9dd582b501892c95909a420421b1b672.ppt
- Количество слайдов: 56
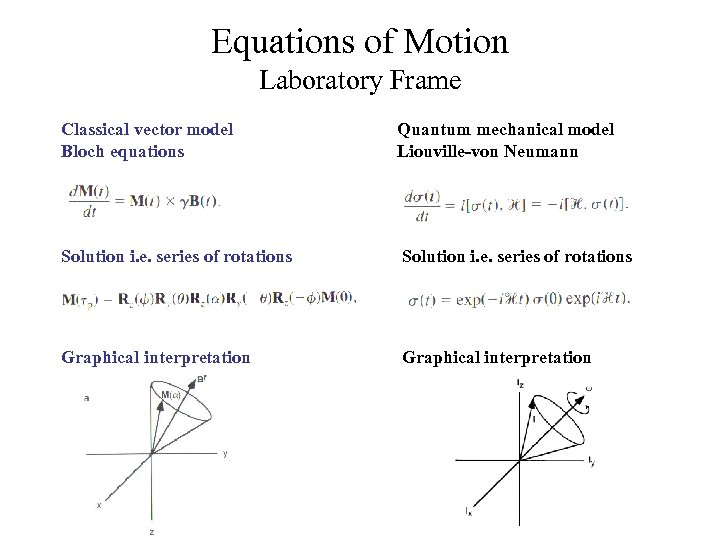
Equations of Motion Laboratory Frame Classical vector model Bloch equations Quantum mechanical model Liouville-von Neumann Solution i. e. series of rotations Graphical interpretation
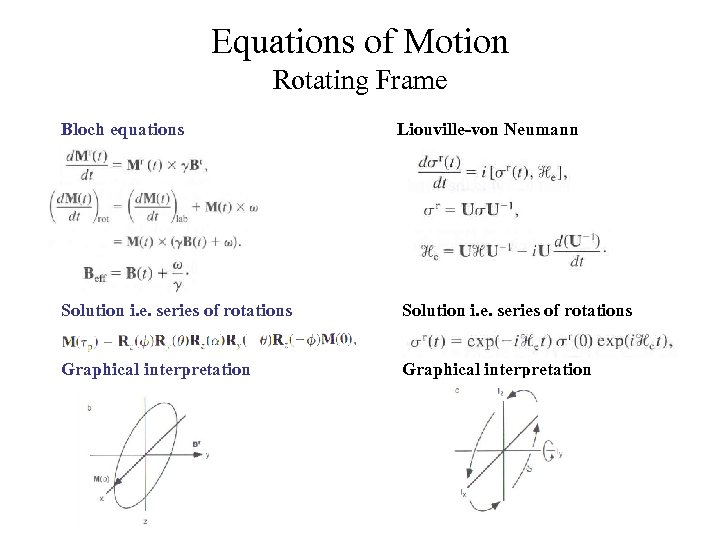
Equations of Motion Rotating Frame Bloch equations Liouville-von Neumann Solution i. e. series of rotations Graphical interpretation
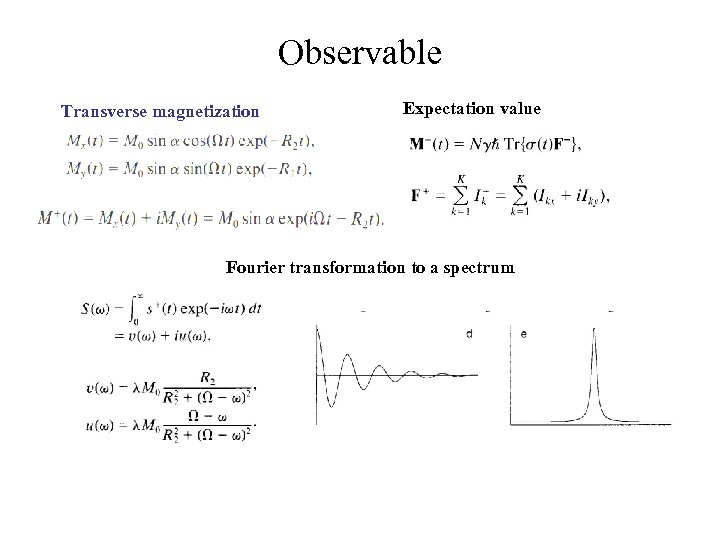
Observable Transverse magnetization Expectation value Fourier transformation to a spectrum
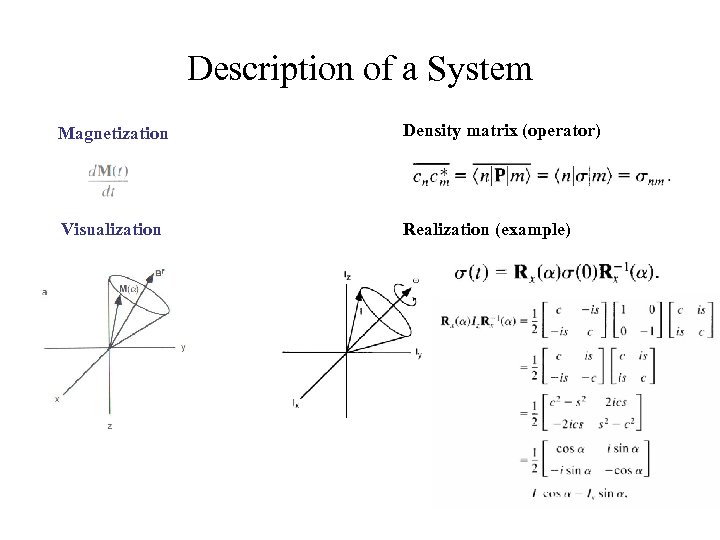
Description of a System Magnetization Density matrix (operator) Visualization Realization (example)
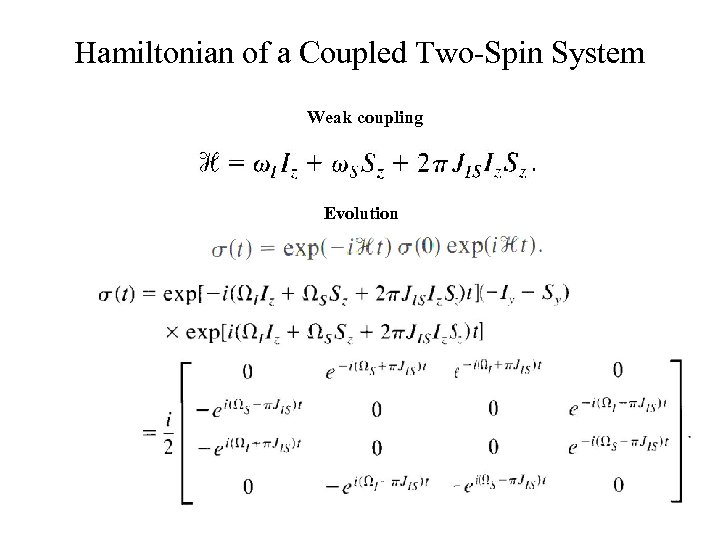
Hamiltonian of a Coupled Two-Spin System Weak coupling Evolution
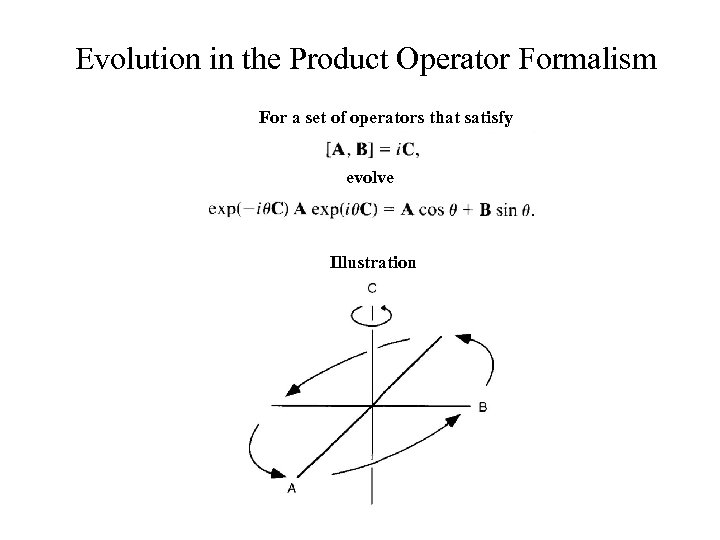
Evolution in the Product Operator Formalism For a set of operators that satisfy evolve Illustration
