ea66e487ffbc39b4c6b012abcc40bfc9.ppt
- Количество слайдов: 38
 Etiology of Autism: A Role for Epigenetics? Rosanna Weksberg Neuro. Dev. Net September 21, 2012 University of Toronto
Etiology of Autism: A Role for Epigenetics? Rosanna Weksberg Neuro. Dev. Net September 21, 2012 University of Toronto
 Complex Etiology of Autism Spectrum Disorders
Complex Etiology of Autism Spectrum Disorders
 Genetics in ASD etiology • Until recently ASD was considered one of the most heritable of neurodevelopmental disorders (~90% heritability) • Rare genetic variants : gene mutations/CNVs/chromosome abnormalities or genetic syndromes account for ~10 -20% of ASD cases • Each individual rare variant is found not in more than ~1 -2% of ASD cases – 15 q 11 -13 maternal duplication, 16 p 11 -12 deletion – SHANK 3, NRXN 1, NLGN 3&4, PTCHD 1 • Common variants: SNPs have been identified through several genome-wide association studies – MACROD 2, CDH 9, PITX 1 – Lack of replication among studies points to high genetic heterogeneity and small effect size of the risk factor alleles
Genetics in ASD etiology • Until recently ASD was considered one of the most heritable of neurodevelopmental disorders (~90% heritability) • Rare genetic variants : gene mutations/CNVs/chromosome abnormalities or genetic syndromes account for ~10 -20% of ASD cases • Each individual rare variant is found not in more than ~1 -2% of ASD cases – 15 q 11 -13 maternal duplication, 16 p 11 -12 deletion – SHANK 3, NRXN 1, NLGN 3&4, PTCHD 1 • Common variants: SNPs have been identified through several genome-wide association studies – MACROD 2, CDH 9, PITX 1 – Lack of replication among studies points to high genetic heterogeneity and small effect size of the risk factor alleles
 Environment in ASD etiology • Recent twin study - estimated that heritability of ASD is only 38%, whereas shared environment contributes to 58% of the liability (Hallmayer et al. 2011) • Environmental risk factors: • Sub-fertility/assisted reproduction? • In utero exposure to antiepileptic drugs – Exposure to VPA increases ASD risk ~10 times • • Pregnancy complications? Viral infections? Ecology? Nutrition? • Exposures can occur at different stages of development – Gametes – Prenatal development – Postnatal development
Environment in ASD etiology • Recent twin study - estimated that heritability of ASD is only 38%, whereas shared environment contributes to 58% of the liability (Hallmayer et al. 2011) • Environmental risk factors: • Sub-fertility/assisted reproduction? • In utero exposure to antiepileptic drugs – Exposure to VPA increases ASD risk ~10 times • • Pregnancy complications? Viral infections? Ecology? Nutrition? • Exposures can occur at different stages of development – Gametes – Prenatal development – Postnatal development
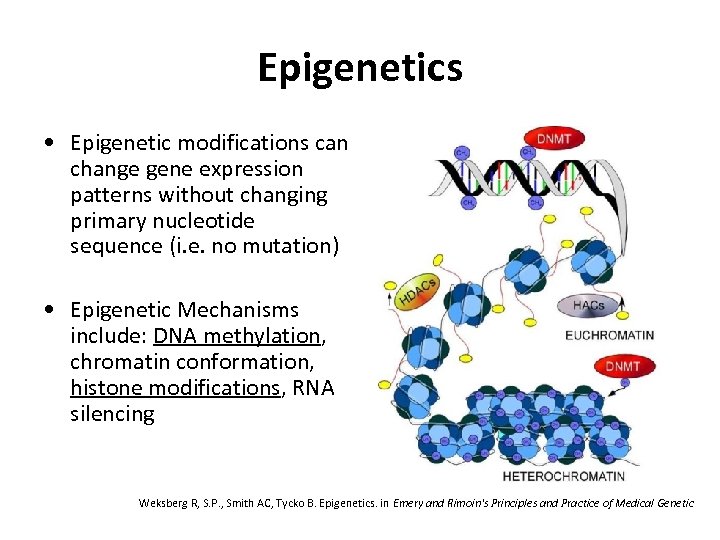 Epigenetics • Epigenetic modifications can change gene expression patterns without changing primary nucleotide sequence (i. e. no mutation) • Epigenetic Mechanisms include: DNA methylation, chromatin conformation, histone modifications, RNA silencing Weksberg R, S. P. , Smith AC, Tycko B. Epigenetics. in Emery and Rimoin's Principles and Practice of Medical Genetic
