ac439beac35769bbf362c4185b54d55d.ppt
- Количество слайдов: 43

Essential Bioinformatics and Biocomputing (LSM 2104: Section I) Biological Databases and Bioinformatics Software Prof. Chen Yu Zong Tel: 6874 -6877 Email: csccyz@nus. edu. sg http: //xin. cz 3. nus. edu. sg Room 07 -24, level 7, SOC 1, NUS January 2003 Essential Bioinformatics and Biocomputing (LSM 2104), NUS
Essential Bioinformatics and Biocomputing (LSM 2104: Section I) Four lectures Part 1: Biological databases: Lecture 2. Biological information and databases Lecture 3. More databases, retrieval systems, and database searching Part 2: Software: Lecture 4. Examples of the applications of bioinformatics software and basic principles Lecture 5. Overview of bioinformatics software Essential Bioinformatics and Biocomputing (LSM 2104), NUS

Part 1: Biological databases Part 1 outline: 1. Biological information and databases – Overview and definition, types of biological databases 2. Popular databases, records, data format – Genbank, Swiss. Prot, OMIM, PDB, KEGG, BIND, Pfam, PROSITE, Pub. Med 3. Accessing biological databases, retrieval systems – Entrez, SRS 4. Searching biological databases – Data quality, coverage, redundancy, errors Textbook: --T. K. Atwood and D. J. Parry Smith, Introduction to Bioinformatics. Biological databases: chapters 3 and 4 Essential Bioinformatics and Biocomputing (LSM 2104), NUS 3
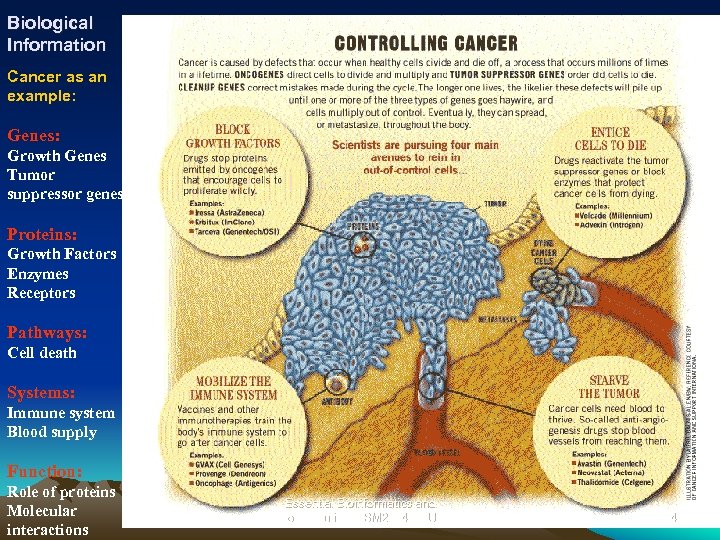
Biological Information Cancer as an example: Genes: Growth Genes Tumor suppressor genes Proteins: Growth Factors Enzymes Receptors Pathways: Cell death Systems: Immune system Blood supply Function: Role of proteins Molecular interactions Essential Bioinformatics and Biocomputing (LSM 2104), NUS 4
Biological Information Nucleic acids: • DNA sequence, genes, gene products (proteins), mutation, gene coding, distribution patterns, motifs • Genomics: genome, gene structure and expression, genetic map, genetic disorder • RNA sequence, secondary structure, 3 D structure, interactions Proteins: • Protein sequence, corresponding gene, secondary structure, 3 D structure, function, motifs, homology, interactions • Proteomics: expression profile, proteins in disease processes etc. • Ligands and drugs (inhibitors, activators, substrates, metabolites) Essential Bioinformatics and Biocomputing (LSM 2104), NUS 5
Biological Information Pathways: • Molecular networks, biological chain events, regulation, feedback, kinetic data Function: • Binding sites, interactions, molecular action (binding, chemical reaction, etc. ) • Biological effect (signaling, transport, feedback, regulation, modification, etc. ) • Functional relationship, protein families, motifs, and homologs Essential Bioinformatics and Biocomputing (LSM 2104), NUS 6