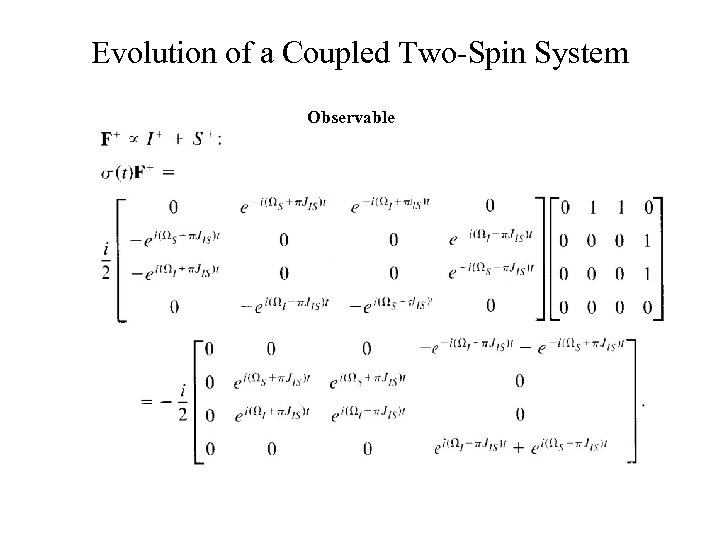
Evolution of a Coupled Two-Spin System Observable
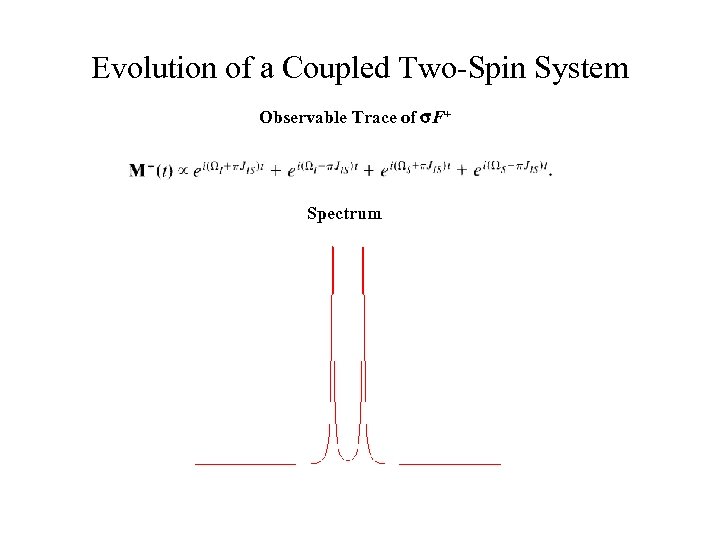
Evolution of a Coupled Two-Spin System Observable Trace of s. F+ Spectrum
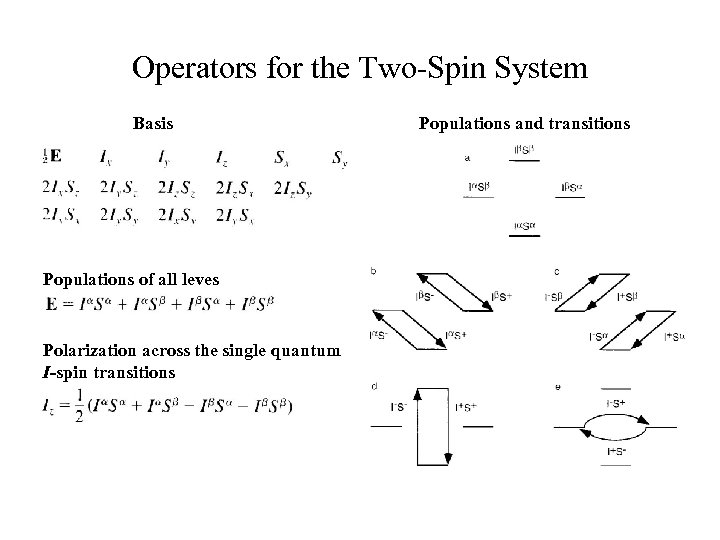
Operators for the Two-Spin System Basis Populations of all leves Polarization across the single quantum I-spin transitions Populations and transitions
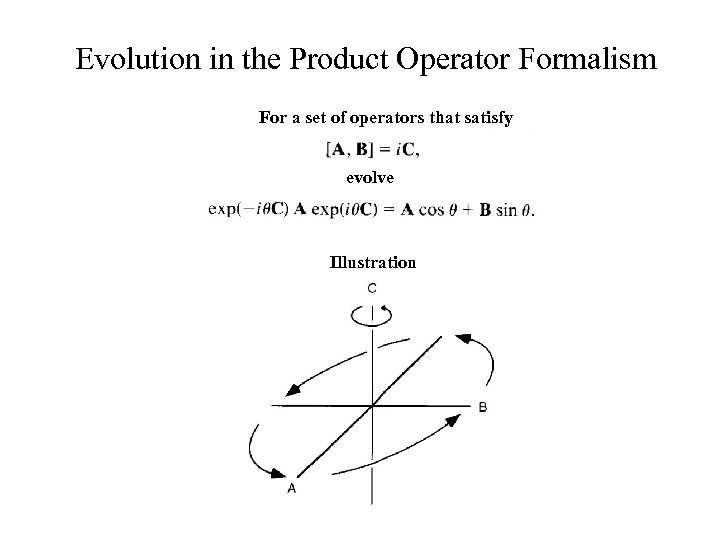
Evolution in the Product Operator Formalism For a set of operators that satisfy evolve Illustration
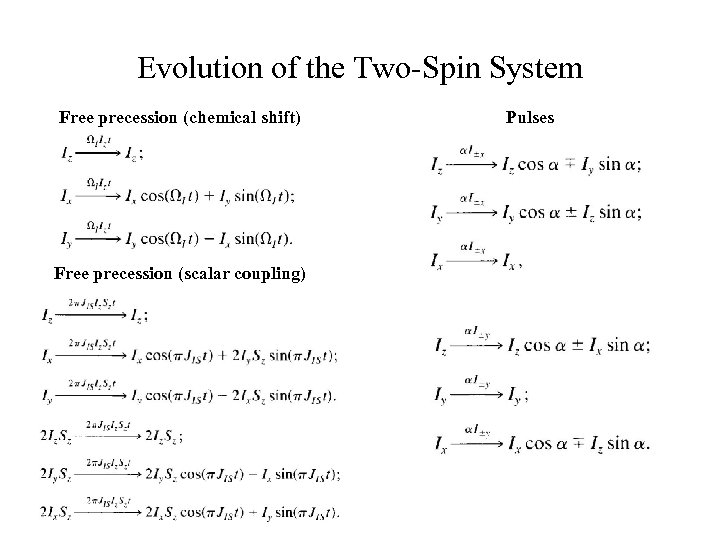
Evolution of the Two-Spin System Free precession (chemical shift) Free precession (scalar coupling) Pulses
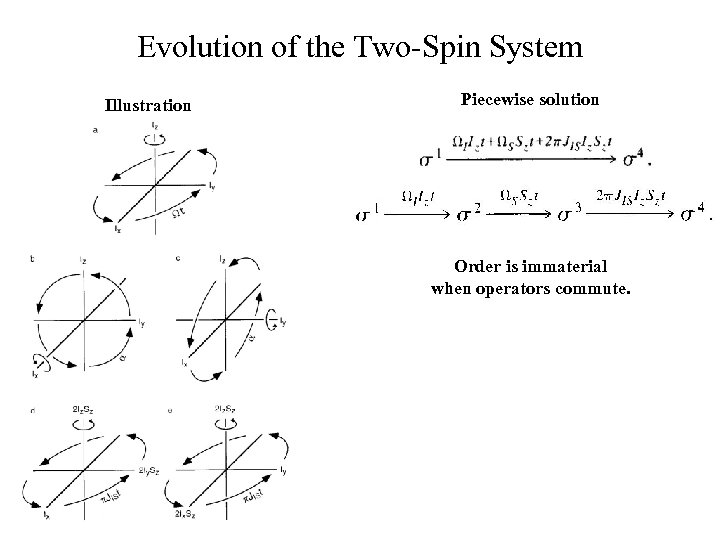
Evolution of the Two-Spin System Illustration Piecewise solution Order is immaterial when operators commute.
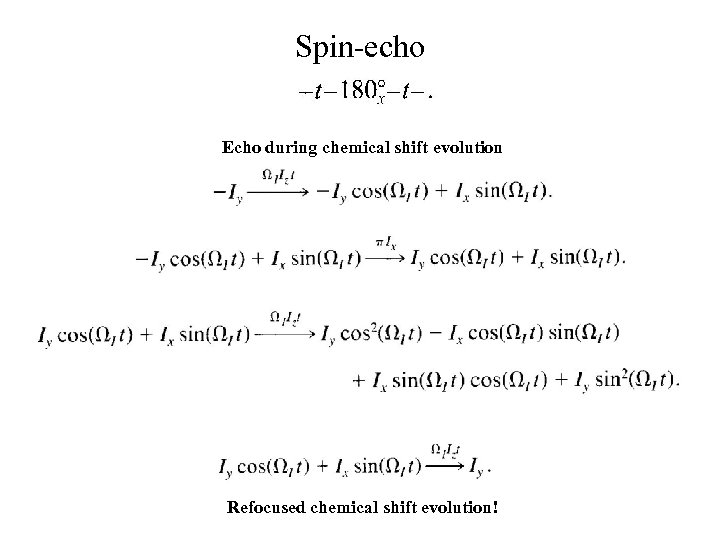
Spin-echo Echo during chemical shift evolution Refocused chemical shift evolution!