Epigenetics • Epigenetic modifications can change gene expression patterns without changing primary nucleotide sequence (i. e. no mutation) • Epigenetic Mechanisms include: DNA methylation, chromatin conformation, histone modifications, RNA silencing Weksberg R, S. P. , Smith AC, Tycko B. Epigenetics. in Emery and Rimoin's Principles and Practice of Medical Genetic
 Epigenetic Modifications • CAN BE STABLE: transmitted through mitotic cell division • CAN BE DYNAMIC: sensitive to environmental stimuli (internal and external) § Cell differentiation § Memory § Circadian cycle § Nutrition (e. g. folic acid supplementation) § Medications (e. g. valproate) • CAN BE CELL TYPE-SPECIFIC and DEVELOPMENTALLY-SPECIFIC
Epigenetic Modifications • CAN BE STABLE: transmitted through mitotic cell division • CAN BE DYNAMIC: sensitive to environmental stimuli (internal and external) § Cell differentiation § Memory § Circadian cycle § Nutrition (e. g. folic acid supplementation) § Medications (e. g. valproate) • CAN BE CELL TYPE-SPECIFIC and DEVELOPMENTALLY-SPECIFIC
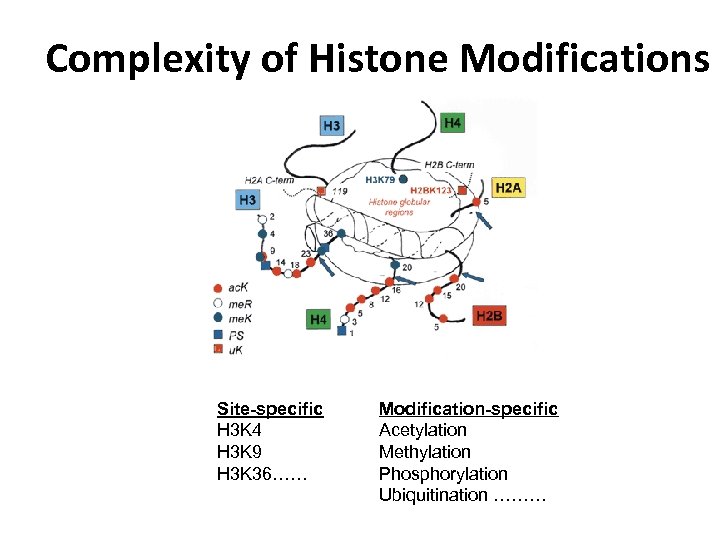 Complexity of Histone Modifications Site-specific H 3 K 4 H 3 K 9 H 3 K 36…… Modification-specific Acetylation Methylation Phosphorylation Ubiquitination ………
Complexity of Histone Modifications Site-specific H 3 K 4 H 3 K 9 H 3 K 36…… Modification-specific Acetylation Methylation Phosphorylation Ubiquitination ………
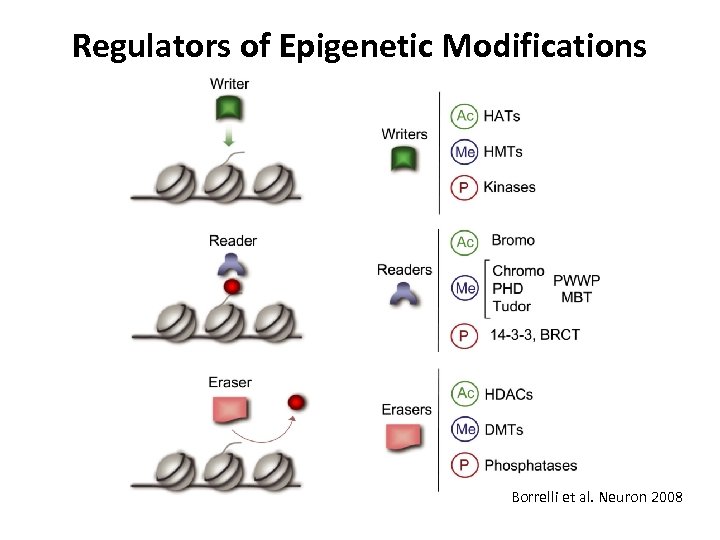 Regulators of Epigenetic Modifications Borrelli et al. Neuron 2008
Regulators of Epigenetic Modifications Borrelli et al. Neuron 2008
 Epigenetics in ASD etiology • Evidence for role of epigenetics in ASD comes from both genetic and environmental risk factors
Epigenetics in ASD etiology • Evidence for role of epigenetics in ASD comes from both genetic and environmental risk factors
 Genetic syndromes co-morbid with ASD and idiopathic ASD are caused by mutations in genes involved in epigenetic regulation
Genetic syndromes co-morbid with ASD and idiopathic ASD are caused by mutations in genes involved in epigenetic regulation
 Environmental Factors Associated with Epigenetic Regulation: Valproate and Autism • The risk of ASD in children exposed in utero to valporate during pregnancy is estimated to be ~9% vs 0. 9% in general population (Moore et al. 2000, J Med Genet ; Rasalam et al. 2005 Dev Med Child Neurol) • Possible mechanisms of valproate teratogenic action: o oxidative stress and cell toxicity caused by metabolites of valproate o interference with folate metabolism o inhibition of histone deacetylases
Environmental Factors Associated with Epigenetic Regulation: Valproate and Autism • The risk of ASD in children exposed in utero to valporate during pregnancy is estimated to be ~9% vs 0. 9% in general population (Moore et al. 2000, J Med Genet ; Rasalam et al. 2005 Dev Med Child Neurol) • Possible mechanisms of valproate teratogenic action: o oxidative stress and cell toxicity caused by metabolites of valproate o interference with folate metabolism o inhibition of histone deacetylases
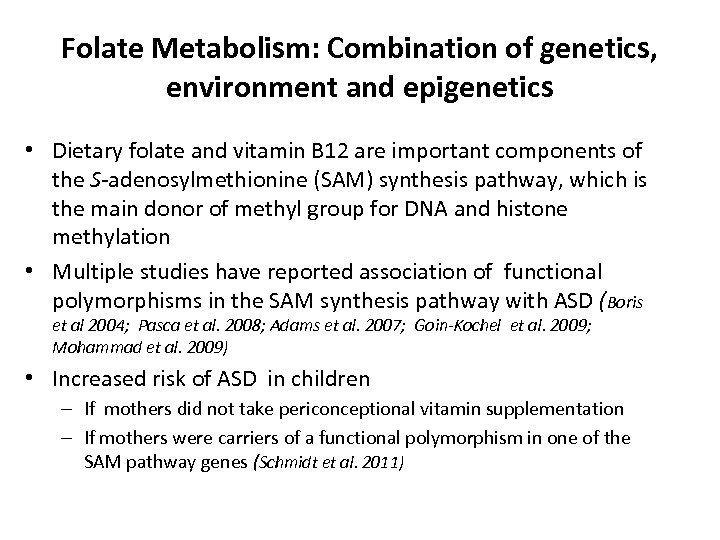 Folate Metabolism: Combination of genetics, environment and epigenetics • Dietary folate and vitamin B 12 are important components of the S-adenosylmethionine (SAM) synthesis pathway, which is the main donor of methyl group for DNA and histone methylation • Multiple studies have reported association of functional polymorphisms in the SAM synthesis pathway with ASD (Boris et al 2004; Pasca et al. 2008; Adams et al. 2007; Goin-Kochel et al. 2009; Mohammad et al. 2009) • Increased risk of ASD in children – If mothers did not take periconceptional vitamin supplementation – If mothers were carriers of a functional polymorphism in one of the SAM pathway genes (Schmidt et al. 2011)
