6cb447bcd60b273983187903ae521b2b.ppt
- Количество слайдов: 35
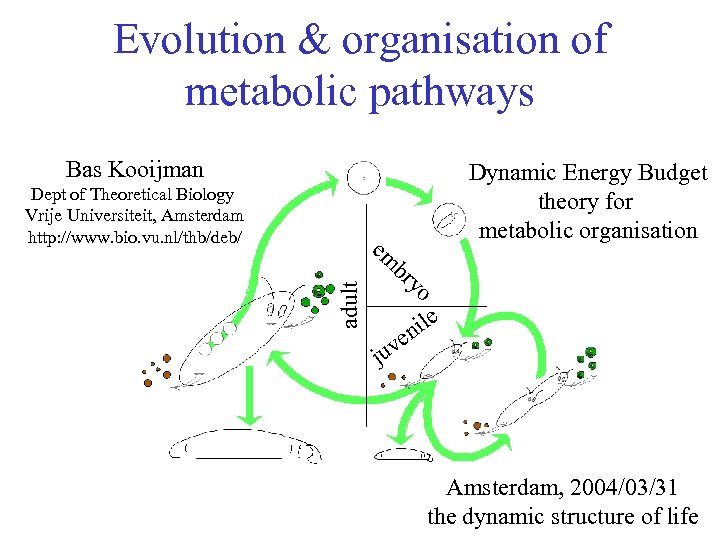
Evolution & organisation of metabolic pathways Bas Kooijman Dept of Theoretical Biology Vrije Universiteit, Amsterdam http: //www. bio. vu. nl/thb/deb/ Dynamic Energy Budget theory for metabolic organisation adult em br yo ile n ve ju Amsterdam, 2004/03/31 the dynamic structure of life
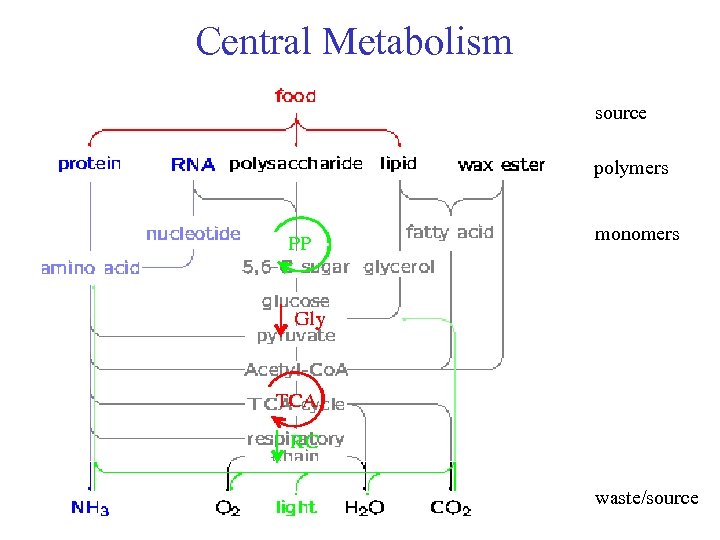
Central Metabolism source polymers monomers waste/source
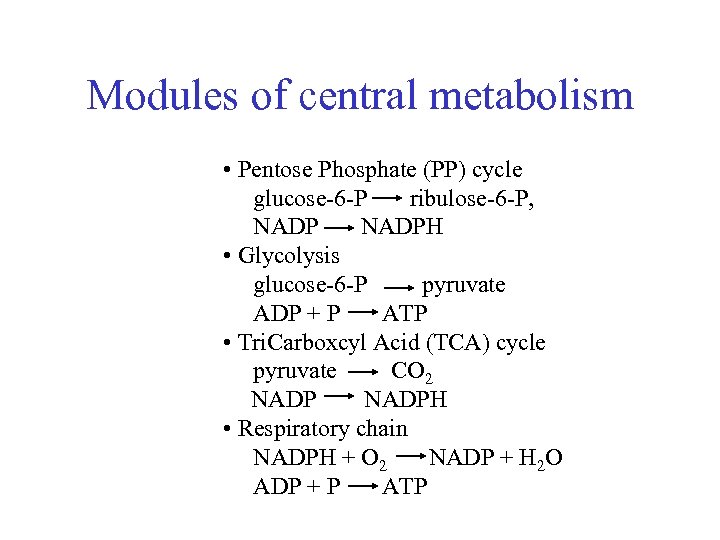
Modules of central metabolism • Pentose Phosphate (PP) cycle glucose-6 -P ribulose-6 -P, NADPH • Glycolysis glucose-6 -P pyruvate ADP + P ATP • Tri. Carboxcyl Acid (TCA) cycle pyruvate CO 2 NADPH • Respiratory chain NADPH + O 2 NADP + H 2 O ADP + P ATP
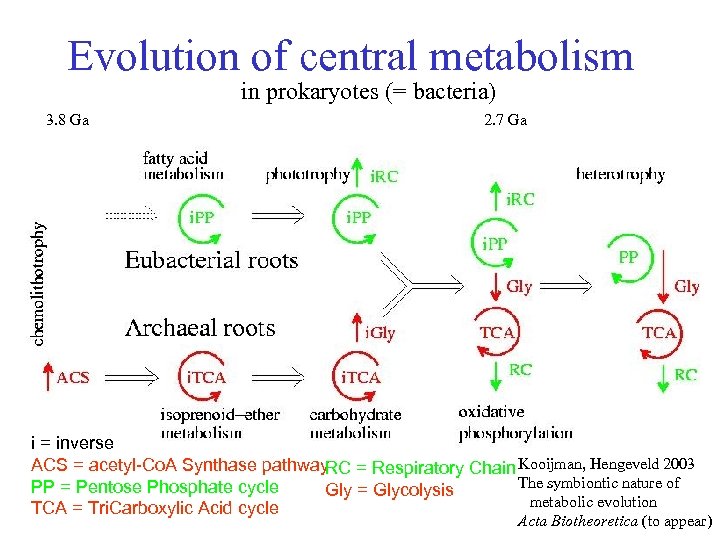
Evolution of central metabolism in prokaryotes (= bacteria) 3. 8 Ga 2. 7 Ga i = inverse ACS = acetyl-Co. A Synthase pathway RC = Respiratory Chain Kooijman, Hengeveld 2003 The symbiontic nature of PP = Pentose Phosphate cycle Gly = Glycolysis metabolic evolution TCA = Tri. Carboxylic Acid cycle Acta Biotheoretica (to appear)
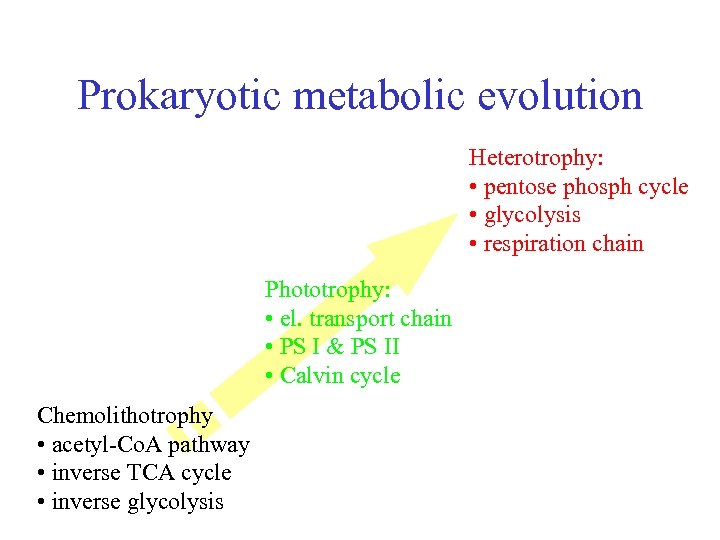
Prokaryotic metabolic evolution Heterotrophy: • pentose phosph cycle • glycolysis • respiration chain Phototrophy: • el. transport chain • PS I & PS II • Calvin cycle Chemolithotrophy • acetyl-Co. A pathway • inverse TCA cycle • inverse glycolysis
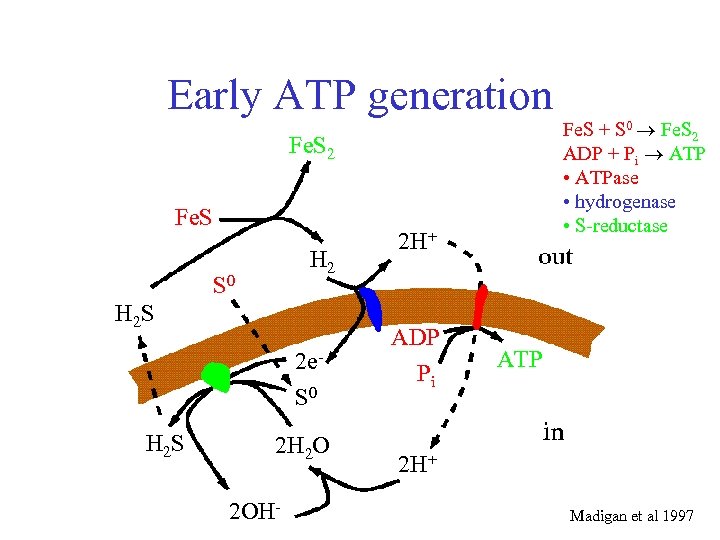
Early ATP generation Fe. S 2 Fe. S H 2 S 0 H 2 S 2 e. S 0 H 2 S 2 H 2 O 2 OH- 2 H+ ADP Pi Fe. S + S 0 Fe. S 2 ADP + Pi ATP • ATPase • hydrogenase • S-reductase ATP 2 H+ Madigan et al 1997
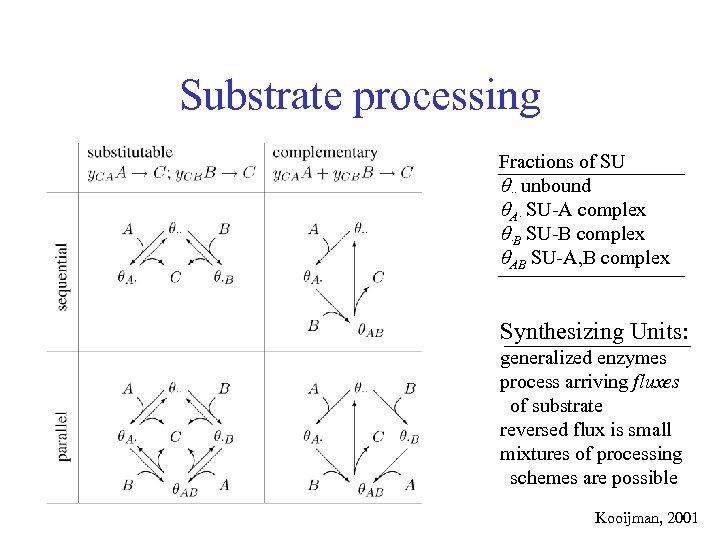