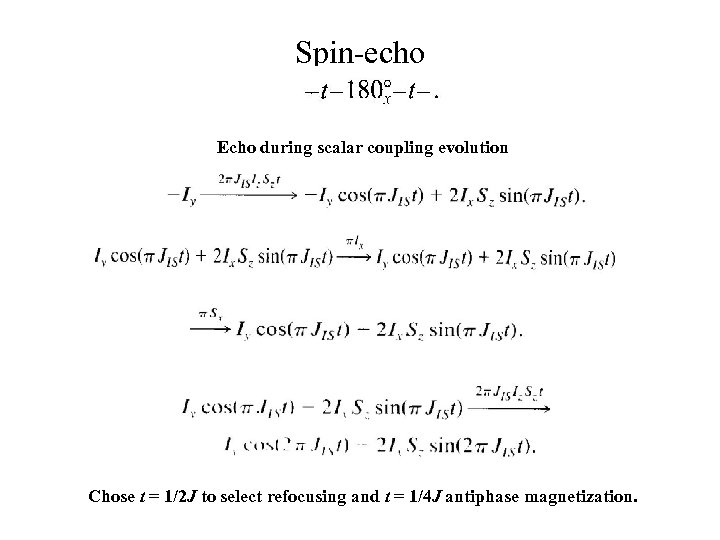
Spin-echo Echo during scalar coupling evolution Chose t = 1/2 J to select refocusing and t = 1/4 J antiphase magnetization.
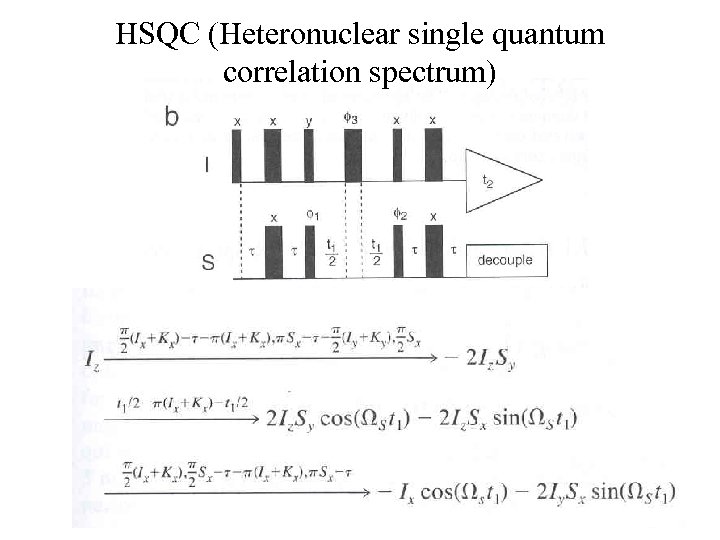
HSQC (Heteronuclear single quantum correlation spectrum)
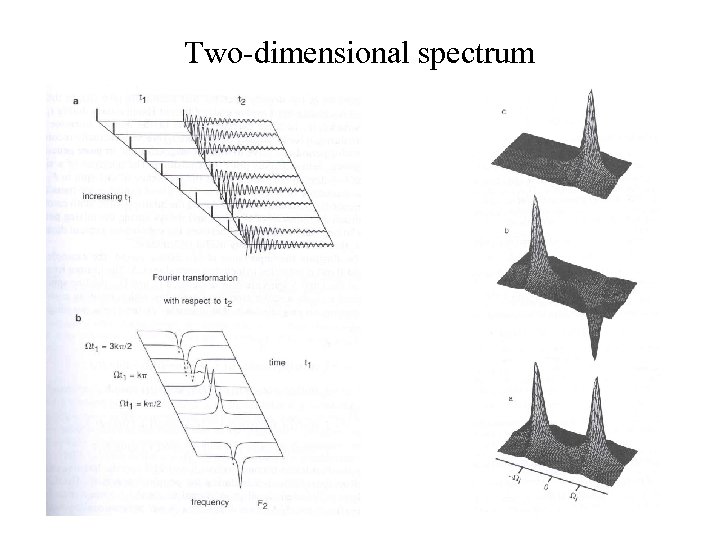
Two-dimensional spectrum
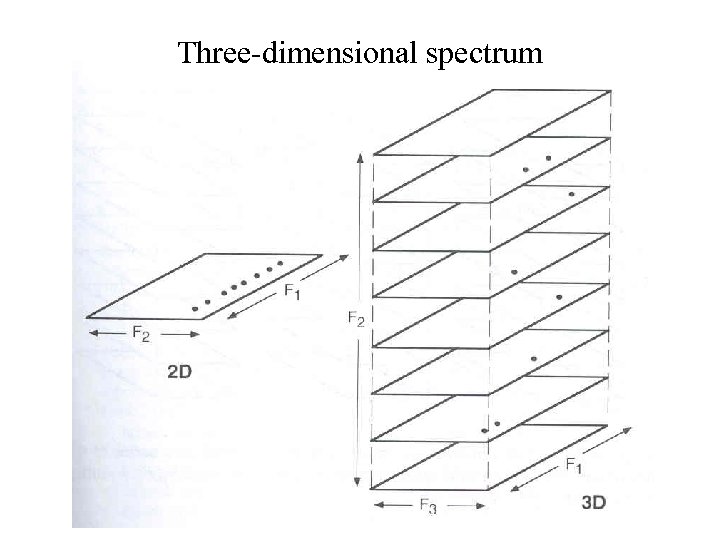
Three-dimensional spectrum
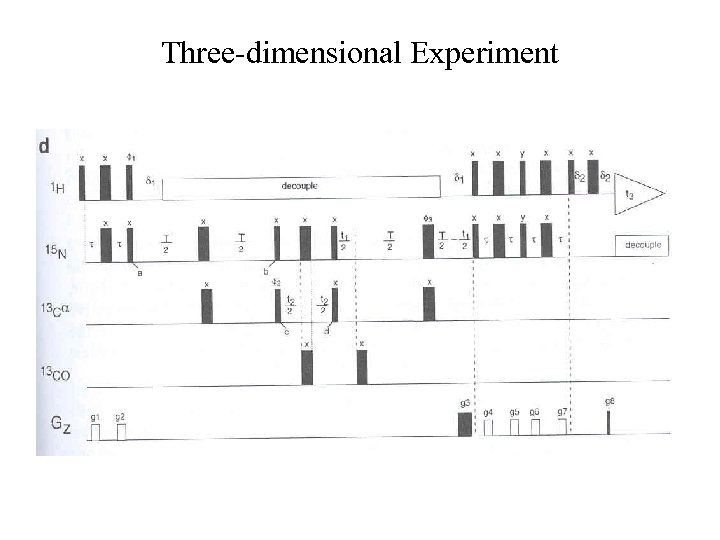
Three-dimensional Experiment
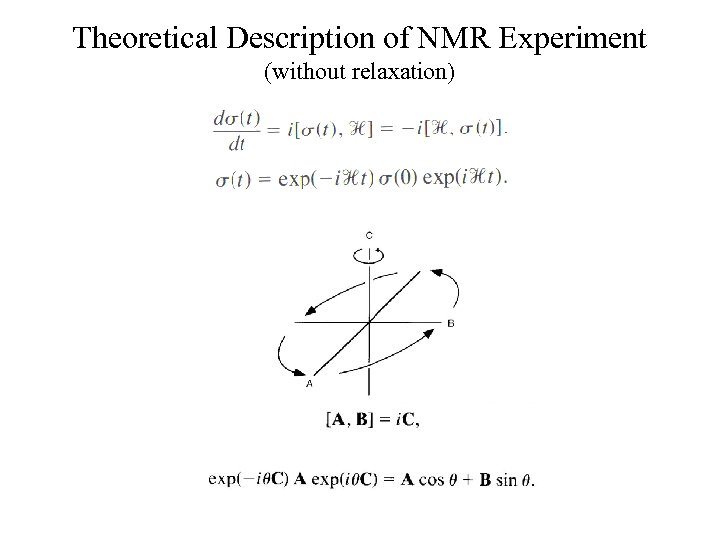
Theoretical Description of NMR Experiment (without relaxation)
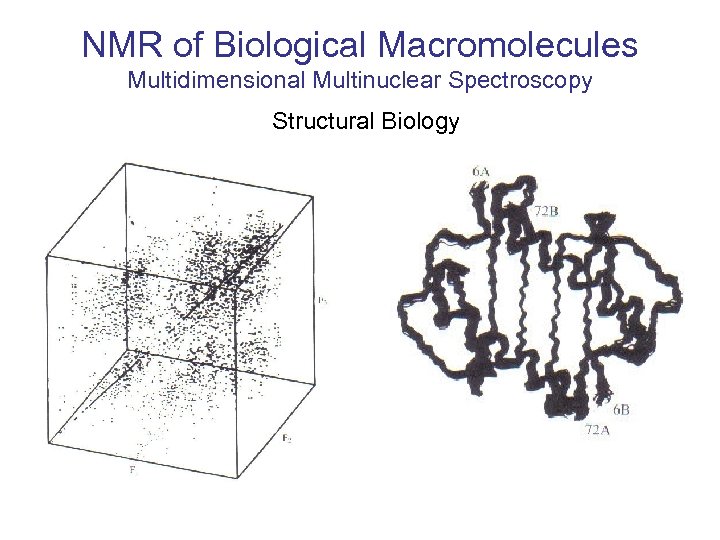
NMR of Biological Macromolecules Multidimensional Multinuclear Spectroscopy Structural Biology
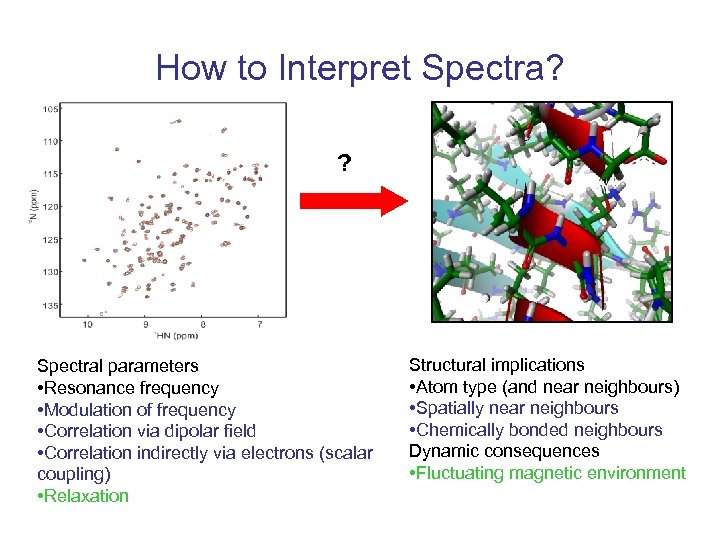
How to Interpret Spectra? ? Spectral parameters • Resonance frequency • Modulation of frequency • Correlation via dipolar field • Correlation indirectly via electrons (scalar coupling) • Relaxation Structural implications • Atom type (and near neighbours) • Spatially near neighbours • Chemically bonded neighbours Dynamic consequences • Fluctuating magnetic environment