Folate Metabolism: Combination of genetics, environment and epigenetics • Dietary folate and vitamin B 12 are important components of the S-adenosylmethionine (SAM) synthesis pathway, which is the main donor of methyl group for DNA and histone methylation • Multiple studies have reported association of functional polymorphisms in the SAM synthesis pathway with ASD (Boris et al 2004; Pasca et al. 2008; Adams et al. 2007; Goin-Kochel et al. 2009; Mohammad et al. 2009) • Increased risk of ASD in children – If mothers did not take periconceptional vitamin supplementation – If mothers were carriers of a functional polymorphism in one of the SAM pathway genes (Schmidt et al. 2011)
 Epigenetic alterations in ASD • Epigenetic alterations in ASD could occur due to: – Genetic alterations in genes involved in epigenetic regulation – Environmental exposures causing epigenetic alterations – Unknown/stochastic factors • Challenges of identifying epigenetic alterations in ASD: – Etiological heterogeneity – Tissue specificity of epigenetic marks
Epigenetic alterations in ASD • Epigenetic alterations in ASD could occur due to: – Genetic alterations in genes involved in epigenetic regulation – Environmental exposures causing epigenetic alterations – Unknown/stochastic factors • Challenges of identifying epigenetic alterations in ASD: – Etiological heterogeneity – Tissue specificity of epigenetic marks
 Interplay of genetic and epigenetic factors: lessons from KDM 5 C mutation
Interplay of genetic and epigenetic factors: lessons from KDM 5 C mutation
 KDM 5 C • Mutations in X-linked gene KDM 5 C cause intellectual disability (mild to severe) • More than 20 mutations are identified to date (Rujirabanjerd et al. 2010) • KDM 5 C encodes H 3 lysine 4 (K 4) demethylase specific for demethylating H 3 K 4 me 3/2 (Iwase et al. 2007) • KDM 5 C escapes X-inactivation, and has a Y-linked functional homologue KDM 5 D • All forms of H 3 K 4 methylation protect DNA from de novo methylation in embryonic development by blocking DNMT 3 A/L binding (Ooi et al. 2007)
KDM 5 C • Mutations in X-linked gene KDM 5 C cause intellectual disability (mild to severe) • More than 20 mutations are identified to date (Rujirabanjerd et al. 2010) • KDM 5 C encodes H 3 lysine 4 (K 4) demethylase specific for demethylating H 3 K 4 me 3/2 (Iwase et al. 2007) • KDM 5 C escapes X-inactivation, and has a Y-linked functional homologue KDM 5 D • All forms of H 3 K 4 methylation protect DNA from de novo methylation in embryonic development by blocking DNMT 3 A/L binding (Ooi et al. 2007)
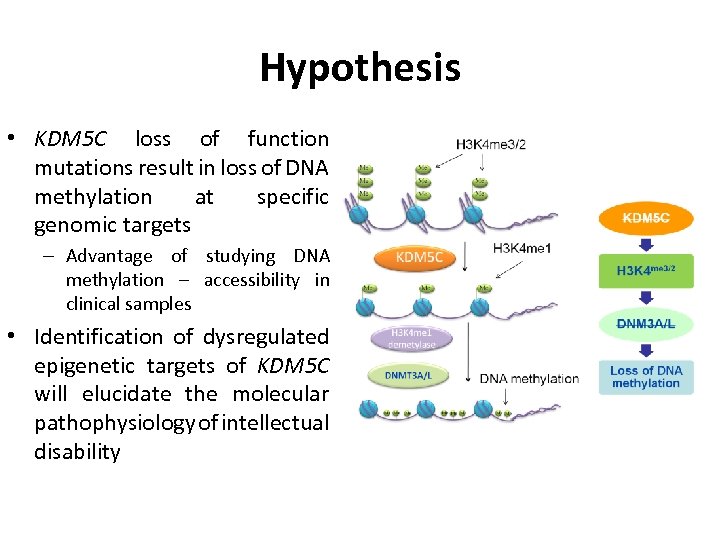 Hypothesis • KDM 5 C loss of function mutations result in loss of DNA methylation at specific genomic targets – Advantage of studying DNA methylation – accessibility in clinical samples • Identification of dysregulated epigenetic targets of KDM 5 C will elucidate the molecular pathophysiology of intellectual disability
