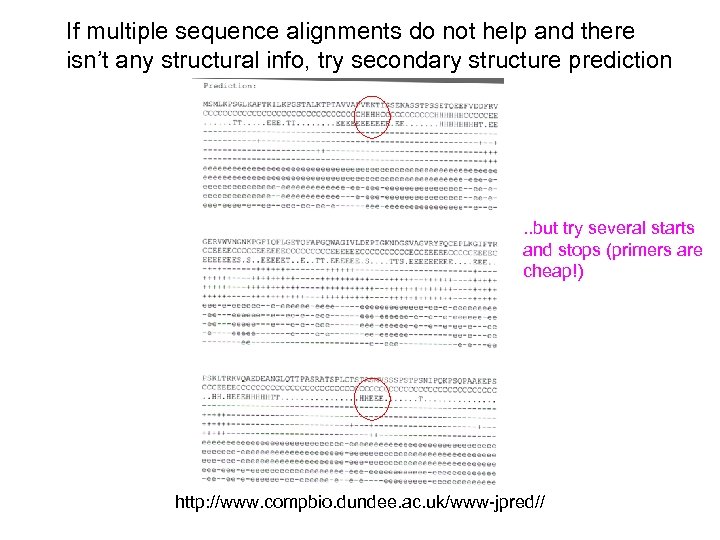
If multiple sequence alignments do not help and there isn’t any structural info, try secondary structure prediction . . but try several starts and stops (primers are cheap!) http: //www. compbio. dundee. ac. uk/www-jpred//
Do you have the gene?
Lots of output! Here’s what you want

buy
Shopping cart – price varies with order size
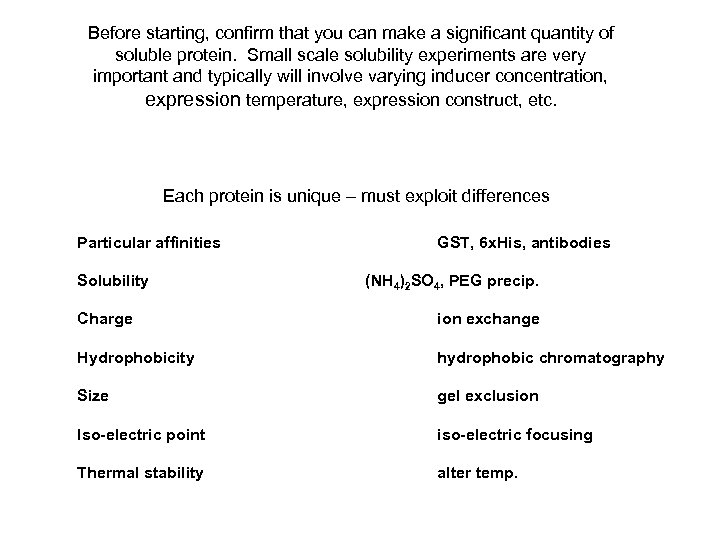
Before starting, confirm that you can make a significant quantity of soluble protein. Small scale solubility experiments are very important and typically will involve varying inducer concentration, expression temperature, expression construct, etc. Each protein is unique – must exploit differences Particular affinities Solubility GST, 6 x. His, antibodies (NH 4)2 SO 4, PEG precip. Charge ion exchange Hydrophobicity hydrophobic chromatography Size gel exclusion Iso-electric point iso-electric focusing Thermal stability alter temp.