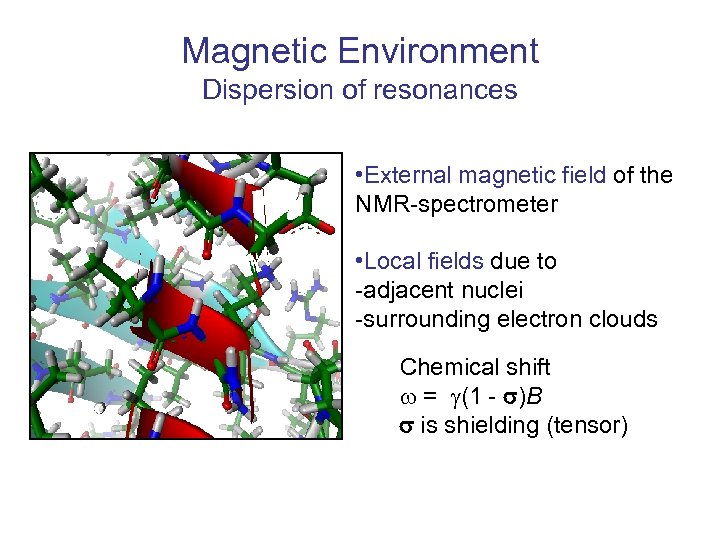
Magnetic Environment Dispersion of resonances • External magnetic field of the NMR-spectrometer • Local fields due to -adjacent nuclei -surrounding electron clouds Chemical shift w = g(1 - s)B s is shielding (tensor)
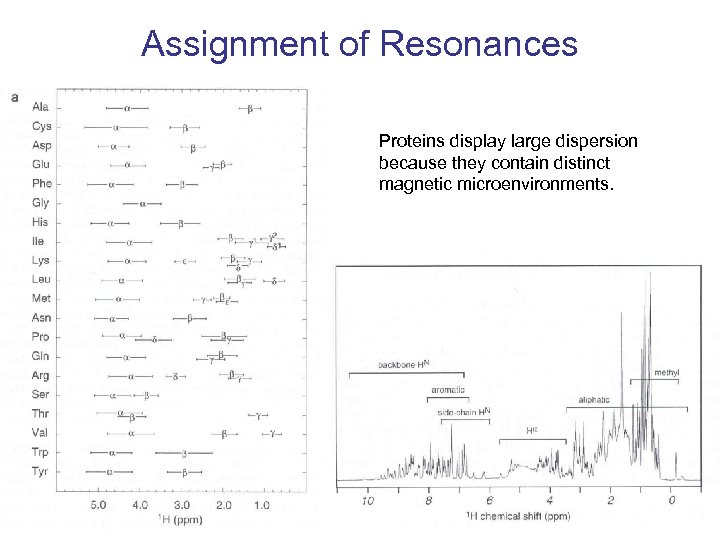
Assignment of Resonances Proteins display large dispersion because they contain distinct magnetic microenvironments.
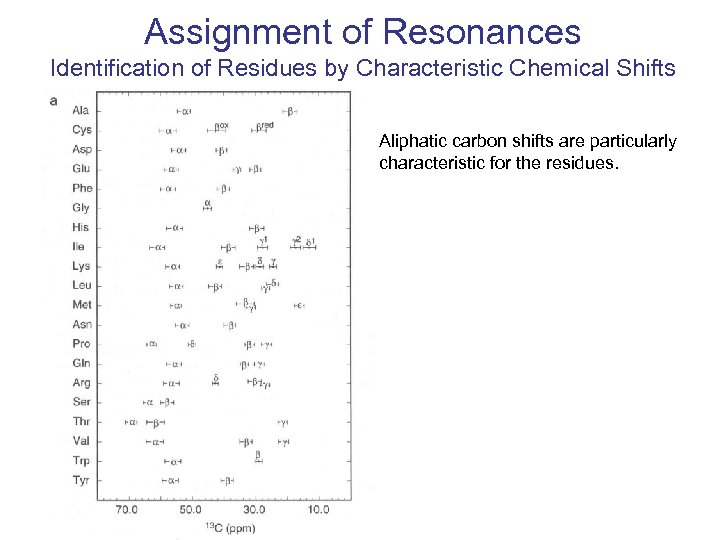
Assignment of Resonances Identification of Residues by Characteristic Chemical Shifts Aliphatic carbon shifts are particularly characteristic for the residues.
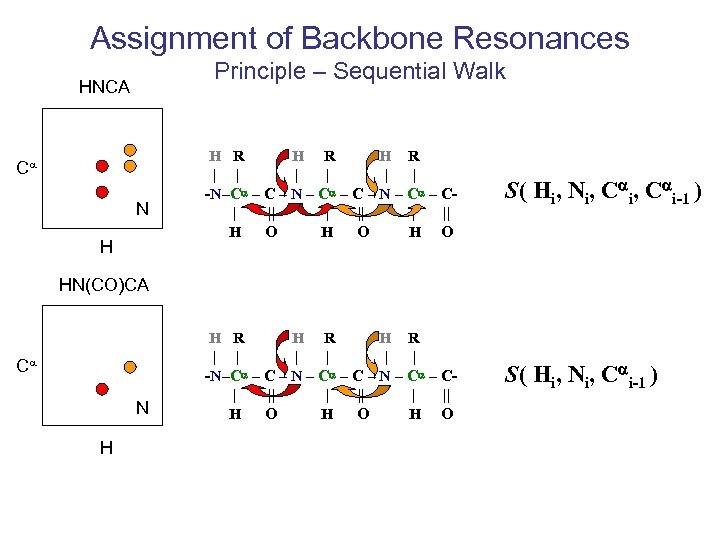
Assignment of Backbone Resonances Principle – Sequential Walk HNCA Ca N H H R H R | | | -N–Ca – C – N – Ca – C| || | || H O H O S( Hi, Ni, Cai-1 ) HN(CO)CA Ca N H
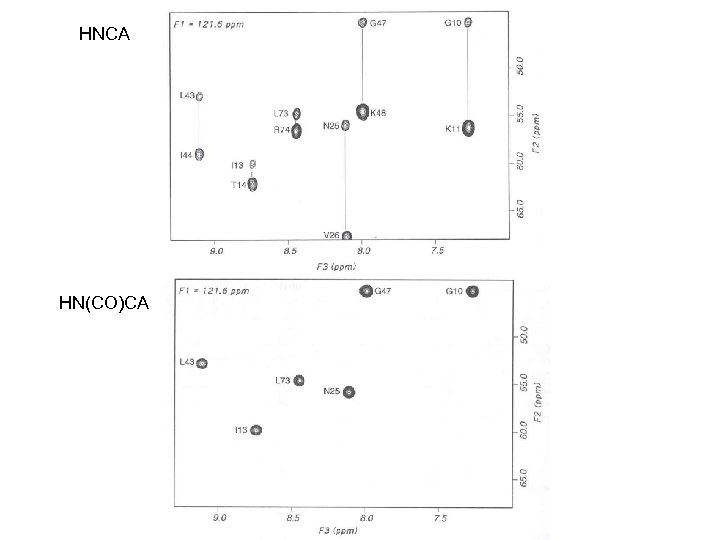
HNCA HN(CO)CA
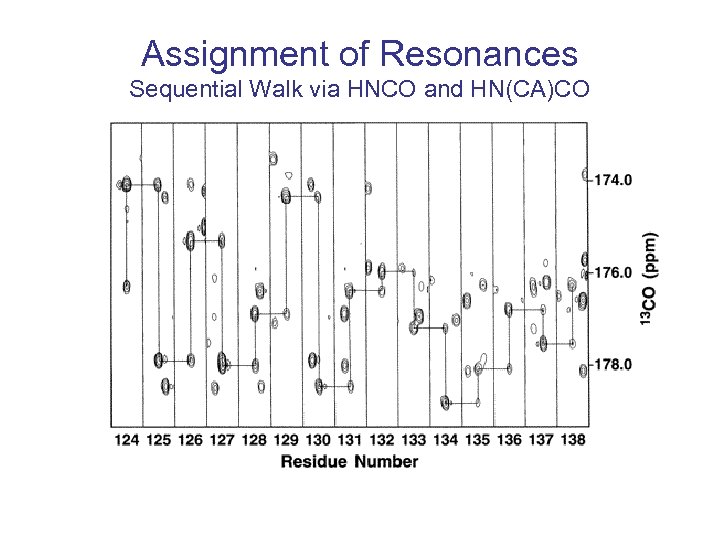
Assignment of Resonances Sequential Walk via HNCO and HN(CA)CO

The redundancy in many alternatives for sequential assignment is important for automated assignment.