Hypothesis • KDM 5 C loss of function mutations result in loss of DNA methylation at specific genomic targets – Advantage of studying DNA methylation – accessibility in clinical samples • Identification of dysregulated epigenetic targets of KDM 5 C will elucidate the molecular pathophysiology of intellectual disability
 Study Design • Illumina Methylation 27 array containing 27, 578 Cp. G sites covering >14, 000 genes was used for genome-wide comparison of Cp. G methylation patterns in blood samples of 10 patients with KDM 5 C mutations vs 19 male controls • Mann-Whitney U test with permutation analysis was used for group comparisons • Targeted validation by Sodium bisulfite pyrosequencing • DNA methylation analysis of top candidate Cp. G sites using publically available control datasets • Comparison of DNA methylation in KDM 5 C targets between normal XY males (KDM 5 C/KDM 5 D) and XX females (KDM 5 C/KDM 5 C) in blood and brain using published dataset
Study Design • Illumina Methylation 27 array containing 27, 578 Cp. G sites covering >14, 000 genes was used for genome-wide comparison of Cp. G methylation patterns in blood samples of 10 patients with KDM 5 C mutations vs 19 male controls • Mann-Whitney U test with permutation analysis was used for group comparisons • Targeted validation by Sodium bisulfite pyrosequencing • DNA methylation analysis of top candidate Cp. G sites using publically available control datasets • Comparison of DNA methylation in KDM 5 C targets between normal XY males (KDM 5 C/KDM 5 D) and XX females (KDM 5 C/KDM 5 C) in blood and brain using published dataset
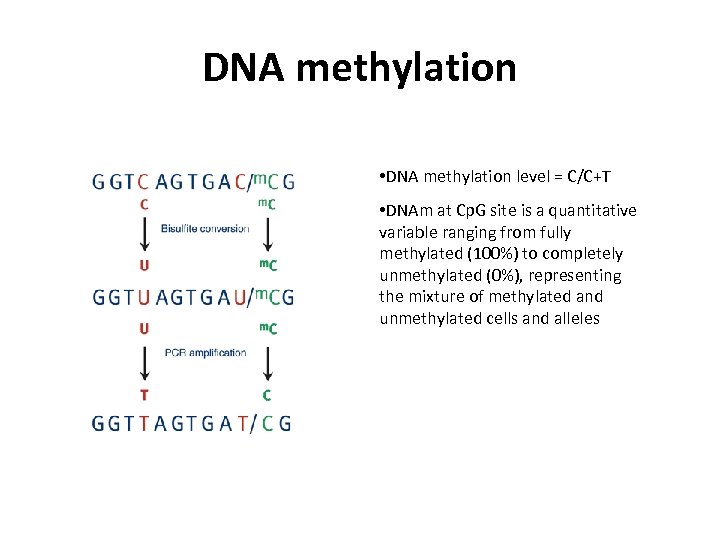 DNA methylation • DNA methylation level = C/C+T • DNAm at Cp. G site is a quantitative variable ranging from fully methylated (100%) to completely unmethylated (0%), representing the mixture of methylated and unmethylated cells and alleles
DNA methylation • DNA methylation level = C/C+T • DNAm at Cp. G site is a quantitative variable ranging from fully methylated (100%) to completely unmethylated (0%), representing the mixture of methylated and unmethylated cells and alleles
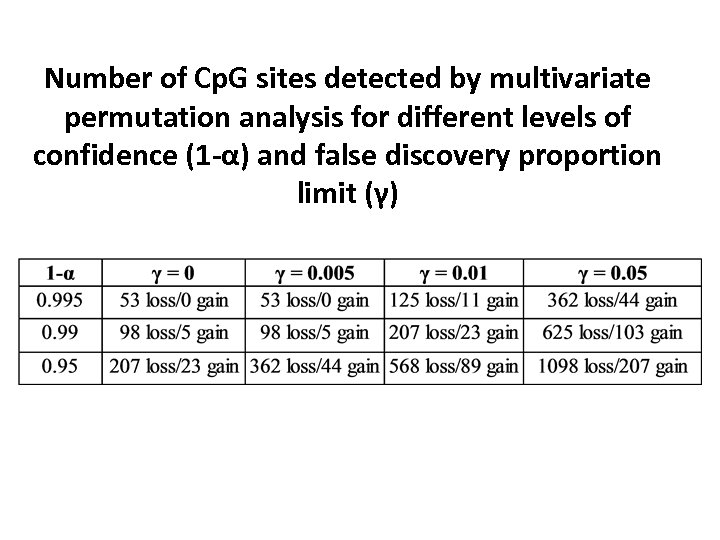 Number of Cp. G sites detected by multivariate permutation analysis for different levels of confidence (1 -α) and false discovery proportion limit (γ)
Number of Cp. G sites detected by multivariate permutation analysis for different levels of confidence (1 -α) and false discovery proportion limit (γ)
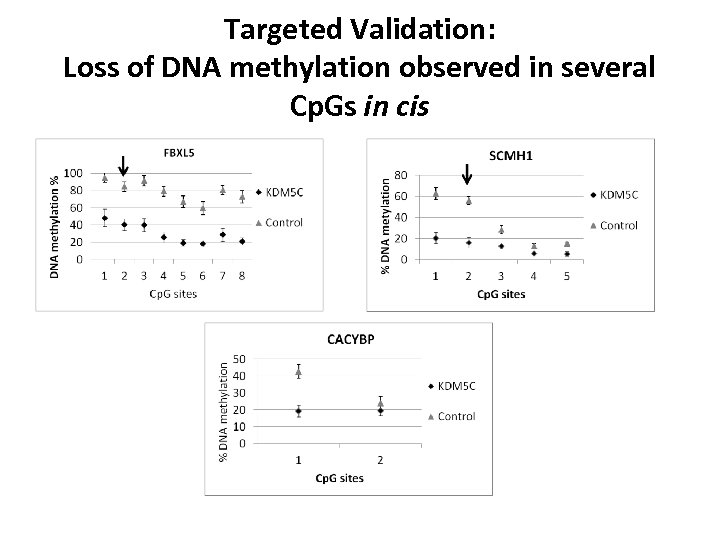 Targeted Validation: Loss of DNA methylation observed in several Cp. Gs in cis
Targeted Validation: Loss of DNA methylation observed in several Cp. Gs in cis
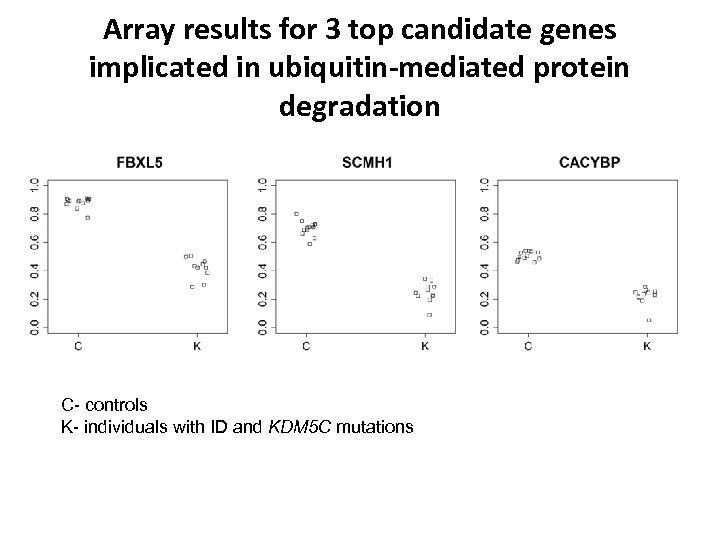 Array results for 3 top candidate genes implicated in ubiquitin-mediated protein degradation C- controls K- individuals with ID and KDM 5 C mutations
Array results for 3 top candidate genes implicated in ubiquitin-mediated protein degradation C- controls K- individuals with ID and KDM 5 C mutations
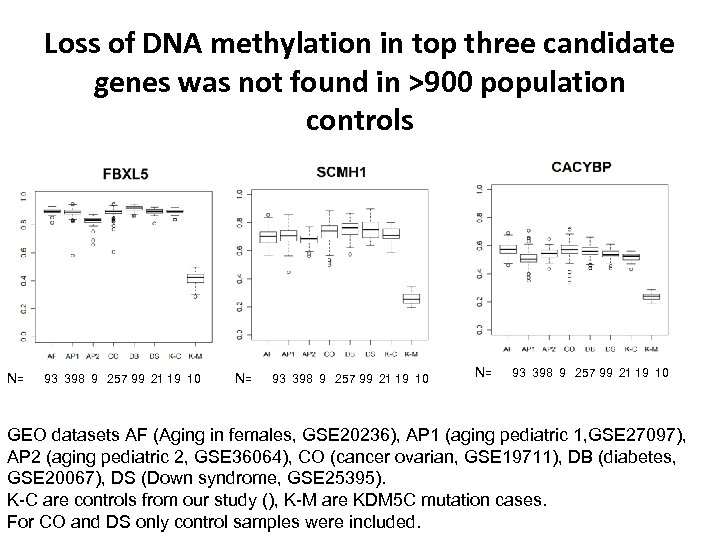 Loss of DNA methylation in top three candidate genes was not found in >900 population controls N= 93 398 9 257 99 21 19 10 GEO datasets AF (Aging in females, GSE 20236), AP 1 (aging pediatric 1, GSE 27097), AP 2 (aging pediatric 2, GSE 36064), CO (cancer ovarian, GSE 19711), DB (diabetes, GSE 20067), DS (Down syndrome, GSE 25395). K-C are controls from our study (), K-M are KDM 5 C mutation cases. For CO and DS only control samples were included.