Substrate processing Fractions of SU ·· unbound A· SU-A complex ·B SU-B complex AB SU-A, B complex Synthesizing Units: generalized enzymes process arriving fluxes of substrate reversed flux is small mixtures of processing schemes are possible Kooijman, 2001
Biomass: reserve(s) + structure(s) Reserve(s), structure(s): generalized compounds, mixtures of proteins, lipids, carbohydrates: fixed composition Reserve(s) do complicate model & implications & testing Reasons to delineate reserve, distinct from structure • metabolic memory • biomass composition depends on growth rate • explanation of respiration patterns (freshly laid eggs don’t respire) method of indirect calorimetry fluxes are linear sums of assimilation, dissipation and growth inter-species body size scaling relationships • fate of metabolites (e. g. conversion into energy vs buiding blocks)
Reserve vs structure Reserve does not mean: “set apart for later use” compounds in reserve can have active functions Life span of compounds in • reserve: limited due to turnover of reserve all reserve compounds have the same mean life span • structure: controlled by somatic maintenance structure compounds can differ in mean life span Important difference between reserve and structure: no maintenance costs for reserve Empirical evidence: freshly laid eggs consist of reserve and do not respire
Homeostasis: constant body composition in varying environments Strong homeostasis generalized compounds applies to reserve(s) and structure(s) separately Weak homeostasis: ratio reserve/structure becomes and remains constant if food or substrate is constant (while the individual is growing) applies to juvenile and adult stages, not to embryos Implication: stoichiometric constraints on growth
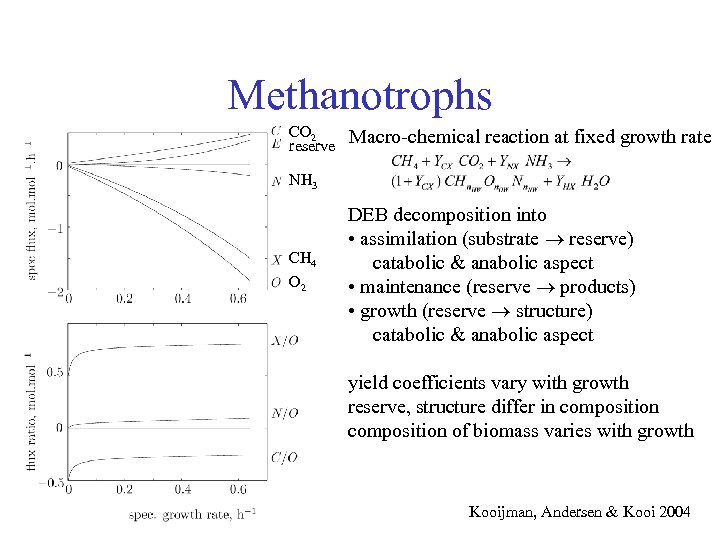