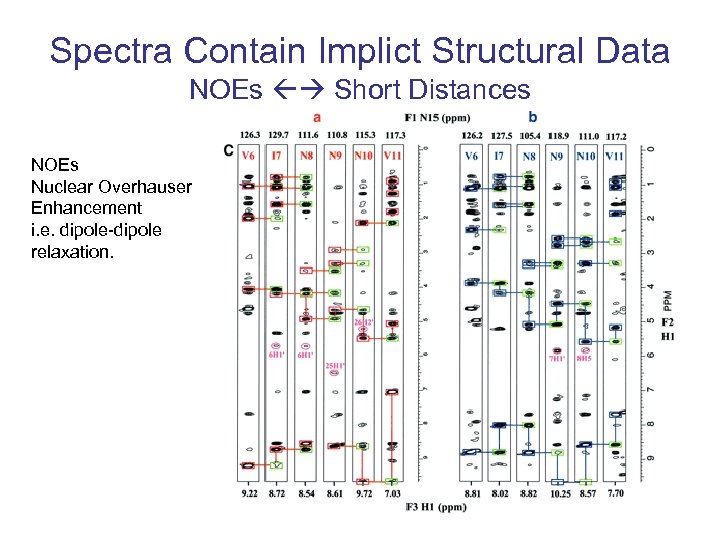
Spectra Contain Implict Structural Data NOEs Short Distances NOEs Nuclear Overhauser Enhancement i. e. dipole-dipole relaxation.
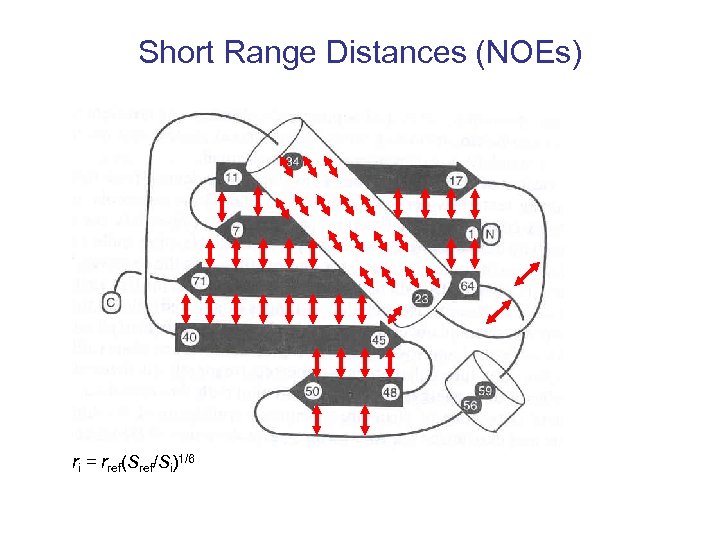
Short Range Distances (NOEs) ri = rref(Sref/Si)1/6
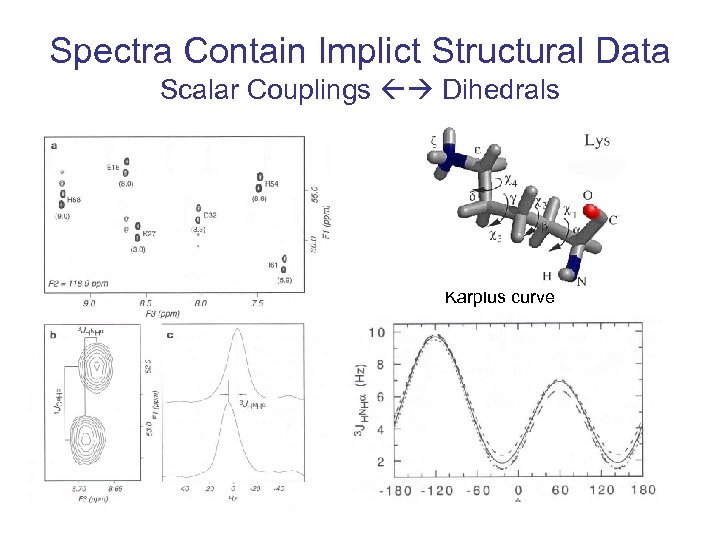
Spectra Contain Implict Structural Data Scalar Couplings Dihedrals Karplus curve
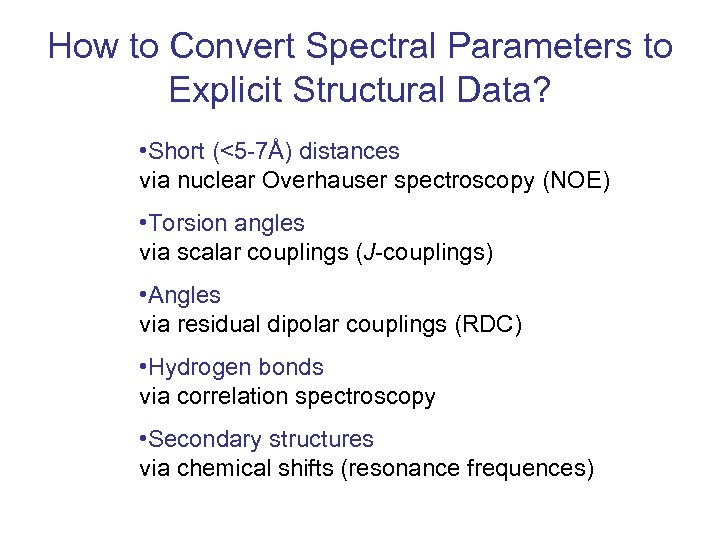
How to Convert Spectral Parameters to Explicit Structural Data? • Short (<5 -7Å) distances via nuclear Overhauser spectroscopy (NOE) • Torsion angles via scalar couplings (J-couplings) • Angles via residual dipolar couplings (RDC) • Hydrogen bonds via correlation spectroscopy • Secondary structures via chemical shifts (resonance frequences)
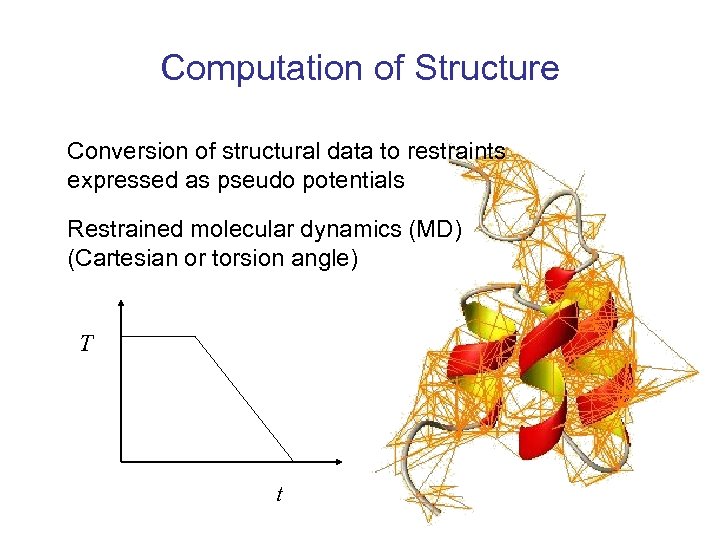
Computation of Structure Conversion of structural data to restraints expressed as pseudo potentials Restrained molecular dynamics (MD) (Cartesian or torsion angle) T t
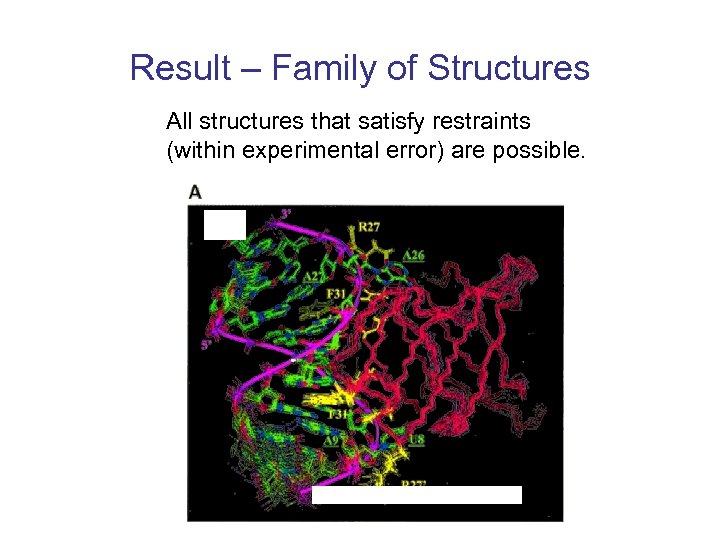
Result – Family of Structures All structures that satisfy restraints (within experimental error) are possible.