Loss of DNA methylation in top three candidate genes was not found in >900 population controls N= 93 398 9 257 99 21 19 10 GEO datasets AF (Aging in females, GSE 20236), AP 1 (aging pediatric 1, GSE 27097), AP 2 (aging pediatric 2, GSE 36064), CO (cancer ovarian, GSE 19711), DB (diabetes, GSE 20067), DS (Down syndrome, GSE 25395). K-C are controls from our study (), K-M are KDM 5 C mutation cases. For CO and DS only control samples were included.
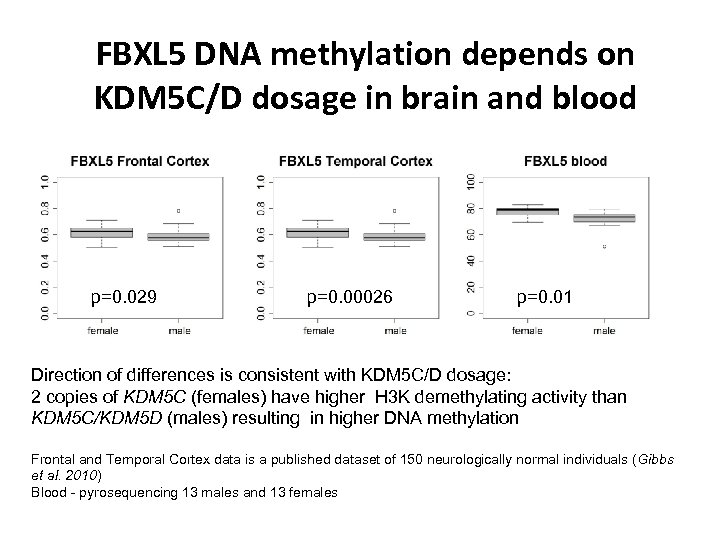 FBXL 5 DNA methylation depends on KDM 5 C/D dosage in brain and blood p=0. 029 p=0. 00026 p=0. 01 Direction of differences is consistent with KDM 5 C/D dosage: 2 copies of KDM 5 C (females) have higher H 3 K demethylating activity than KDM 5 C/KDM 5 D (males) resulting in higher DNA methylation Frontal and Temporal Cortex data is a published dataset of 150 neurologically normal individuals (Gibbs et al. 2010) Blood - pyrosequencing 13 males and 13 females
FBXL 5 DNA methylation depends on KDM 5 C/D dosage in brain and blood p=0. 029 p=0. 00026 p=0. 01 Direction of differences is consistent with KDM 5 C/D dosage: 2 copies of KDM 5 C (females) have higher H 3 K demethylating activity than KDM 5 C/KDM 5 D (males) resulting in higher DNA methylation Frontal and Temporal Cortex data is a published dataset of 150 neurologically normal individuals (Gibbs et al. 2010) Blood - pyrosequencing 13 males and 13 females
 Conclusions: KDM 5 C • Loss of DNA methylation at specific genes was found to be associated with KDM 5 C mutation: – Large degree of change 20 -50% similar to changes seen in imprinting disorders – Supports interplay between H 3 K 4 methylation and DNA methylation in humans – Supports the feasibility of studying DNA methylation in neurodevelopmental disorders, including ASD • Dependence of FBXL 5 and CACYBP DNA methylation on KDM 5 C/D dosage in normal males and females – Suggest that loss of DNA methylation at FBXL 5 and CACYBP promoters in blood of patients with KDM 5 C mutations could be a biomarker of similar changes occurring in brain
Conclusions: KDM 5 C • Loss of DNA methylation at specific genes was found to be associated with KDM 5 C mutation: – Large degree of change 20 -50% similar to changes seen in imprinting disorders – Supports interplay between H 3 K 4 methylation and DNA methylation in humans – Supports the feasibility of studying DNA methylation in neurodevelopmental disorders, including ASD • Dependence of FBXL 5 and CACYBP DNA methylation on KDM 5 C/D dosage in normal males and females – Suggest that loss of DNA methylation at FBXL 5 and CACYBP promoters in blood of patients with KDM 5 C mutations could be a biomarker of similar changes occurring in brain
 Interplay of environmental and epigenetic factors in ASD etiology
Interplay of environmental and epigenetic factors in ASD etiology
 DOES ART/SUBFERTILITY INCREASE RISK OF ASD? • Cohort study: – California, University of California, San Francisco: 4 fold increase of ASD in children born following assisted reproduction (Croughan et al. American Society for Reproductive Medicine conference 2006, 1699 naturally conceived/1008 conceived using ART or FT) • Case control studies: – Denmark: 2. 3 decrease of ART in ASD(N=461) compared to controls (n-461) (Maimburg and Vaeth, Human Reproduction, 2007) – California: 2. 2 increase of history of infertility in ASD twins (N=21) compared to twin controls (N=54), no association in singletons (349 cases vs 1. 