Biological databases Purpose 1. To disseminate biological data and information 2. To provide biological data in computer-readable form 3. To allow analysis of biological data A database needs to have at minimum a specific tool for searching and data extraction. – • Web pages, books, journal articles, tables, text files, and spreadsheet files cannot be considered as databases Reading materials: – Baxevanis AD. The Molecular Biology Database Collection: 2002 update. Nucleic Acids Res. 2002 Jan 1; 30(1): 1 -12. Essential Bioinformatics and Biocomputing (LSM 2104), NUS 7

Biological databases Lists of biological databases • INFOBIOGEN Catalog of Databases • Nucleic Acids Research Database Listing http: //www. infobiogen. fr/services/dbcat/ http: //nar. oupjournals. org/cgi/content/full/30/1/1/DC 1 – These serve as starting point of biological databases. – More than 500 databases have been catalogued to date and those from the two listings satisfy minimal criteria for the content, access, and quality. – Other sites as a starting point. Essential Bioinformatics and Biocomputing (LSM 2104), NUS 8
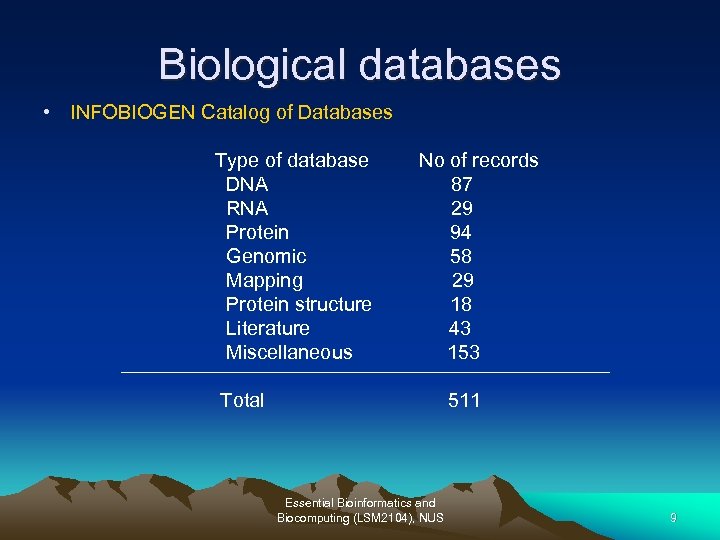
Biological databases • INFOBIOGEN Catalog of Databases Type of database DNA RNA Protein Genomic Mapping Protein structure Literature Miscellaneous No of records 87 29 94 58 29 18 43 153 Total 511 Essential Bioinformatics and Biocomputing (LSM 2104), NUS 9

Biological databases- in Nucleic Acids Research Type of database No of records Major Sequence Repositories 7 Comparative Genomics 7 Gene Expression 20 Gene Identification and Structure 30 Genetic and Physical Maps 10 Genomic Databases 48 Intermolecular Interactions 5 Metabolic Pathways and Cellular Regulation 12 Mutation Databases 33 Pathology 8 Protein Databases 50 Protein Sequence Motifs 18 Proteome Resources 7 RNA Sequences 26 Retrieval Systems and Database Structure 32 Transgenics 2 Varied Biomedical Content 18 TOTAL 336 Essential Bioinformatics and Biocomputing (LSM 2104), NUS 10
Literature databases – Pub. Med (Med. Line) 1. It contains entries for more than 11 million abstracts of scientific publications. 2. It enables user to do keyword searches, provides links to a selection of full articles, and has text mining capabilities, e. g. provides links to related articles, and Gen. Bank entries, among others. 3. Efficient searching Pub. Med requires some skill. For example, searching with a keyword “interleukin” returns 108, 366 matches. Essential Bioinformatics and Biocomputing (LSM 2104), NUS 11
Pub. Med web-site (http: //www 3. ncbi. nlm. nih. gov/entrez/query. fcgi? db=Pub. Med ) Essential Bioinformatics and Biocomputing (LSM 2104), NUS 12