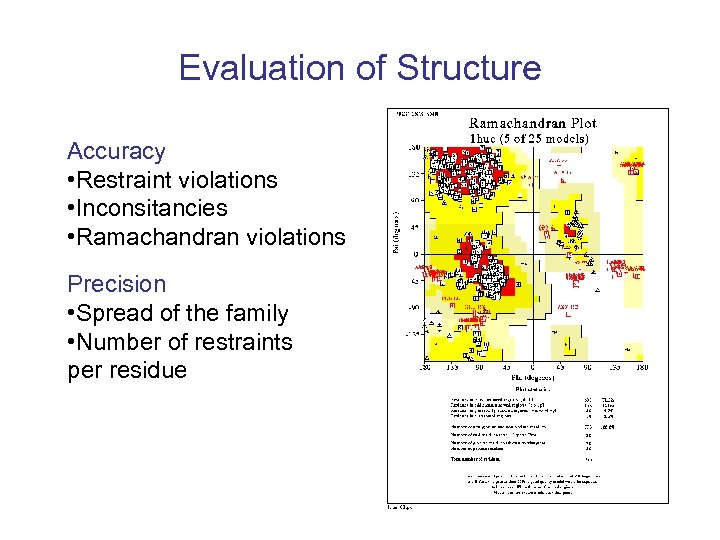
Evaluation of Structure Accuracy • Restraint violations • Inconsitancies • Ramachandran violations Precision • Spread of the family • Number of restraints per residue
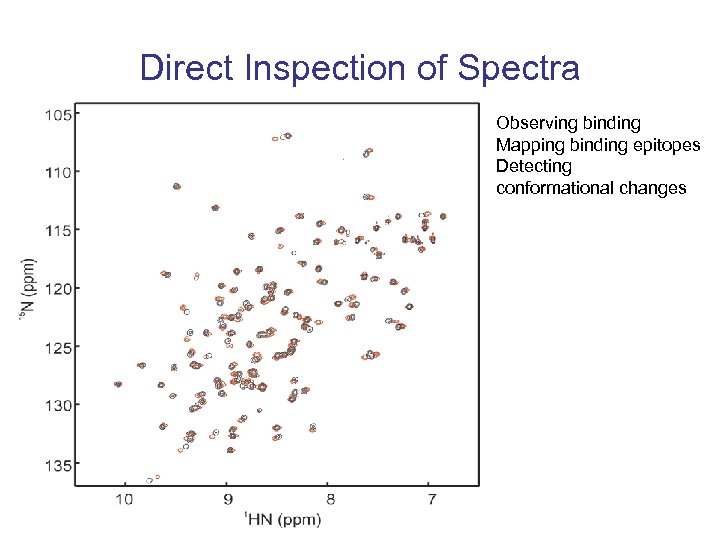
Direct Inspection of Spectra Observing binding Mapping binding epitopes Detecting conformational changes
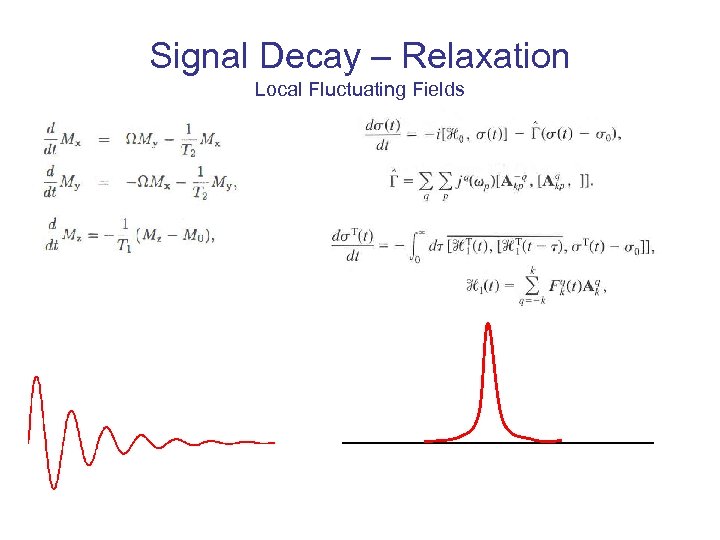
Signal Decay – Relaxation Local Fluctuating Fields
About Field Fluctuations ”Reasons” • Bond vibrations from pico to nano seconds ”Spectral Manifestations” • Relaxation measurements -> rate constants, order parameters, correlation times • Conformational changes from micro to milli seconds • Relaxation measurements -> dispersion of parameters • Chemical exchange from micro seconds to days • Line width analysis -> rate constants Motional model
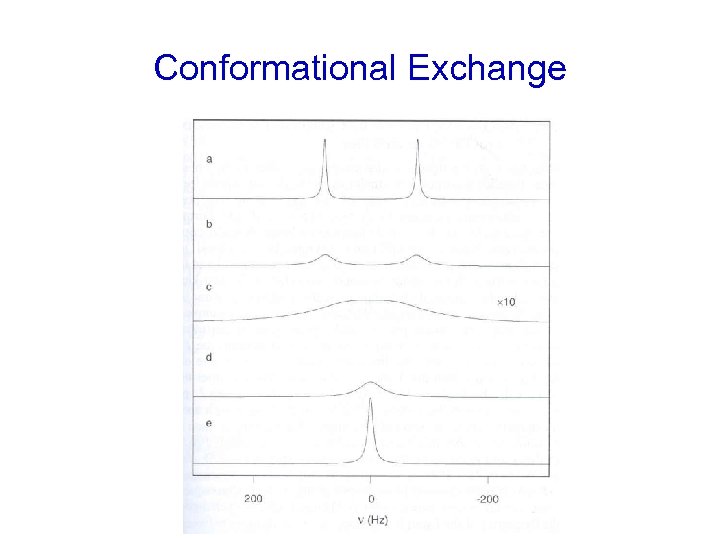
Conformational Exchange