Methanotrophs CO 2 reserve Macro-chemical reaction at fixed growth rate NH 3 CH 4 O 2 DEB decomposition into • assimilation (substrate reserve) catabolic & anabolic aspect • maintenance (reserve products) • growth (reserve structure) catabolic & anabolic aspect yield coefficients vary with growth reserve, structure differ in composition of biomass varies with growth Kooijman, Andersen & Kooi 2004
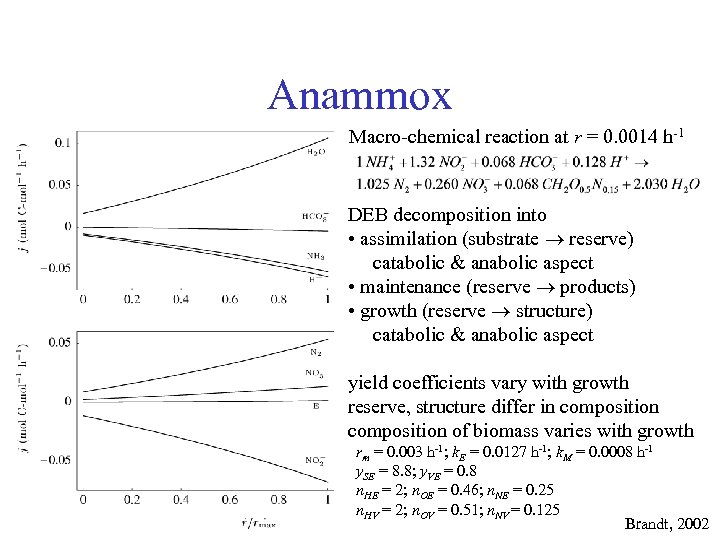
Anammox Macro-chemical reaction at r = 0. 0014 h-1 DEB decomposition into • assimilation (substrate reserve) catabolic & anabolic aspect • maintenance (reserve products) • growth (reserve structure) catabolic & anabolic aspect yield coefficients vary with growth reserve, structure differ in composition of biomass varies with growth rm = 0. 003 h-1; k. E = 0. 0127 h-1; k. M = 0. 0008 h-1 y. SE = 8. 8; y. VE = 0. 8 n. HE = 2; n. OE = 0. 46; n. NE = 0. 25 n. HV = 2; n. OV = 0. 51; n. NV = 0. 125 Brandt, 2002
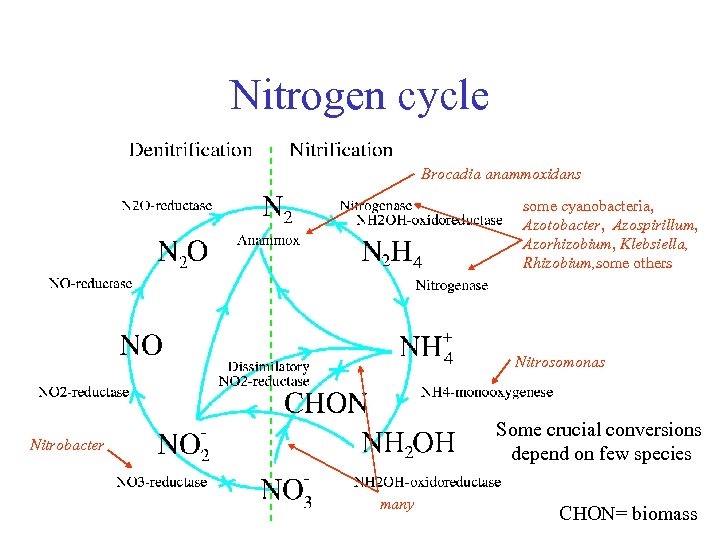
Nitrogen cycle Brocadia anammoxidans some cyanobacteria, Azotobacter, Azospirillum, Azorhizobium, Klebsiella, Rhizobium, some others Nitrosomonas Some crucial conversions depend on few species Nitrobacter many CHON= biomass
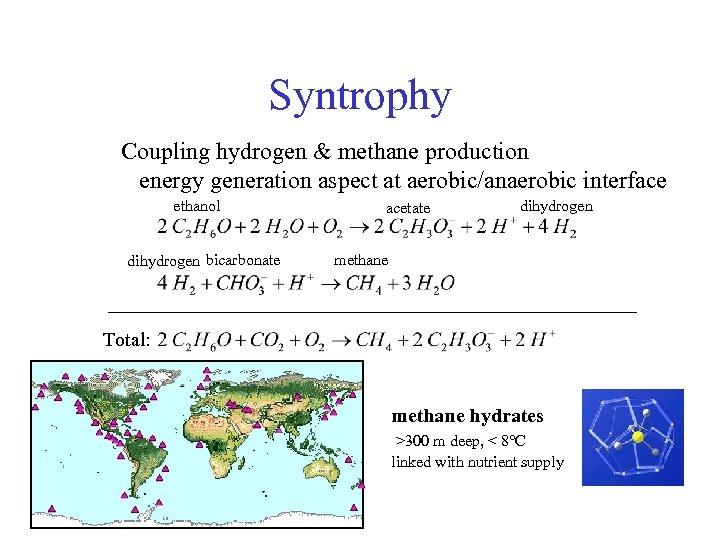
Syntrophy Coupling hydrogen & methane production energy generation aspect at aerobic/anaerobic interface ethanol dihydrogen bicarbonate acetate dihydrogen methane Total: methane hydrates >300 m deep, < 8 C linked with nutrient supply
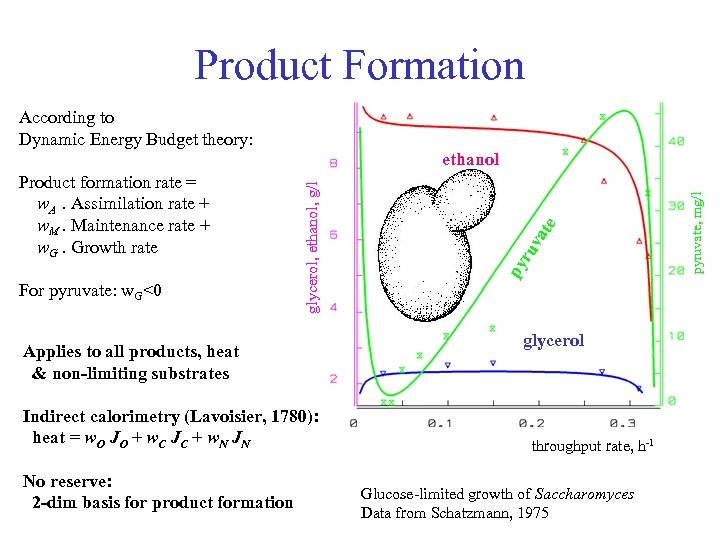