847 controls) (Grether et al. 2012, J Autism Dev Disord) – Boston: 1. 7 increased rated of fertility treatments in ASD born to advanced age mothers>35 (N=164) compared to controls born to advanced age mothers (N=857) )(Lyall, et al. , Paediatric and Perinatal Epidemiology • Comparison to general population: – Israel: 10. 7% rate of ART in 507 ASD cases vs 3% rate of ART in Israeli population (Zachor and Itzchak, Research in Developmental Disabilities, 2011) – Japan: 4. 5% rate of ART in in 466 ASD cases vs 2, 5% of ART in Japan population (Shimada et al. Research in Autism Spectrum Disorders, 2012)
DOES ART/SUBFERTILITY INCREASE RISK OF ASD? • Cohort study: – California, University of California, San Francisco: 4 fold increase of ASD in children born following assisted reproduction (Croughan et al. American Society for Reproductive Medicine conference 2006, 1699 naturally conceived/1008 conceived using ART or FT) • Case control studies: – Denmark: 2. 3 decrease of ART in ASD(N=461) compared to controls (n-461) (Maimburg and Vaeth, Human Reproduction, 2007) – California: 2. 2 increase of history of infertility in ASD twins (N=21) compared to twin controls (N=54), no association in singletons (349 cases vs 1. 847 controls) (Grether et al. 2012, J Autism Dev Disord) – Boston: 1. 7 increased rated of fertility treatments in ASD born to advanced age mothers>35 (N=164) compared to controls born to advanced age mothers (N=857) )(Lyall, et al. , Paediatric and Perinatal Epidemiology • Comparison to general population: – Israel: 10. 7% rate of ART in 507 ASD cases vs 3% rate of ART in Israeli population (Zachor and Itzchak, Research in Developmental Disabilities, 2011) – Japan: 4. 5% rate of ART in in 466 ASD cases vs 2, 5% of ART in Japan population (Shimada et al. Research in Autism Spectrum Disorders, 2012)
 Sources of inconsistency • Parental age • Twinning (number of transferred embryos) • Variability in ART/FT procedures: type/dose of medications and embryo culture • ASD diagnosis
Sources of inconsistency • Parental age • Twinning (number of transferred embryos) • Variability in ART/FT procedures: type/dose of medications and embryo culture • ASD diagnosis
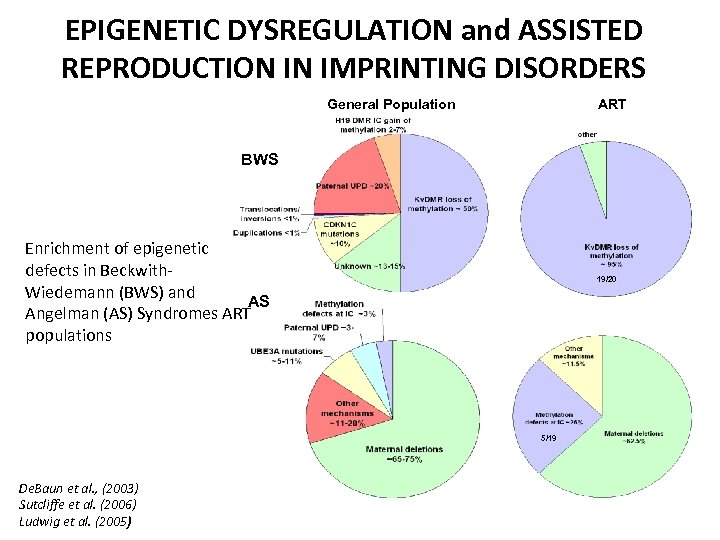 EPIGENETIC DYSREGULATION and ASSISTED REPRODUCTION IN IMPRINTING DISORDERS General Population ART BWS Enrichment of epigenetic defects in Beckwith. Wiedemann (BWS) and AS Angelman (AS) Syndromes ART populations 19/20 5/19 De. Baun et al. , (2003) Sutcliffe et al. (2006) Ludwig et al. (2005)
EPIGENETIC DYSREGULATION and ASSISTED REPRODUCTION IN IMPRINTING DISORDERS General Population ART BWS Enrichment of epigenetic defects in Beckwith. Wiedemann (BWS) and AS Angelman (AS) Syndromes ART populations 19/20 5/19 De. Baun et al. , (2003) Sutcliffe et al. (2006) Ludwig et al. (2005)
 Fertility Treatments Can Change DNA Methylation Patterns • Ovulation stimulation (FSH/clomid): – Maturation and ovulation of oocytes with incomplete/aberrant DNA methylation • In vitro fertilization (IVF): – In vitro embryo culture disrupts proper imprint maintenance during global genome demethylation • Intracytoplasmic sperm injection (ICSI): – Sperm with incomplete/aberrant methylation bypass natural selection
Fertility Treatments Can Change DNA Methylation Patterns • Ovulation stimulation (FSH/clomid): – Maturation and ovulation of oocytes with incomplete/aberrant DNA methylation • In vitro fertilization (IVF): – In vitro embryo culture disrupts proper imprint maintenance during global genome demethylation • Intracytoplasmic sperm injection (ICSI): – Sperm with incomplete/aberrant methylation bypass natural selection