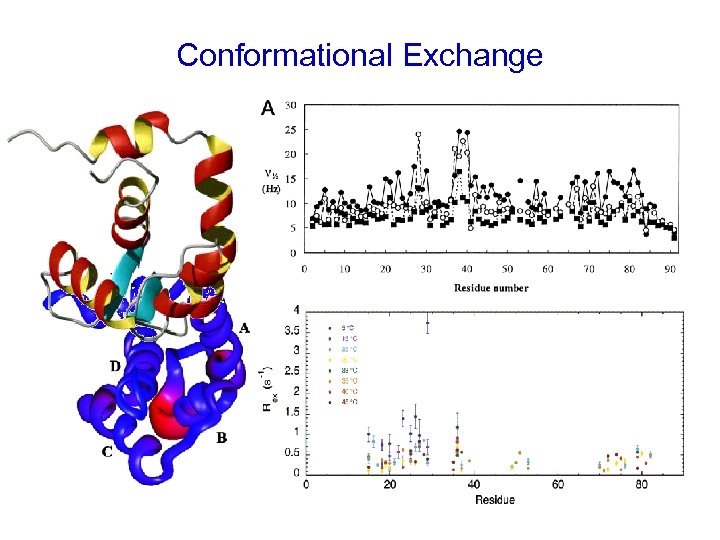
Conformational Exchange
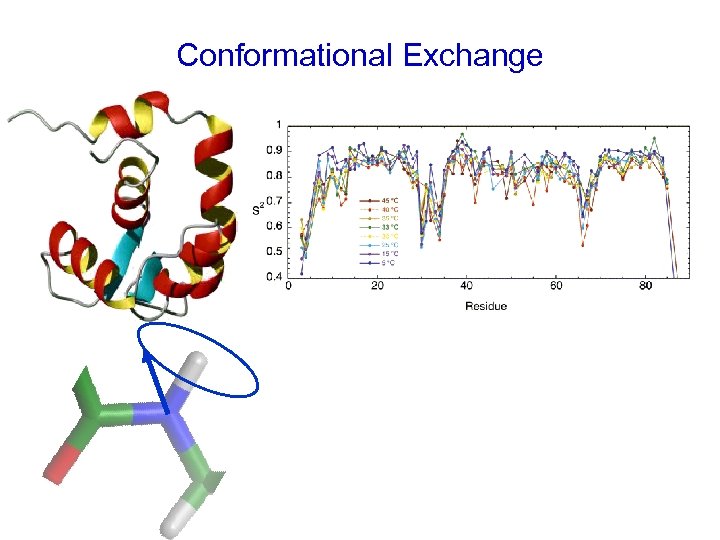
Conformational Exchange
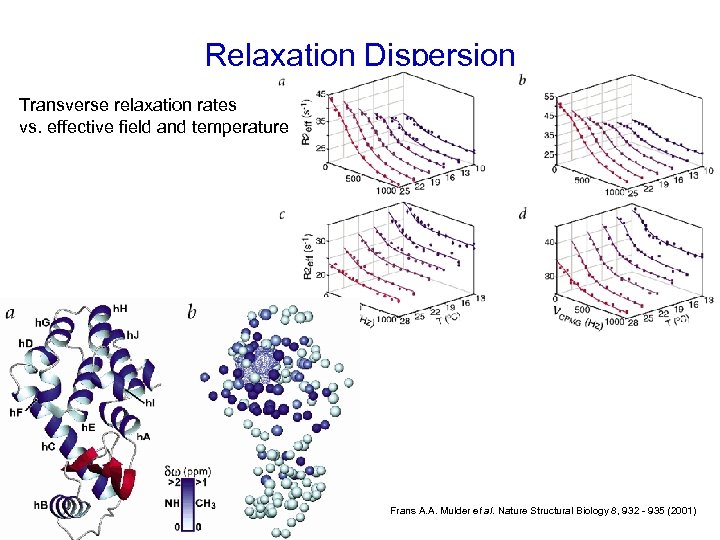
Relaxation Dispersion Transverse relaxation rates vs. effective field and temperature Frans A. A. Mulder et al. Nature Structural Biology 8, 932 - 935 (2001)
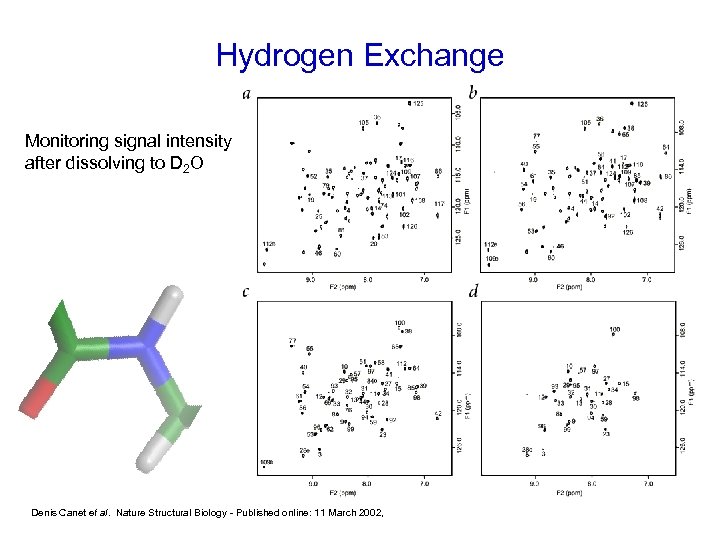
Hydrogen Exchange Monitoring signal intensity after dissolving to D 2 O Denis Canet et al. Nature Structural Biology - Published online: 11 March 2002,
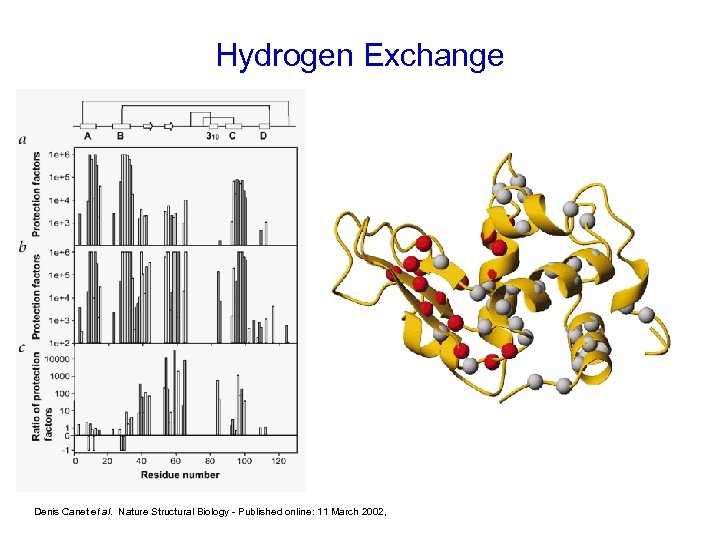
Hydrogen Exchange Denis Canet et al. Nature Structural Biology - Published online: 11 March 2002,
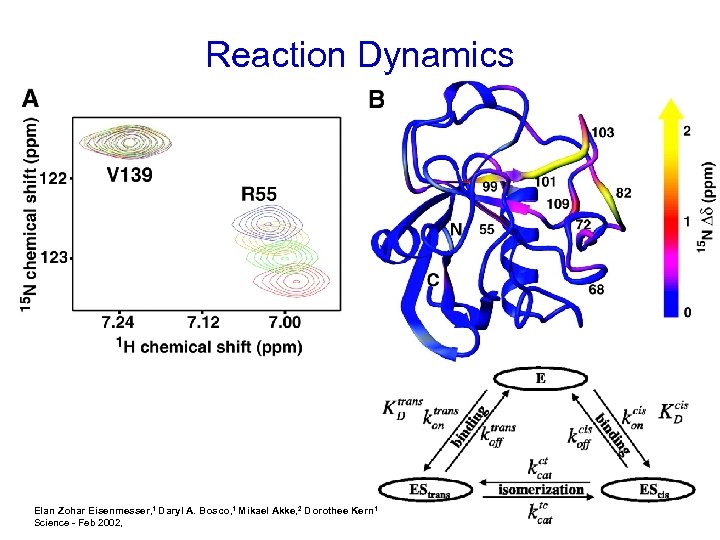
Reaction Dynamics Elan Zohar Eisenmesser, 1 Daryl A. Bosco, 1 Mikael Akke, 2 Dorothee Kern 1* Science - Feb 2002,
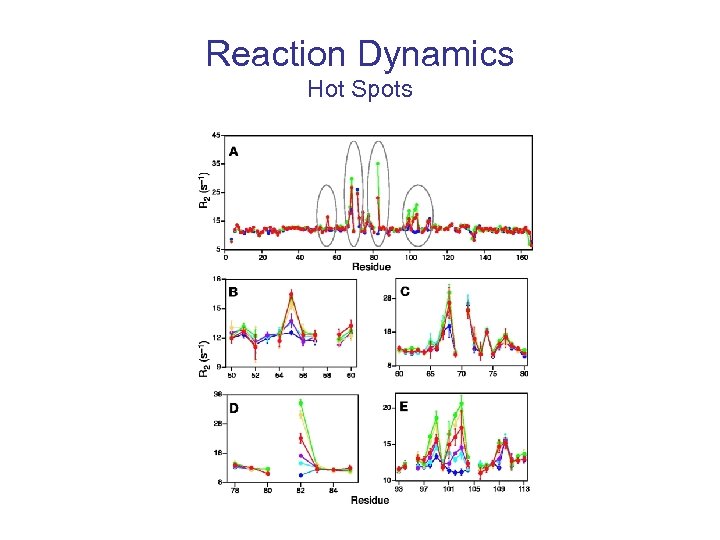
Reaction Dynamics Hot Spots
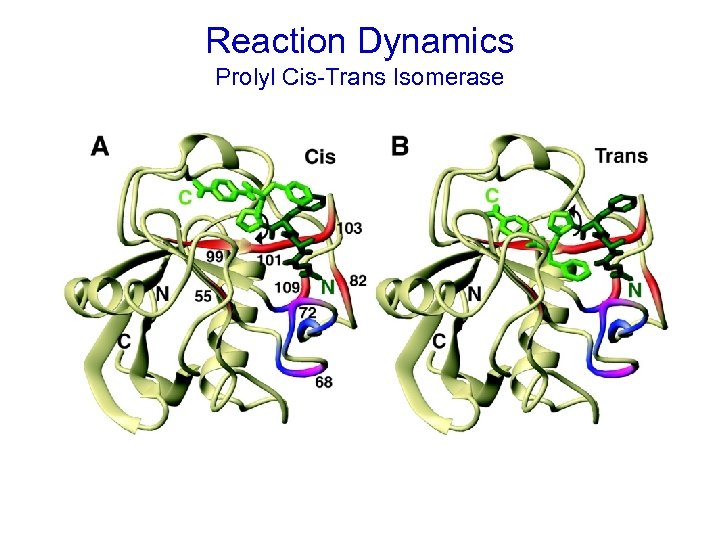