Pub. Med Search (http: //www 3. ncbi. nlm. nih. gov/entrez/query. fcgi? db=Pub. Med ) Key Word Cancer treatment by targeting blood supply: Cancer growth depends on blood supply (why? ) and thus requires the growth of new blood vessels – angiogenesis Proteins involved in angiogenesis may be potential anticancer targets You can find some of these targets by searching Pubmed Key word “cancer angiogenesis enzyme drug” produces 856 entries No. of Entries 1. 45 M Cancer Blood supply 22 K Cancer Blood supply Protein 3. 9 K Cancer Blood supply Enzyme 1. 5 K Cancer Blood supply Enzyme Drug 500 Essential Bioinformatics and Biocomputing (LSM 2104), NUS 13
Nucleic Acids databases What info are in these databases: • DNA sequence, genes, gene products (proteins), mutation, gene coding, distribution patterns, motifs • Genomics: genome, gene structure and expression, genetic map, genetic disorder • RNA sequence, secondary structure, 3 D structure, interactions Essential Bioinformatics and Biocomputing (LSM 2104), NUS 14
Nucleic Acids databases DNA databases – Gen. Bank, EMBL, DDBJ 1. General purpose databases focusing on DNA sequences and their properties 2. Gen. Bank, EMBL-bank and DDBJ exchange data to ensure comprehensive worldwide coverage and accession numbers are managed consistently between the three centers. Reading materials: – Textbook, chapter 4 Essential Bioinformatics and Biocomputing (LSM 2104), NUS 15
DNA databases • Gen. Bank database (http: //www. ncbi. nih. gov/Genbank/) – Contains publicly available DNA sequences from more than 100, 000 organisms. – Also contains derived protein sequences, and annotations describing biological, structural, and other relevant features. – Accessible through Entrez, NCBI’s integrated retrieval system (studied later) – Sequence similarity search tools: BLAST (studied later) • EMBL nucleotide sequence database (http: //www. ebi. ac. uk/embl/) – Contains nucleotide sequences collected from all public sources. – Accessible through Sequence Retrieval System (SRS) which allows keyword searching (studied later) – Sequence similarity search tools: Blitz, Fasta, and BLAST (studied later) Essential Bioinformatics and Biocomputing (LSM 2104), NUS 16
DNA databases: Gen. Bank Web page Essential Bioinformatics and Biocomputing (LSM 2104), NUS 17
DNA databases • An Example from Gen. Bank– flat file – Human Alpha-Lactalbumin gene This protein is a complex of 2 proteins A and B. In the absence of the B protein, the enzyme catalyzes the transfer of galactose from UDP-galactose to Nacetylglucosamine (cf. EC 2. 4. 1. 90). Essential Bioinformatics and Biocomputing (LSM 2104), NUS 18
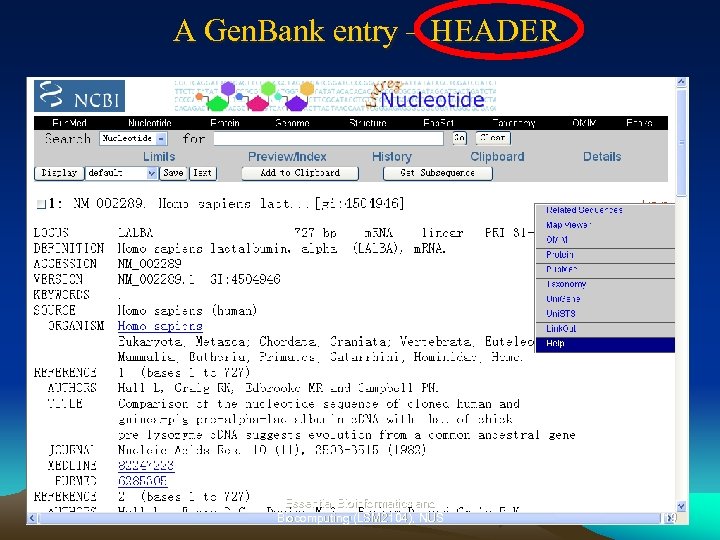
A Gen. Bank entry – HEADER Essential Bioinformatics and Biocomputing (LSM 2104), NUS 19