Product Formation According to Dynamic Energy Budget theory: For pyruvate: w. G<0 Applies to all products, heat & non-limiting substrates Indirect calorimetry (Lavoisier, 1780): heat = w. O JO + w. C JC + w. N JN No reserve: 2 -dim basis for product formation glycerol throughput rate, h-1 Glucose-limited growth of Saccharomyces Data from Schatzmann, 1975 pyruvate, mg/l te va ru py Product formation rate = w. A. Assimilation rate + w. M. Maintenance rate + w. G. Growth rate glycerol, ethanol, g/l ethanol
Symbiosis substrate product
Symbiosis substrate
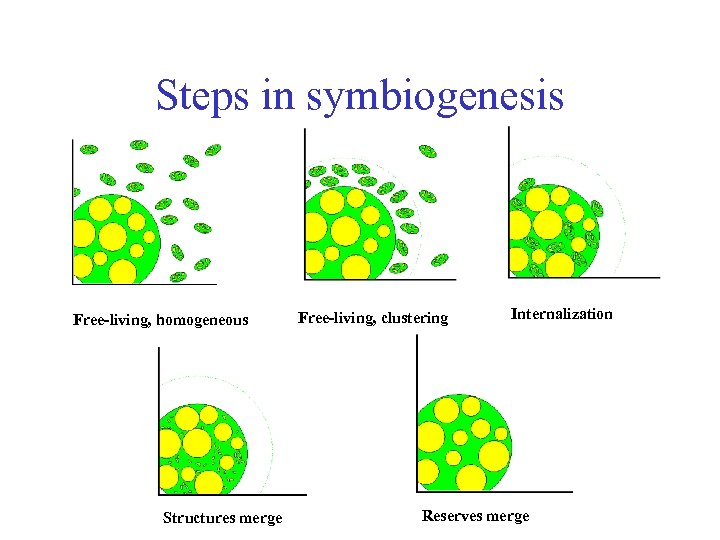
Steps in symbiogenesis Free-living, homogeneous Structures merge Free-living, clustering Internalization Reserves merge
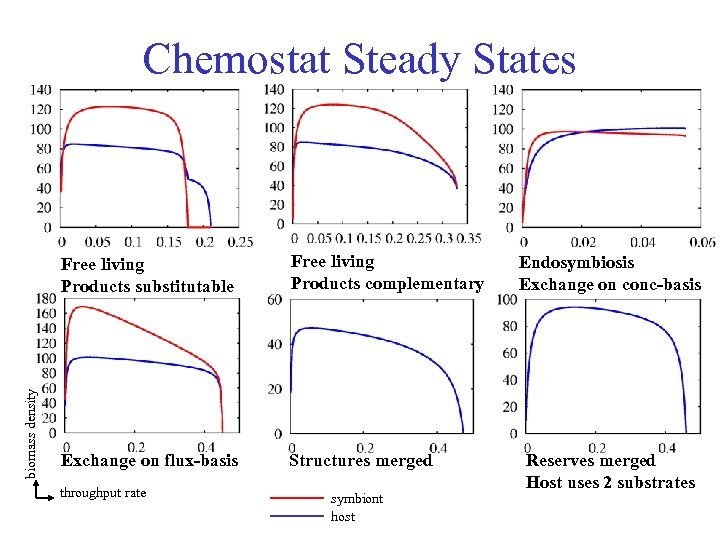
Chemostat Steady States biomass density Free living Products substitutable Free living Products complementary Exchange on flux-basis Structures merged throughput rate symbiont host Endosymbiosis Exchange on conc-basis Reserves merged Host uses 2 substrates
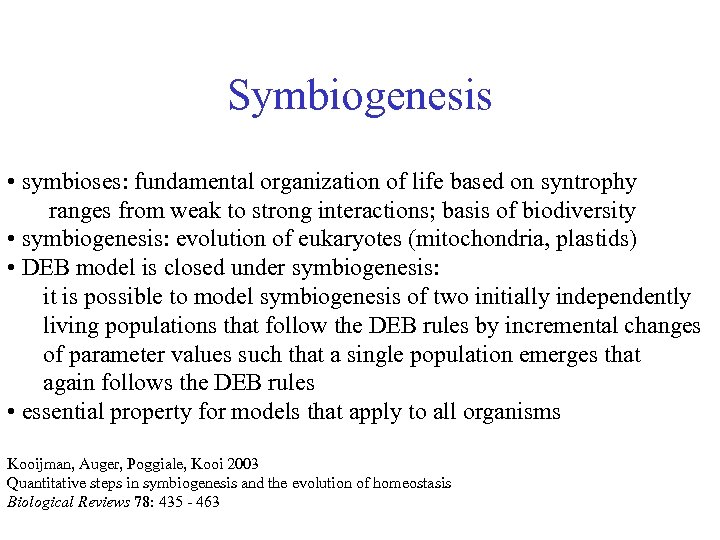
Symbiogenesis • symbioses: fundamental organization of life based on syntrophy ranges from weak to strong interactions; basis of biodiversity • symbiogenesis: evolution of eukaryotes (mitochondria, plastids) • DEB model is closed under symbiogenesis: it is possible to model symbiogenesis of two initially independently living populations that follow the DEB rules by incremental changes of parameter values such that a single population emerges that again follows the DEB rules • essential property for models that apply to all organisms Kooijman, Auger, Poggiale, Kooi 2003 Quantitative steps in symbiogenesis and the evolution of homeostasis Biological Reviews 78: 435 - 463
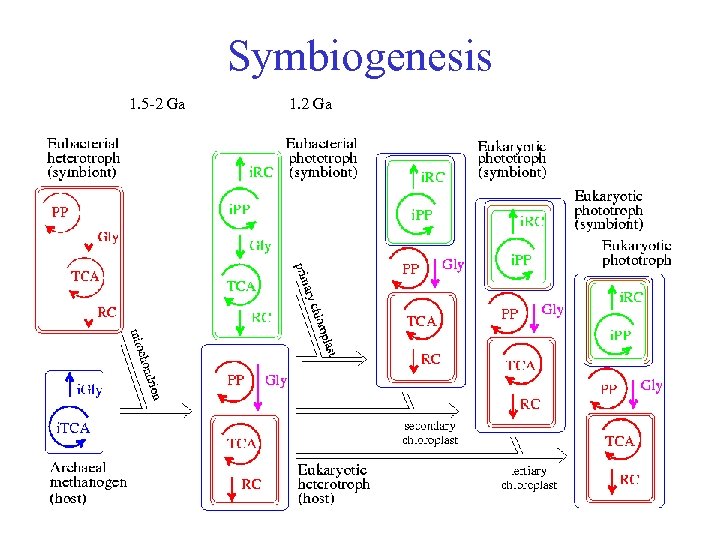
Symbiogenesis 1. 5 -2 Ga 1. 2 Ga