Reaction Dynamics Prolyl Cis-Trans Isomerase

Dilute Liquid Crystal Anisotropic Medium
Distribution of Molecular Orientations
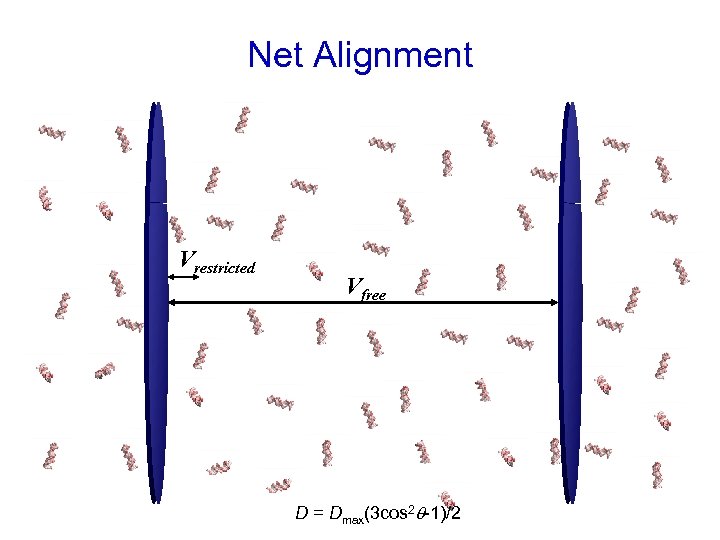
Net Alignment Vrestricted Vfree D = Dmax(3 cos 2 q-1)/2
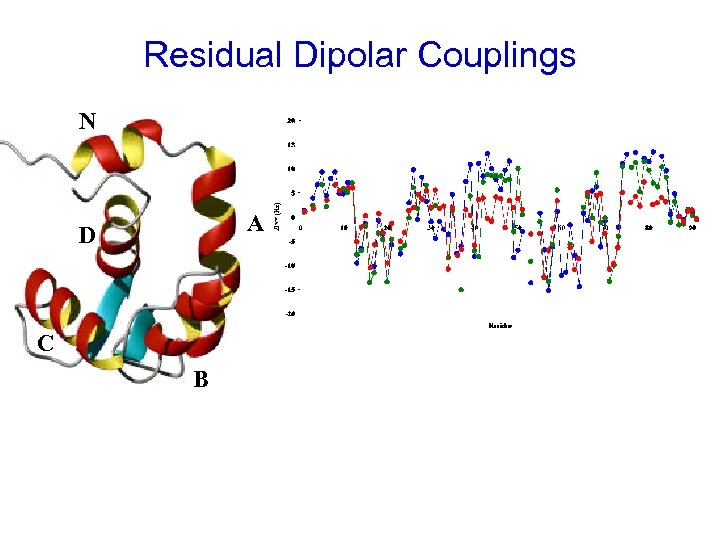
Residual Dipolar Couplings N A D C B
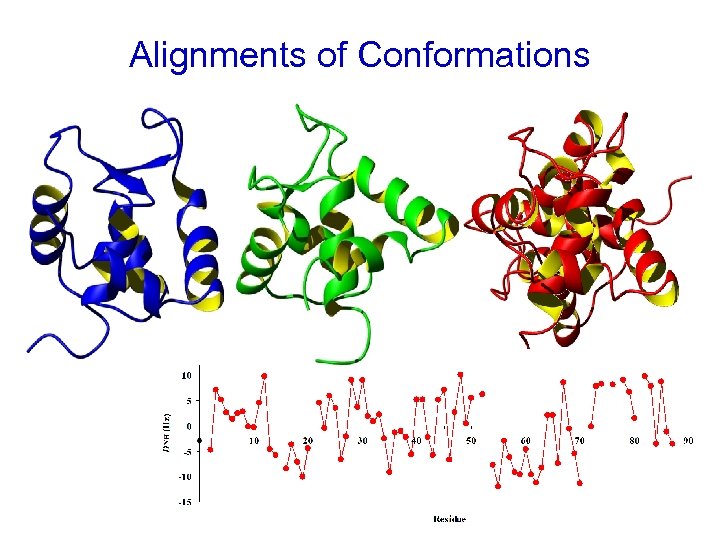
Alignments of Conformations
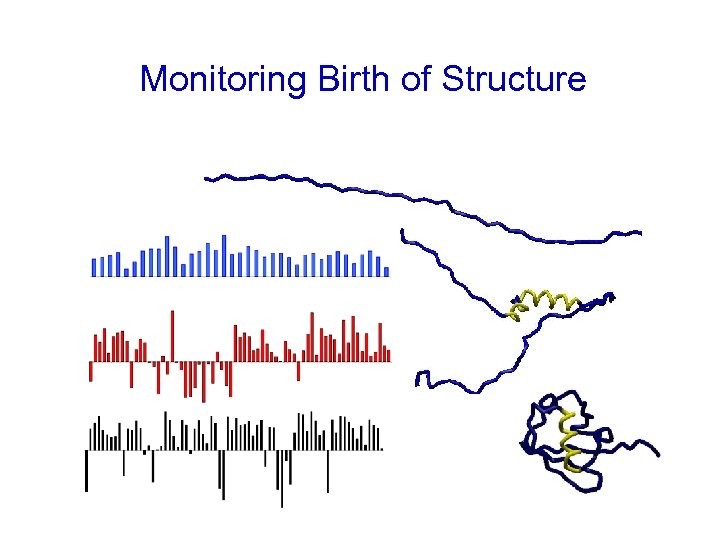
Monitoring Birth of Structure
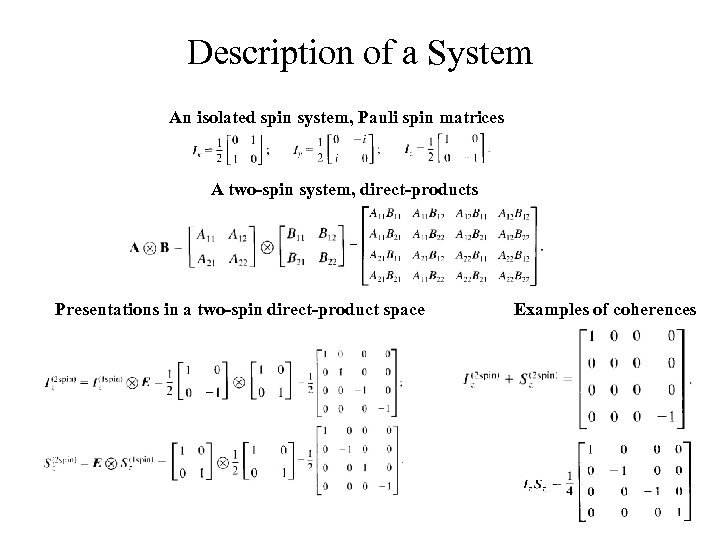
Description of a System An isolated spin system, Pauli spin matrices A two-spin system, direct-products Presentations in a two-spin direct-product space Examples of coherences
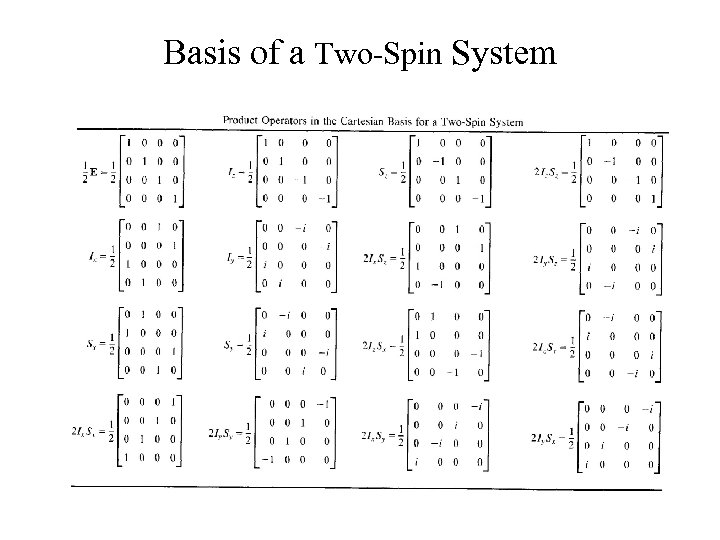
Basis of a Two-Spin System A two-spin system, direct-products
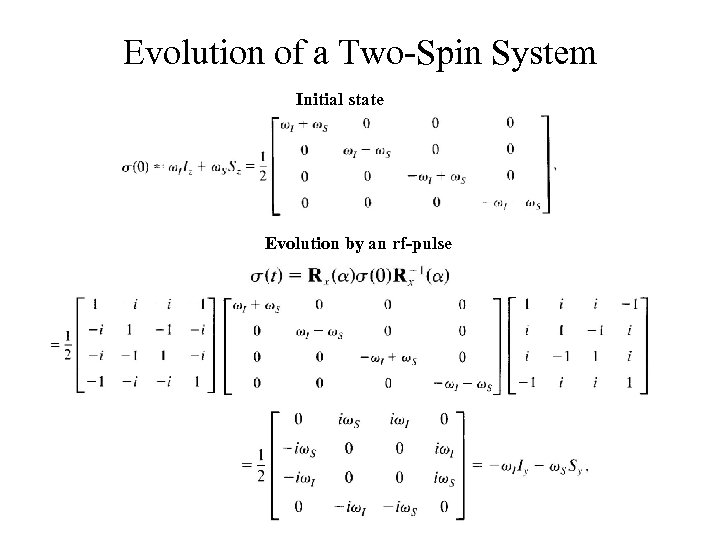
Evolution of a Two-Spin System Initial state Evolution by an rf-pulse
9dd582b501892c95909a420421b1b672.ppt