Gen. Bank Entry – Links provided in the Header • • • Map. Viewer – find the gene position in chromosome Related Sequences – other entries related to this gene (or sequence) OMIM– link to catalog of human genes and genetic disorders Protein – retrieve protein record from Gen. Pept Medline and Pub. Med –literature abstracts related to this gene Taxonomy – Classification of organisms Uni. Gene – Unified gene data Uni. STS – Unified sequence tagged sites, marker and mapping data Link. Out – links to publishers, aggregators libraries, biological databases, sequence centers, and other Web resources • REFSEQ – reference sequence standards Note: These links are representative. Other links may also be found in Gen. Bank entries. Essential Bioinformatics and Biocomputing (LSM 2104), NUS 20
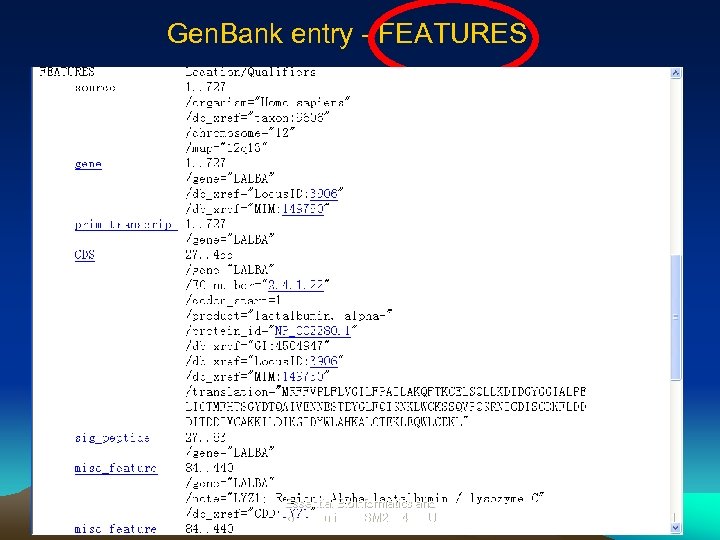
Gen. Bank entry - FEATURES Essential Bioinformatics and Biocomputing (LSM 2104), NUS 21
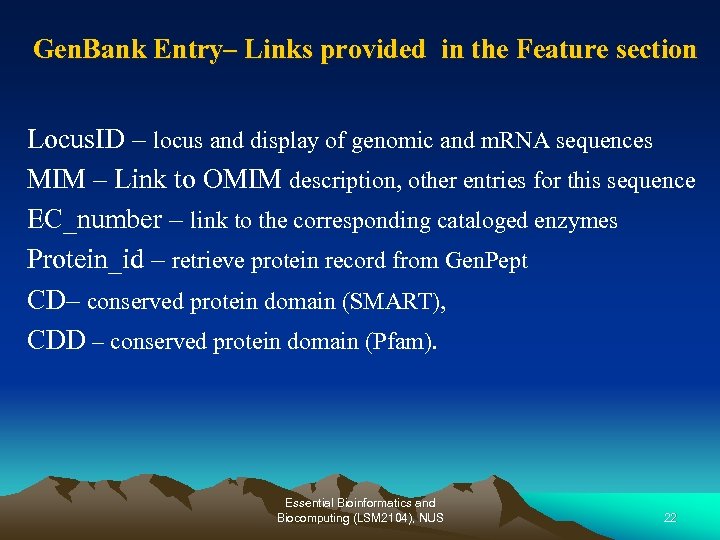
Gen. Bank Entry– Links provided in the Feature section Locus. ID – locus and display of genomic and m. RNA sequences MIM – Link to OMIM description, other entries for this sequence EC_number – link to the corresponding cataloged enzymes Protein_id – retrieve protein record from Gen. Pept CD– conserved protein domain (SMART), CDD – conserved protein domain (Pfam). Essential Bioinformatics and Biocomputing (LSM 2104), NUS 22

Biological databases: Gen. Bank - SEQUENCE Essential Bioinformatics and Biocomputing (LSM 2104), NUS 23