Eukaryote metabolic evolution First eukaryotes: heterotrophs by symbiogenesis compartmental cellular organisation Acquisition of phototrophy frequently did not result in loss of heterotrophy Acquisition of membrane transport between internalization of mitochondria and plastids No phagocytosis in fungi & plants; loss? pinocytosis in animals = phagocytosis in e. g. amoeba? Direct link between phagocytosis and membrane transport?
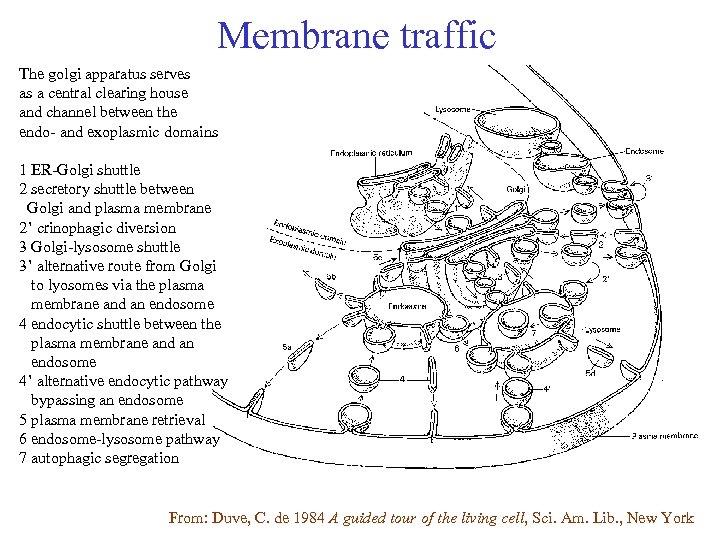
Membrane traffic The golgi apparatus serves as a central clearing house and channel between the endo- and exoplasmic domains 1 ER-Golgi shuttle 2 secretory shuttle between Golgi and plasma membrane 2’ crinophagic diversion 3 Golgi-lysosome shuttle 3’ alternative route from Golgi to lyosomes via the plasma membrane and an endosome 4 endocytic shuttle between the plasma membrane and an endosome 4’ alternative endocytic pathway bypassing an endosome 5 plasma membrane retrieval 6 endosome-lysosome pathway 7 autophagic segregation From: Duve, C. de 1984 A guided tour of the living cell, Sci. Am. Lib. , New York
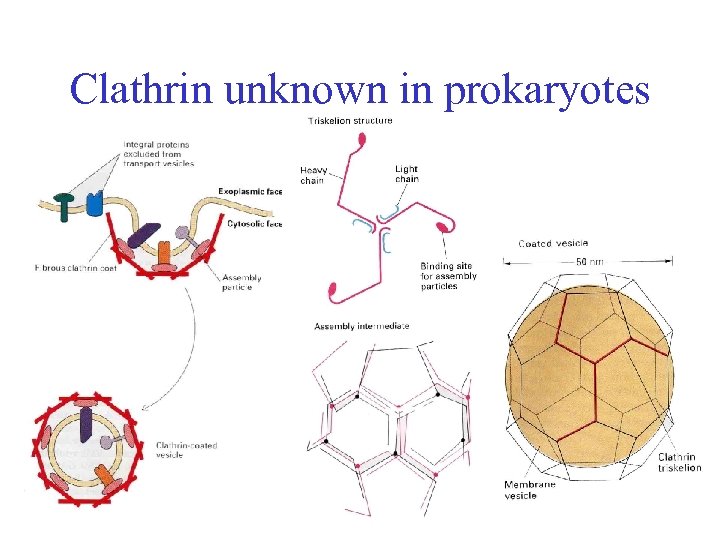
Clathrin unknown in prokaryotes

Chloroplast dynamics Coordinated movement of chloroplasts through cells
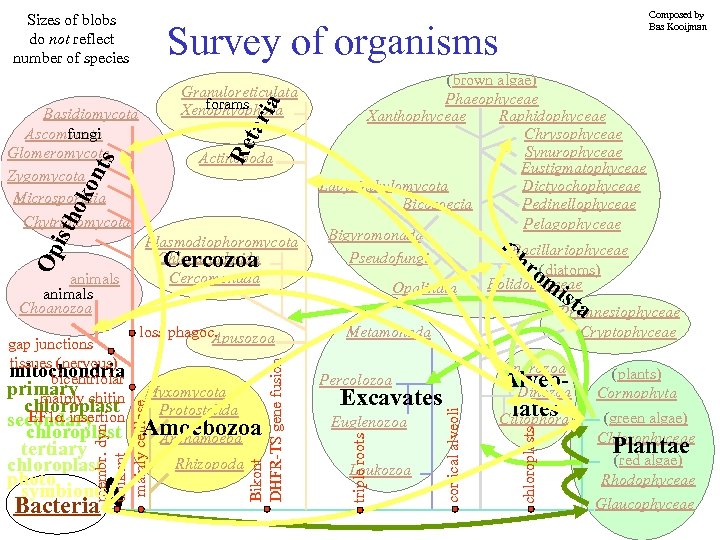
Sizes of blobs do not reflect number of species Survey of organisms chloroplast Amoebozoa Archamoeba tertiary chloroplast photo symbionts Bacteria Rhizopoda Excavates Euglenozoa Loukozoa Alveo. Dinozoa lates Ciliophora chloroplasts Myxomycota Protostelida Sporozoa Percolozoa cortical alveoli Re loss phagoc. Apusozoa membr. dyn unikont mainly celllose gap junctions tissues (nervous) mitochondria bicentriolar primary chitin mainly chloroplast EF 1 insertion secondary Plasmodiophoromycota Chlorarachnida Cercozoa Cercomonada Bikont DHFR-TS gene fusion Op isth ok on ts Actinopoda (brown algae) Phaeophyceae