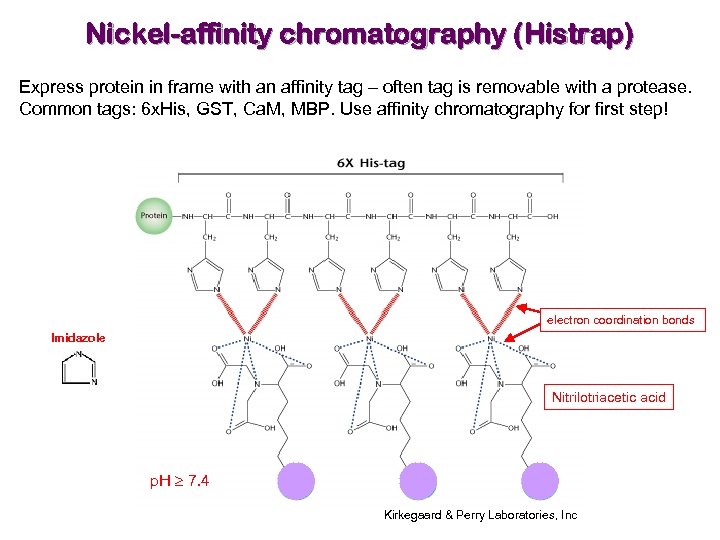
Nickel-affinity chromatography (Histrap) Express protein in frame with an affinity tag – often tag is removable with a protease. Common tags: 6 x. His, GST, Ca. M, MBP. Use affinity chromatography for first step! electron coordination bonds Imidazole Nitrilotriacetic acid p. H 7. 4 Kirkegaard & Perry Laboratories, Inc
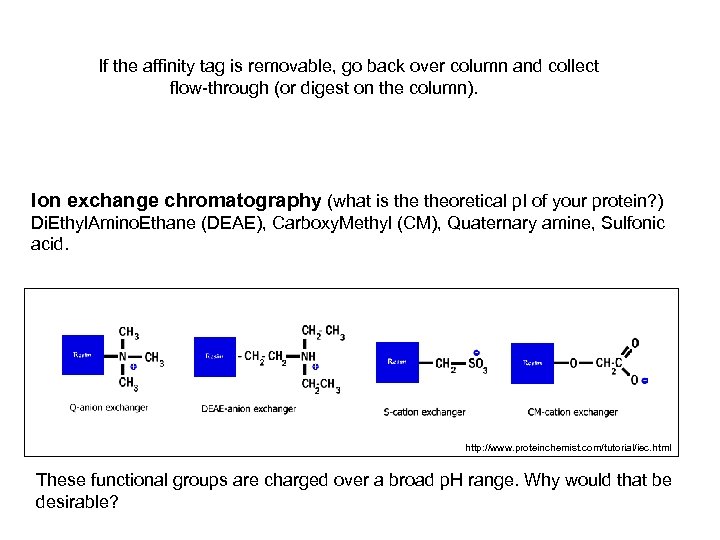
If the affinity tag is removable, go back over column and collect flow-through (or digest on the column). Ion exchange chromatography (what is theoretical p. I of your protein? ) Di. Ethyl. Amino. Ethane (DEAE), Carboxy. Methyl (CM), Quaternary amine, Sulfonic acid. http: //www. proteinchemist. com/tutorial/iec. html These functional groups are charged over a broad p. H range. Why would that be desirable?
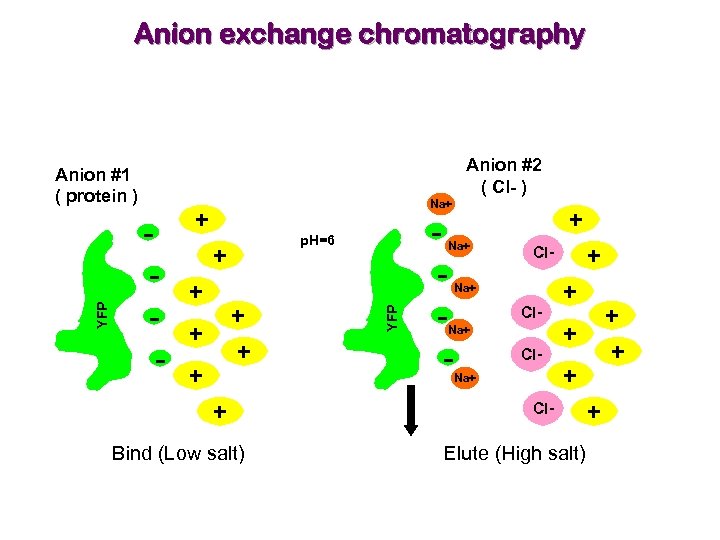
Anion exchange chromatography Anion #2 ( Cl- ) - Na+ + p. H=6 + + + + Bind (Low salt) - + Na+ YFP Anion #1 ( protein ) + Cl- Na+ Cl. Na+ + + Cl- Elute (High salt) +
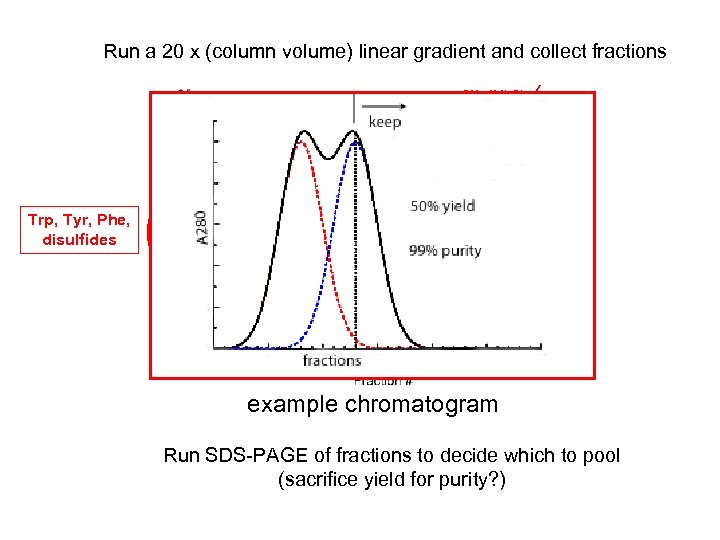
Run a 20 x (column volume) linear gradient and collect fractions 500 m. M Na. Cl Linear gradient (also step) Trp, Tyr, Phe, disulfides 50 m. M Na. Cl example chromatogram Run SDS-PAGE of fractions to decide which to pool (sacrifice yield for purity? )