Gen. Bank - NOTES Majority of Gen. Bank entries have similar form to our example. When accessing the database, the following needs to be noticed: • Some entries are huge, containing as much as 30, 000 lines. (NT_021877 Homo sapiens chromosome 1 working draft sequence segment) • Some entries have contig information instead of sequence information. (NT_021877 Homo sapiens chromosome 1 working draft sequence segment) • Some entries are derived from c. DNA sequences and thus represent putative genes/proteins. These should be used with caution. (AK 007430. Mus musculus 10 d. . . [gi: 12840976]). • Some annotations are predicted using automated analysis. These should also be used with caution. (XM_131483 Mus musculus simi. . . [gi: 20832685]). Essential Bioinformatics and Biocomputing (LSM 2104), NUS 24

Gen. Bank - Statistics Year Base Pairs Sequences 1982 680338 606 1992 101008486 78608 2000 11101066288 10106023 2001 15849921438 14976310 Data size is large and increases fast Essential Bioinformatics and Biocomputing (LSM 2104), NUS 25
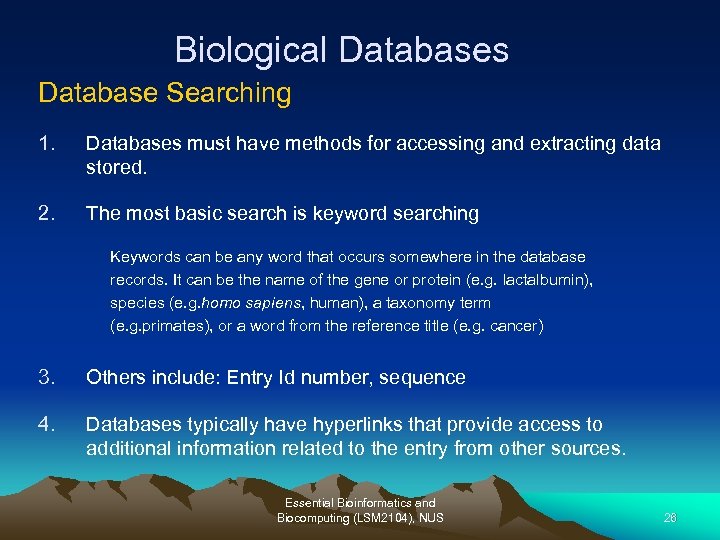
Biological Databases Database Searching 1. Databases must have methods for accessing and extracting data stored. 2. The most basic search is keyword searching Keywords can be any word that occurs somewhere in the database records. It can be the name of the gene or protein (e. g. lactalbumin), species (e. g. homo sapiens, human), a taxonomy term (e. g. primates), or a word from the reference title (e. g. cancer) 3. Others include: Entry Id number, sequence 4. Databases typically have hyperlinks that provide access to additional information related to the entry from other sources. Essential Bioinformatics and Biocomputing (LSM 2104), NUS 26
Biological databases: OMIM Online Mendelian Inheritance in Man (http: //www. ncbi. nlm. nih. gov/Omim/) • The OMIM database contains abstracts and texts describing genetic disorders to support genomics efforts and clinical genetics. It provides gene maps, and known disorder maps in tabular listing formats. Contains keyword search. Hamosh A. et al. Online Mendelian Inheritance in Man (OMIM), a knowledge base of human genes and genetic disorders Nucleic Acids Res. 2002 30: 52 -55. Essential Bioinformatics and Biocomputing (LSM 2104), NUS 27
Biological databases: OMIM web-page Essential Bioinformatics and Biocomputing (LSM 2104), NUS 28

Biological databases: OMIM search engine Essential Bioinformatics and Biocomputing (LSM 2104), NUS 29
Biological databases: OMIM statistics All Entries : 14088 Established Gene Locus : 10476 Phenotype Descriptions : 1194 Other Entries : 2418 Essential Bioinformatics and Biocomputing (LSM 2104), NUS 30
Biological databases Protein databases 1. SWISS-PROT (http: //us. expasy. org/sprot-top. html) is a curated database focusing on high level of annotation (sequence, function, structure, post-translational modifications, variants, etc. ) of proteins. 2. Tr. EMBL is Computer-annotated supplement to SWISSPROT Reading materials: Textbook, chapter 3 Essential Bioinformatics and Biocomputing (LSM 2104), NUS 31