 HYPOTHESIS § Epigenetic alterations, specifically DNA methylation, play an important role in ASD etiology § Subfertility/fertility treatments are associated with an increased rate of epigenetic errors that contribute to the ASD phenotype
HYPOTHESIS § Epigenetic alterations, specifically DNA methylation, play an important role in ASD etiology § Subfertility/fertility treatments are associated with an increased rate of epigenetic errors that contribute to the ASD phenotype
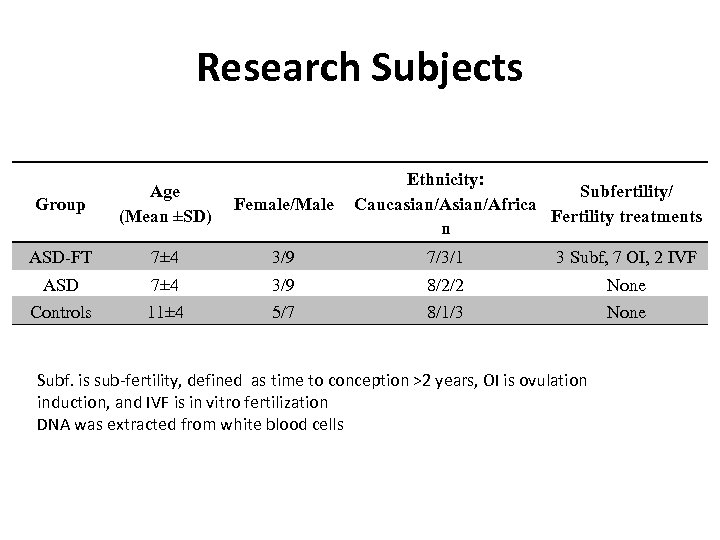 Research Subjects Ethnicity: Subfertility/ Caucasian/Africa Fertility treatments n Group Age (Mean ±SD) Female/Male ASD-FT 7± 4 3/9 7/3/1 3 Subf, 7 OI, 2 IVF ASD 7± 4 3/9 8/2/2 None Controls 11± 4 5/7 8/1/3 None Subf. is sub-fertility, defined as time to conception >2 years, OI is ovulation induction, and IVF is in vitro fertilization DNA was extracted from white blood cells
Research Subjects Ethnicity: Subfertility/ Caucasian/Africa Fertility treatments n Group Age (Mean ±SD) Female/Male ASD-FT 7± 4 3/9 7/3/1 3 Subf, 7 OI, 2 IVF ASD 7± 4 3/9 8/2/2 None Controls 11± 4 5/7 8/1/3 None Subf. is sub-fertility, defined as time to conception >2 years, OI is ovulation induction, and IVF is in vitro fertilization DNA was extracted from white blood cells
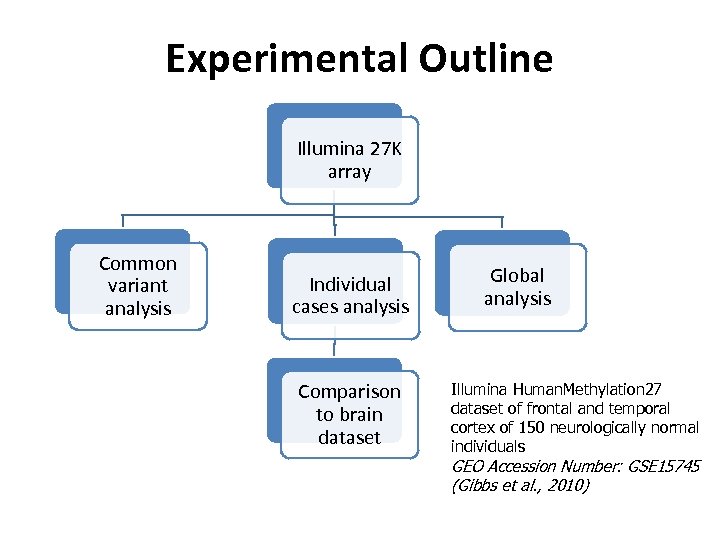 Experimental Outline Illumina 27 K array Common variant analysis Individual cases analysis Comparison to brain dataset Global analysis Illumina Human. Methylation 27 dataset of frontal and temporal cortex of 150 neurologically normal individuals GEO Accession Number: GSE 15745 (Gibbs et al. , 2010)
Experimental Outline Illumina 27 K array Common variant analysis Individual cases analysis Comparison to brain dataset Global analysis Illumina Human. Methylation 27 dataset of frontal and temporal cortex of 150 neurologically normal individuals GEO Accession Number: GSE 15745 (Gibbs et al. , 2010)
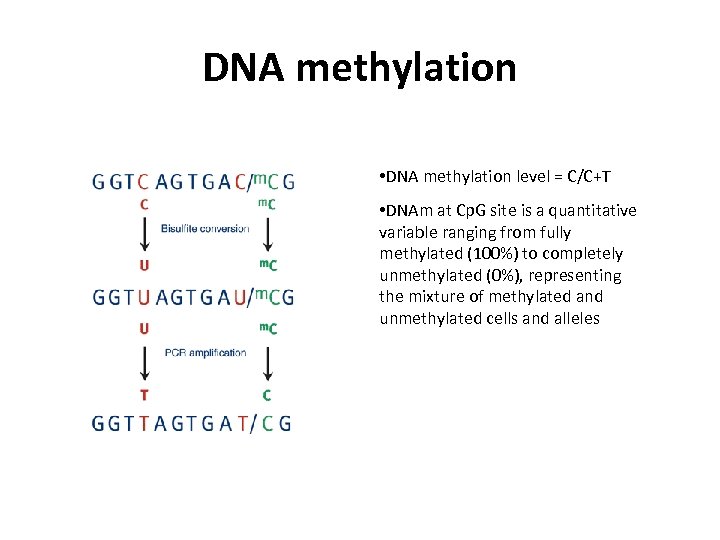 DNA methylation • DNA methylation level = C/C+T • DNAm at Cp. G site is a quantitative variable ranging from fully methylated (100%) to completely unmethylated (0%), representing the mixture of methylated and unmethylated cells and alleles
DNA methylation • DNA methylation level = C/C+T • DNAm at Cp. G site is a quantitative variable ranging from fully methylated (100%) to completely unmethylated (0%), representing the mixture of methylated and unmethylated cells and alleles
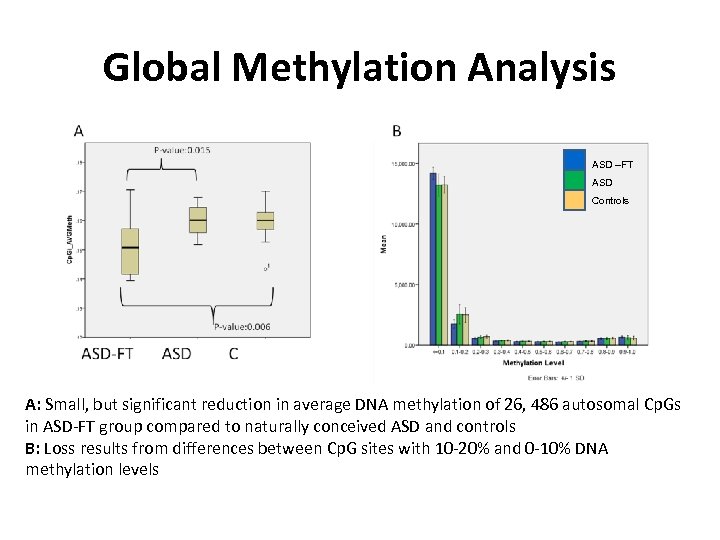 Global Methylation Analysis ASD –FT ASD Controls A: Small, but significant reduction in average DNA methylation of 26, 486 autosomal Cp. Gs in ASD-FT group compared to naturally conceived ASD and controls B: Loss results from differences between Cp. G sites with 10 -20% and 0 -10% DNA methylation levels
Global Methylation Analysis ASD –FT ASD Controls A: Small, but significant reduction in average DNA methylation of 26, 486 autosomal Cp. Gs in ASD-FT group compared to naturally conceived ASD and controls B: Loss results from differences between Cp. G sites with 10 -20% and 0 -10% DNA methylation levels