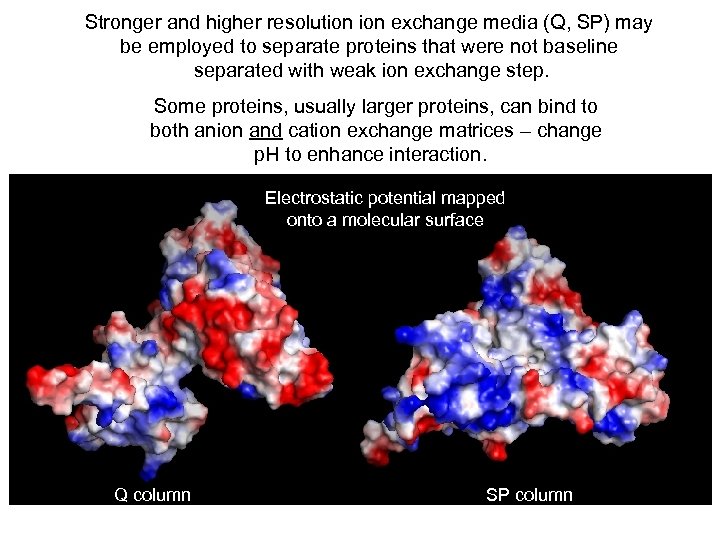
Stronger and higher resolution exchange media (Q, SP) may be employed to separate proteins that were not baseline separated with weak ion exchange step. Some proteins, usually larger proteins, can bind to both anion and cation exchange matrices – change p. H to enhance interaction. Electrostatic potential mapped onto a molecular surface Q column SP column
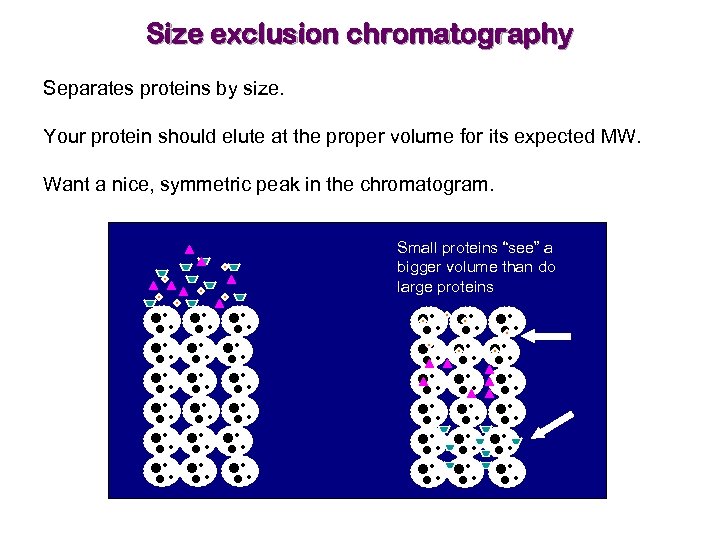
Size exclusion chromatography Separates proteins by size. Your protein should elute at the proper volume for its expected MW. Want a nice, symmetric peak in the chromatogram. Small proteins “see” a bigger volume than do large proteins
Some other chromatographic techniques Salting out – Proteins precipitate differentially in the presence of (NH 4)2 SO 4 or polyethylene glycol - It’s probably worth trying Hydrophobic – Load proteins onto phenyl sepharose in presence of ~1. 5 M (NH 4)2 SO 4 and run decreasing [(NH 4)2 SO 4] gradient. More hydrophobic elutes later. Isoelectric focusing – Electrophorese protein in matrix containing p. H gradient. When the protein reaches that p. H where it has no net charge it ceases to migrate. Retrieve protein from matrix.