Xanthophyceae Raphidophyceae Chrysophyceae Synurophyceae Eustigmatophyceae Labyrinthulomycota Dictyochophyceae Bicosoecia Pedinellophyceae Pelagophyceae Bigyromonada CBacillariophyceae Pseudofungi hr (diatoms) om Bolidophyceae Opalinata ist a Prymnesiophyceae Metamonada Cryptophyceae triple roots tar ia Granuloreticulata forams Xenophyophora Basidiomycota Ascomycota fungi Glomeromycota Zygomycota Microsporidia Chytridiomycota animals Choanozoa Composed by Bas Kooijman (plants) Cormophyta (green algae) Chlorophyceae Plantae (red algae) Rhodophyceae Glaucophyceae

Cells, individuals, colonies vague boundaries • plasmodesmata connect cytoplasm; cells form a symplast: plants • pits and large pores connect cytoplasm: fungi, rhodophytes • multinucleated cells occur; individuals can be unicellular: fungi, Eumycetozoa, Myxozoa, ciliates, Xenophyophores, Actinophryids, Biomyxa, diplomonads, Gymnosphaerida, haplosporids, Microsporidia, nephridiophagids, Nucleariidae, plasmodiophorids, Pseudospora, Xanthophyta (e. g. Vaucheria), most classes of Chlorophyta (Chlorophyceae, Ulvoph Charophyceae (in mature cells) and all Cladophoryceae, Bryopsidophyceae and Dasycladophycea cells inside cells: Paramyxea uni- and multicellular stages: multicellular spores in unicellular myxozoa, gametes individuals can remain connected after vegetative propagation: plants, corals, b • • individuals in colonies can strongly interact and specialize for particular tasks: syphonophorans, insects, mole rats Kooijman, Hengeveld 2003 The symbiontic nature of metabolic evolution Acta Biotheoretica (to appear) Heterocephalus glaber rotifer Conochilus hippocrepis
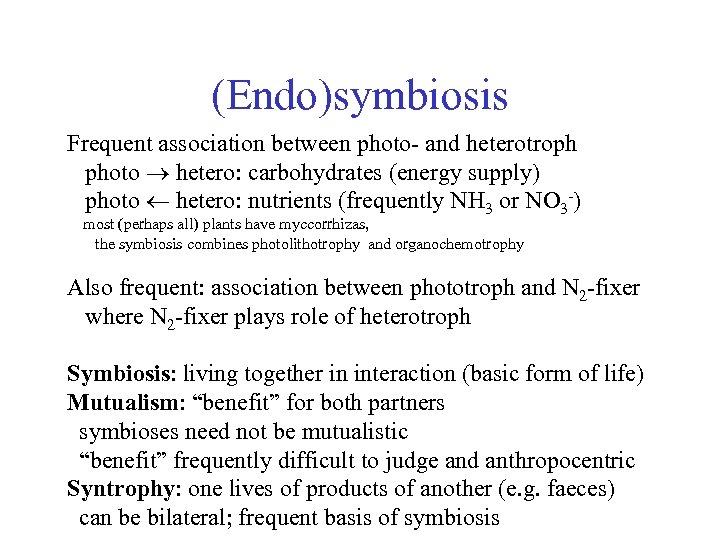