Protein databases What are in these databases: • Protein sequence, corresponding gene, secondary structure, 3 D structure, function, motifs, homology, interactions • Proteomics: expression profile, proteins in disease processes etc. • Ligands and drugs (inhibitors, activators, substrates, metabolites) Essential Bioinformatics and Biocomputing (LSM 2104), NUS 32
Protein databases – SWISS-PROT Notes: • SWISS-PROT provides high-quality annotations and detailed info about sequence, structural, functional, and other properties of proteins. • It provides a rich set of links to other sources of information on SWISS-PROT entries. Unfortunately, some of the links will not work at all times, because of the dynamical change of the Web. • It also provides a rich set of protein analysis tools. Essential Bioinformatics and Biocomputing (LSM 2104), NUS 33
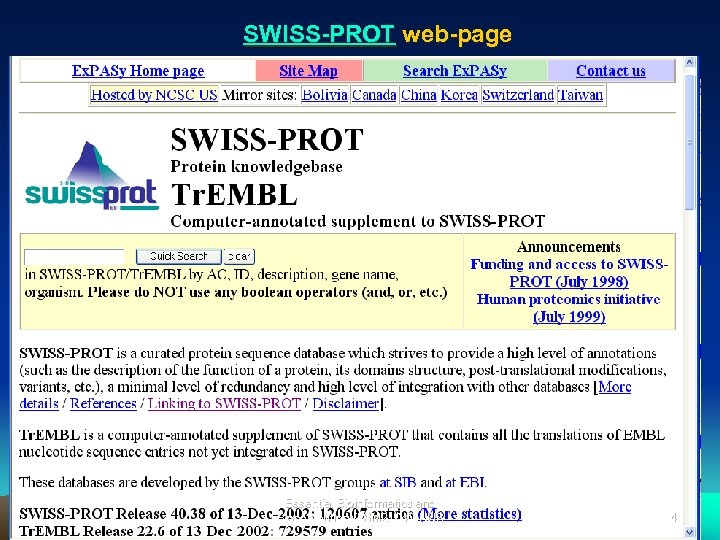
SWISS-PROT web-page Essential Bioinformatics and Biocomputing (LSM 2104), NUS 34
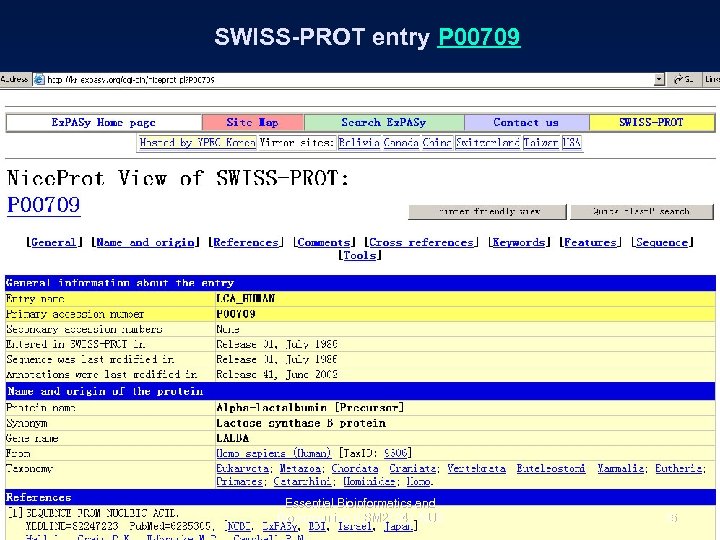
SWISS-PROT entry P 00709 Essential Bioinformatics and Biocomputing (LSM 2104), NUS 35

Essential Bioinformatics and Biocomputing (LSM 2104), NUS 36

SWISS-PROT entry P 00709 Essential Bioinformatics and Biocomputing (LSM 2104), NUS 37