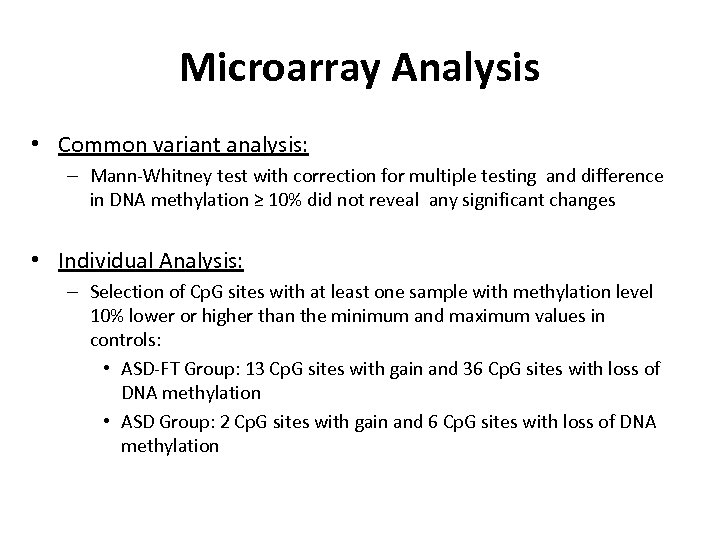 Microarray Analysis • Common variant analysis: – Mann-Whitney test with correction for multiple testing and difference in DNA methylation ≥ 10% did not reveal any significant changes • Individual Analysis: – Selection of Cp. G sites with at least one sample with methylation level 10% lower or higher than the minimum and maximum values in controls: • ASD-FT Group: 13 Cp. G sites with gain and 36 Cp. G sites with loss of DNA methylation • ASD Group: 2 Cp. G sites with gain and 6 Cp. G sites with loss of DNA methylation
Microarray Analysis • Common variant analysis: – Mann-Whitney test with correction for multiple testing and difference in DNA methylation ≥ 10% did not reveal any significant changes • Individual Analysis: – Selection of Cp. G sites with at least one sample with methylation level 10% lower or higher than the minimum and maximum values in controls: • ASD-FT Group: 13 Cp. G sites with gain and 36 Cp. G sites with loss of DNA methylation • ASD Group: 2 Cp. G sites with gain and 6 Cp. G sites with loss of DNA methylation
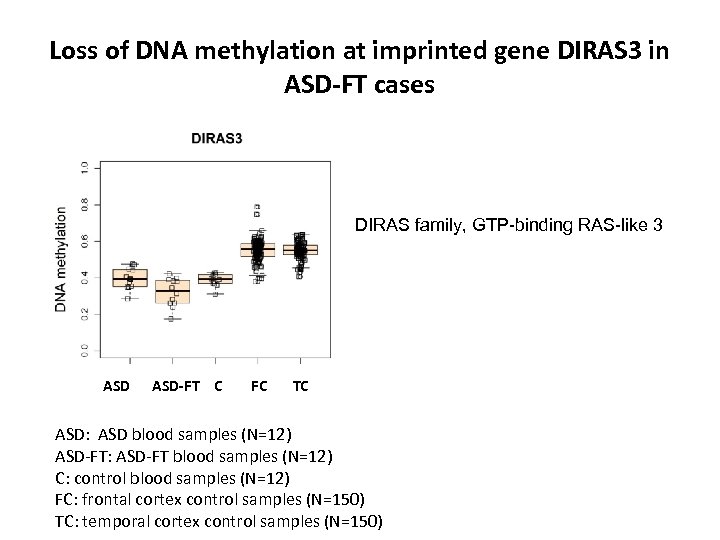 Loss of DNA methylation at imprinted gene DIRAS 3 in ASD-FT cases DIRAS family, GTP-binding RAS-like 3 ASD-FT C FC TC ASD: ASD blood samples (N=12) ASD-FT: ASD-FT blood samples (N=12) C: control blood samples (N=12) FC: frontal cortex control samples (N=150) TC: temporal cortex control samples (N=150)
Loss of DNA methylation at imprinted gene DIRAS 3 in ASD-FT cases DIRAS family, GTP-binding RAS-like 3 ASD-FT C FC TC ASD: ASD blood samples (N=12) ASD-FT: ASD-FT blood samples (N=12) C: control blood samples (N=12) FC: frontal cortex control samples (N=150) TC: temporal cortex control samples (N=150)
 Conclusions Studying cases with 1. neurodevelopmental phenotypes and genetic alterations in epigenetic regulators or 2. certain environmental exposures can identify epigenetic dysregulation – Likely to play important role in molecular pathophysiology of disorder – Could be further studied in idiopathic cases Epigenetics plays at least as important a role in ASD etiology as genetics
Conclusions Studying cases with 1. neurodevelopmental phenotypes and genetic alterations in epigenetic regulators or 2. certain environmental exposures can identify epigenetic dysregulation – Likely to play important role in molecular pathophysiology of disorder – Could be further studied in idiopathic cases Epigenetics plays at least as important a role in ASD etiology as genetics
 Acknowledgments Weksberg Lab Autism project: Daria Grafodatskaya Darci Butcher Brian Chung Rageen Rajendram Sarah Goodman Sanaa Choufani Chunhua Zhao Youliang Lou Jonathan Shapiro Yi-an Chen Tanya Guha Hailey Jin Liis Uuskula Michal Feigenberg Khadine Wiltshire Cinical Genetics Cheryl Cytrynbaum Cheryl Shuman Steve Scherer , The Centre for Applied Genomics Wendy Roberts, Autism Research Unit Evdokia Anagnostou , Bloorview Andrei Turinsky, Centre for Computational Biology KDM 5 C project • C. E. Schwartz • F. E. Abidi • C. Skinn – Greenwood Genetic Center, South Carolina, USA
Acknowledgments Weksberg Lab Autism project: Daria Grafodatskaya Darci Butcher Brian Chung Rageen Rajendram Sarah Goodman Sanaa Choufani Chunhua Zhao Youliang Lou Jonathan Shapiro Yi-an Chen Tanya Guha Hailey Jin Liis Uuskula Michal Feigenberg Khadine Wiltshire Cinical Genetics Cheryl Cytrynbaum Cheryl Shuman Steve Scherer , The Centre for Applied Genomics Wendy Roberts, Autism Research Unit Evdokia Anagnostou , Bloorview Andrei Turinsky, Centre for Computational Biology KDM 5 C project • C. E. Schwartz • F. E. Abidi • C. Skinn – Greenwood Genetic Center, South Carolina, USA