(Endo)symbiosis Frequent association between photo- and heterotroph photo hetero: carbohydrates (energy supply) photo hetero: nutrients (frequently NH 3 or NO 3 -) most (perhaps all) plants have myccorrhizas, the symbiosis combines photolithotrophy and organochemotrophy Also frequent: association between phototroph and N 2 -fixer where N 2 -fixer plays role of heterotroph Symbiosis: living together in interaction (basic form of life) Mutualism: “benefit” for both partners symbioses need not be mutualistic “benefit” frequently difficult to judge and anthropocentric Syntrophy: one lives of products of another (e. g. faeces) can be bilateral; frequent basis of symbiosis
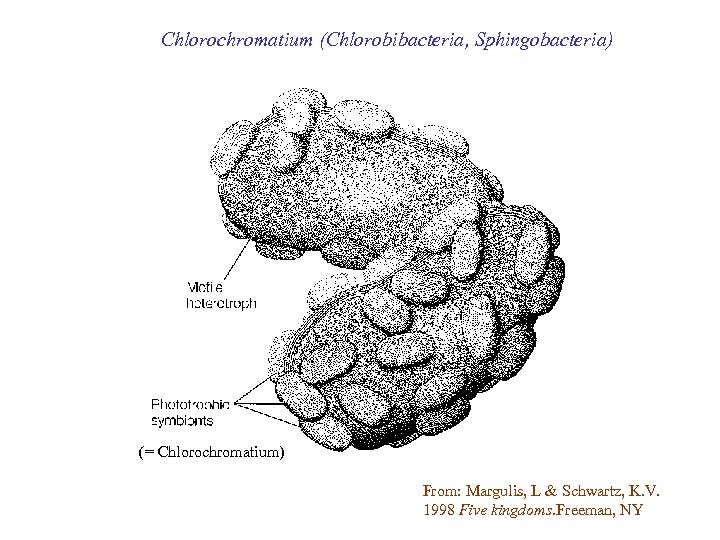
Chlorochromatium (Chlorobibacteria, Sphingobacteria) (= Chlorochromatium) From: Margulis, L & Schwartz, K. V. 1998 Five kingdoms. Freeman, NY
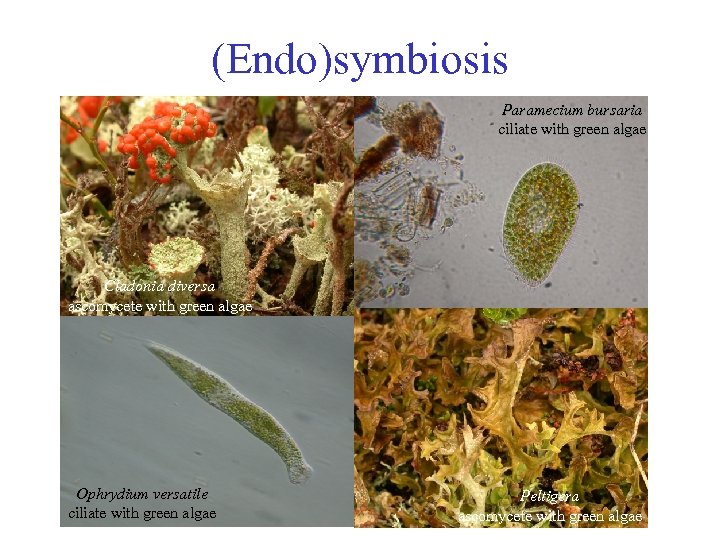
(Endo)symbiosis Paramecium bursaria ciliate with green algae Cladonia diversa ascomycete with green algae Ophrydium versatile ciliate with green algae Peltigera ascomycete with green algae
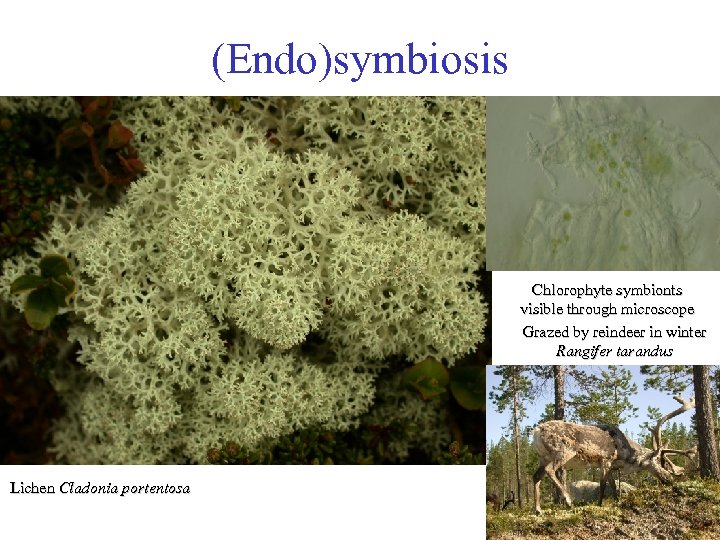
(Endo)symbiosis Chlorophyte symbionts visible through microscope Grazed by reindeer in winter Rangifer tarandus Lichen Cladonia portentosa
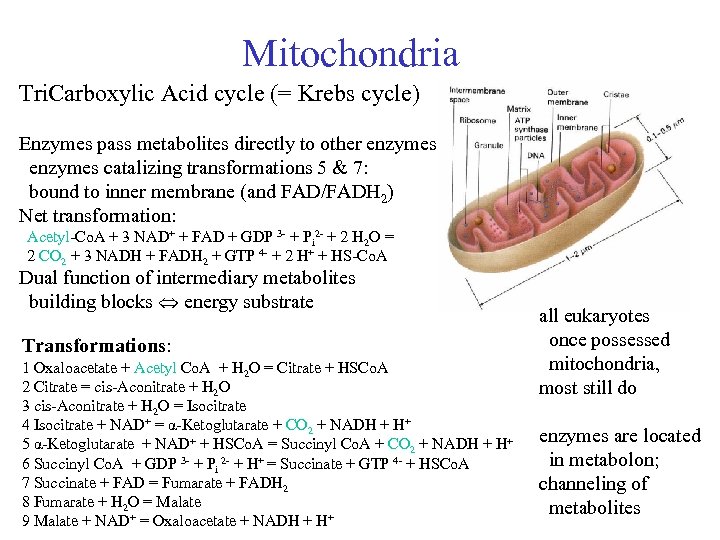
Mitochondria Tri. Carboxylic Acid cycle (= Krebs cycle) Enzymes pass metabolites directly to other enzymes catalizing transformations 5 & 7: bound to inner membrane (and FAD/FADH 2) Net transformation: Acetyl-Co. A + 3 NAD+ + FAD + GDP 3 - + Pi 2 - + 2 H 2 O = 2 CO 2 + 3 NADH + FADH 2 + GTP 4 - + 2 H+ + HS-Co. A Dual function of intermediary metabolites building blocks energy substrate Transformations: 1 Oxaloacetate + Acetyl Co. A + H 2 O = Citrate + HSCo. A 2 Citrate = cis-Aconitrate + H 2 O 3 cis-Aconitrate + H 2 O = Isocitrate 4 Isocitrate + NAD+ = α-Ketoglutarate + CO 2 + NADH + H+ 5 α-Ketoglutarate + NAD+ + HSCo. A = Succinyl Co. A + CO 2 + NADH + H+ 6 Succinyl Co. A + GDP 3 - + Pi 2 - + H+ = Succinate + GTP 4 - + HSCo. A 7 Succinate + FAD = Fumarate + FADH 2 8 Fumarate + H 2 O = Malate 9 Malate + NAD+ = Oxaloacetate + NADH + H+ all eukaryotes once possessed mitochondria, most still do enzymes are located in metabolon; channeling of metabolites
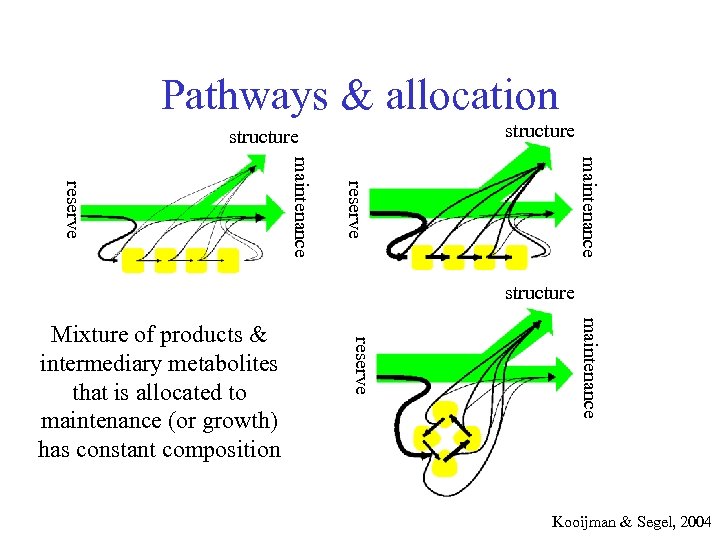
Pathways & allocation structure maintenance reserve Mixture of products & intermediary metabolites that is allocated to maintenance (or growth) has constant composition Kooijman & Segel, 2004
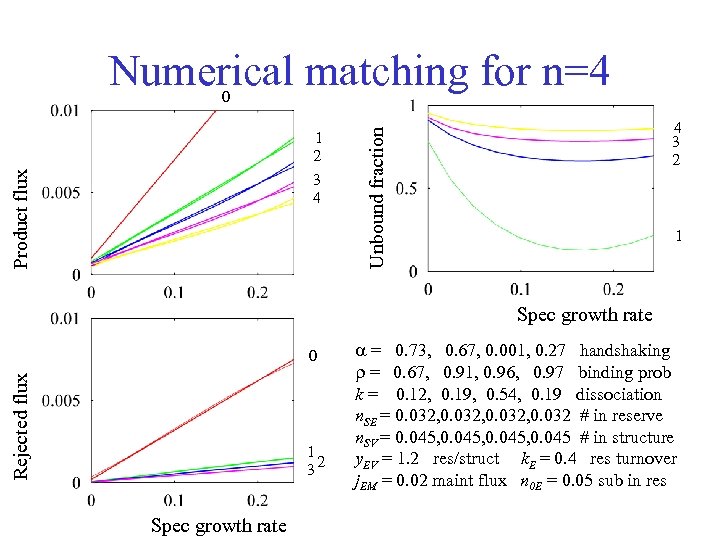
Numerical matching for n=4 0 4 3 2 Unbound fraction Product flux 1 2 3 4 1 Spec growth rate Rejected flux 0 1 2 3 Spec growth rate = 0. 73, 0. 67, 0. 001, 0. 27 handshaking r = 0. 67, 0. 91, 0. 96, 0. 97 binding prob k = 0. 12, 0. 19, 0. 54, 0. 19 dissociation n. SE = 0. 032, 0. 032 # in reserve n. SV = 0. 045, 0. 045 # in structure y. EV = 1. 2 res/struct k. E = 0. 4 res turnover j. EM = 0. 02 maint flux n 0 E = 0. 05 sub in res
Matching pathway whole cell No exact match possible between production of products and intermediary metabolites by pathway and requirements by the cell But very close approximation is possible by tuning abundance parameters and/or binding and handshaking parameters Good approximation requires all four tuning parameters per node growth-dependent reserve abundance plays a key role in tuning Kooijman, S. A. L. M. and Segel, L. A. (2004) How growth affects the fate of cellular substrates. Bull. Math. Biol. (to appear)
6cb447bcd60b273983187903ae521b2b.ppt