SWISS-PROT entry P 00709 Essential Bioinformatics and Biocomputing (LSM 2104), NUS 38
Biological databases: Protein structure database: PDB (http: //www. pdb. org) 1. More than 18, 000 macromolecular structures on proteins, peptides, viruses, protein/nucleic acids complexes, nucleic acids, and carbohydrates. 2. Among the oldest databases – the first structure was deposited in 1972. 3. New deposited structures has been steadily growing (3298 in 2001, and 1486 Jan 1 -June 5, 2002). 4. Determined mainly by the X-ray diffraction and NMR. 5. It Contains tools for keyword search, comprehensive visualization, and information extraction – such as sequence, geometry, and structural neighbors details. Essential Bioinformatics and Biocomputing (LSM 2104), NUS 39

Biological databases: PDB web-page http: //www. rcsb. org/pdb/ Essential Bioinformatics and Biocomputing (LSM 2104), NUS 40
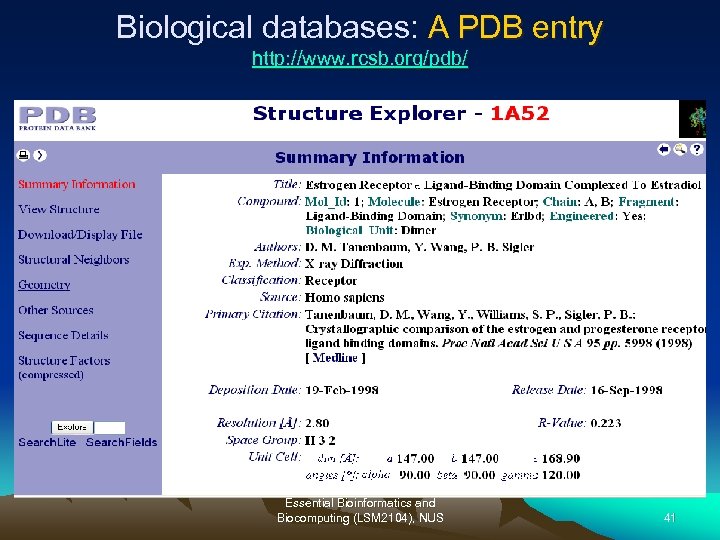
Biological databases: A PDB entry http: //www. rcsb. org/pdb/ Essential Bioinformatics and Biocomputing (LSM 2104), NUS 41
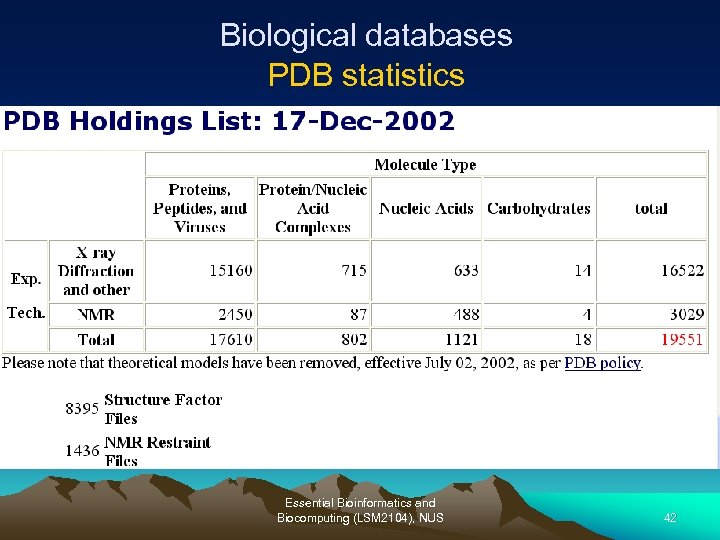
Biological databases PDB statistics Essential Bioinformatics and Biocomputing (LSM 2104), NUS 42
Biological databases Summary of Today’s lecture • Types of Biological information, data and databases • Simple data retrieval method. • Popular databases: Pubmed, Genbank, Swiss. Prot, OMIM, PDB • Statistics: – Large number of publications (MEDLINE: >12 M since 1960) – Large amount of data for sequence (DNA: >14 M, Protein: > 120 K) – Fair amount of data for 3 D structure (Protein >14 K, Nucleic acid >1 K) Essential Bioinformatics and Biocomputing (LSM 2104), NUS 43
ac439beac35769bbf362c4185b54d55d.ppt