Expression of TVMV protease Day 1 Transform Rosetta 2 cells with plasmid containing tvmv gene and plate overnight Day 2 (or -1) In the evening, pick a colony and grown a 10 ml overnight starter culture @ 37 C/200 RPM. (you can save a day by inoculating from a glycerol stock. ) Day 3 (or 1) a) In morning, inoculate 2 L media with overnight starter and grow @37 C/200 PRM. b) After 2 -4 hours (OD 600 ~0. 6 -1), add IPTG to 0. 5 m. M final concentration and induce for 4 hrs @30 C/200 RPM. Expression optimization: at mid-log phase, lower temperature to 12 -20 C, add lower amount of IPTG and induce overnight, or for slow leaky expression, no IPTG for 2 days (membrane proteins) c) Harvest cells (4000 RPM/20’), resuspend in 50 m. L lysis buffer (+ protease inhibitors) and store in -80 C. Day 4 (or 2) a) thaw cells from -80 C and lyse by sonication or with Emulsiflex (cells are squeeze through a small pin-hole by high pressure). b) Start purification
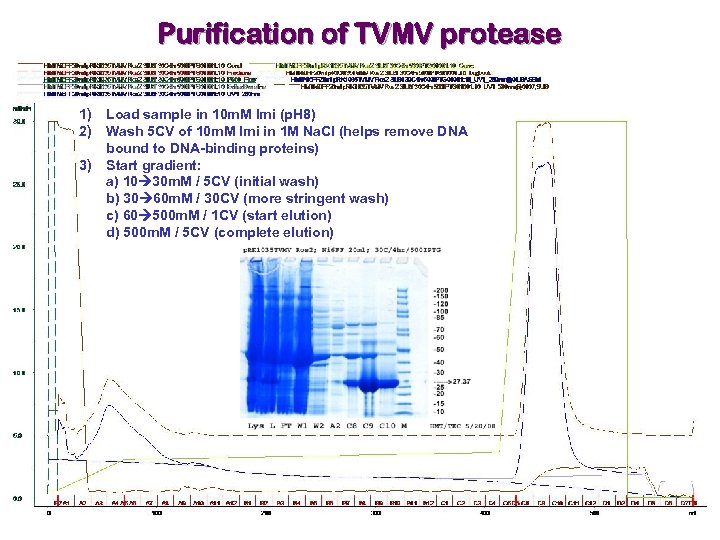
Purification of TVMV protease 1) Load sample in 10 m. M Imi (p. H 8) 2) Wash 5 CV of 10 m. M Imi in 1 M Na. Cl (helps remove DNA bound to DNA-binding proteins) 3) Start gradient: a) 10 30 m. M / 5 CV (initial wash) b) 30 60 m. M / 30 CV (more stringent wash) c) 60 500 m. M / 1 CV (start elution) d) 500 m. M / 5 CV (complete elution)
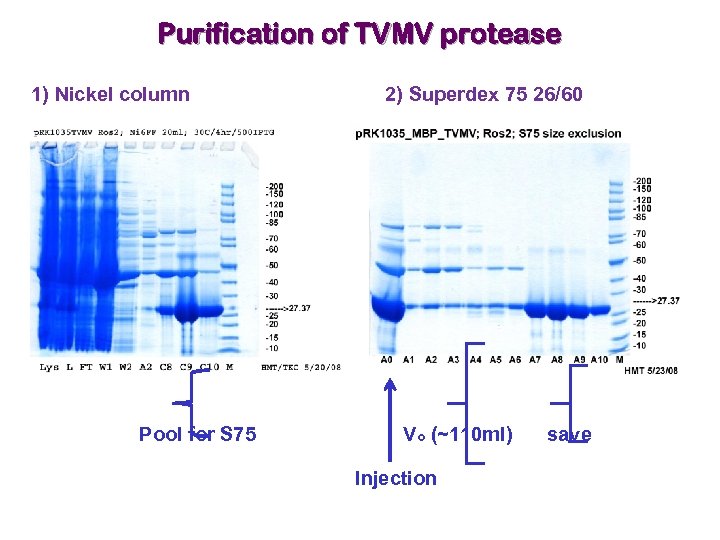
Purification of TVMV protease 1) Nickel column Pool for S 75 2) Superdex 75 26/60 Vo (~110 ml) Injection save
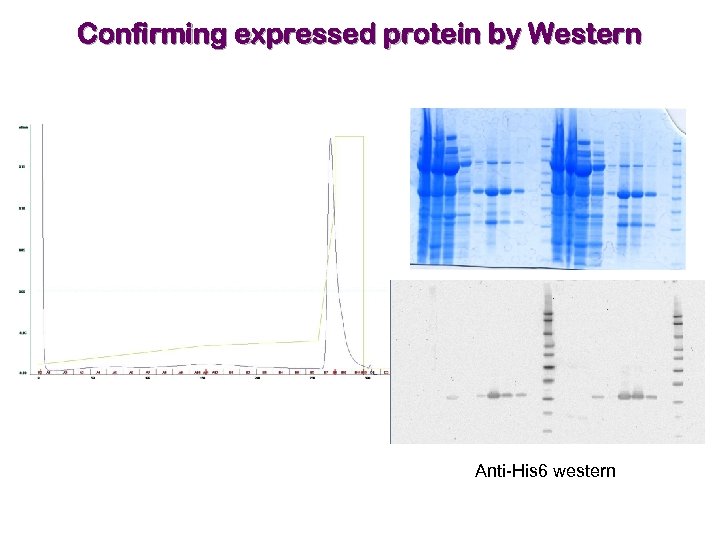
Confirming expressed protein by Western Anti-His 6 western
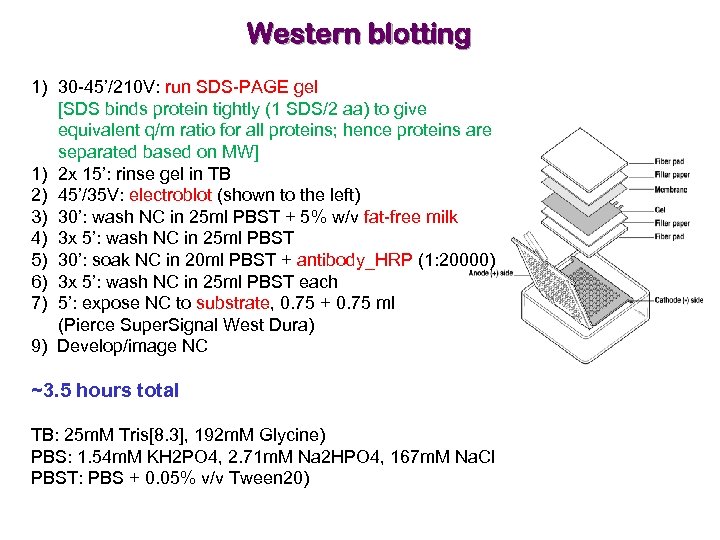
Western blotting 1) 30 -45’/210 V: run SDS-PAGE gel [SDS binds protein tightly (1 SDS/2 aa) to give equivalent q/m ratio for all proteins; hence proteins are separated based on MW] 1) 2 x 15’: rinse gel in TB 2) 45’/35 V: electroblot (shown to the left) 3) 30’: wash NC in 25 ml PBST + 5% w/v fat-free milk 4) 3 x 5’: wash NC in 25 ml PBST 5) 30’: soak NC in 20 ml PBST + antibody_HRP (1: 20000) 6) 3 x 5’: wash NC in 25 ml PBST each 7) 5’: expose NC to substrate, 0. 75 + 0. 75 ml (Pierce Super. Signal West Dura) 9) Develop/image NC ~3. 5 hours total TB: 25 m. M Tris[8. 3], 192 m. M Glycine) PBS: 1. 54 m. M KH 2 PO 4, 2. 71 m. M Na 2 HPO 4, 167 m. M Na. Cl PBST: PBS + 0. 05% v/v Tween 20)

10 things you should know 1) How BL 21(DE 3) cells work (host-encoded T 7 RNAPol and lac. I repressor) 2) General idea about different bacterial cell lines for optimizing expression: a) different promoters b) increasing m. RNA stability c) Lon/Omp. T protease deficiency d) Rosetta 2 cells and codon-optimization 3) Basic idea of plasmid antibiotic selection. 4) Different cloning method (ligation, LIC, Gateway) 5) How PCR works – use of processive DNAPol to save time 6) Bacterial transformation 7) Improving protein solubility by expression as fusion protein, and which partner works best. 8) in-vivo cleavage of protein from fusion partner 9) co-expression with molecular chaperones 10) Basic idea of protein purification a) nickel-affinity (Histrap) b) Size-exclusion c) ion-exchange d) western blot e) hydrophobic
aac5893d64327a70857809ef3fd3a91f.ppt