df034565863cda260cd5c2554dc80172.ppt
- Количество слайдов: 18

EMBL-EBI Dimitris Dimitropoulos Chemistry & the PDB MSDchem

EMBL-EBI The chemical database

EMBL-EBI MSDchem ligand dictionary q Complete, clean, up to date collection of all the chemical species and small molecules in the PDB q A ligand in MSDchem is a complete, distinct stereo isomer of a chemical compound ØAtoms and element types ØBonds and bond orders ØStereo configuration of atoms and bonds in cases of stereo-isomers (R/S – E/Z) q Atom names and coordinates are not fundamental properties
EMBL-EBI Role in the MSD database q An integral component in the core of MSD database q Relational reference from entities where a molecule or atom name is used in the PDB (protein residues and atoms) q. It is not possible for an ATOM line: HETATM 4342 C 2 PLA 86 14. 227 11. 195 -8. 256 1. 00 67. 95 C to be loaded if the “PLA” ligand is not defined or it does not include a “C 2” atom.
EMBL-EBI Chemistry and PDB q. Eliminate chemical inconsistencies from new PDB entries ØStructure and derived properties of a ligand apply automatically to residues and bound molecules that reference it Ø The basic structure is carefully determined during curation, and a rich set of derived attributes is calculated for each ligand Ø Graph isomorphism is being applied to check the consistency of the PDB, taking stereo-configuration into account q Old legacy PDB entries are chemically “corrected” when loaded in the MSD database Ø In thousands of cases errors are identified and corrected, involving most of them times inconsistent naming or different stereoconfiguration q. Exchanged in cooperation with RCSB and the ww. PDB
EMBL-EBI More than just the PDB codes q All ligands are modelled as separate inter-related ligands and the appropriate one is referenced ØNo distinction is made in the PDB between ribo- and deoxyribonucleotides (all are identified with the same residue name i. e. , A, C, G, T, U, I) ØModified nucleic acids are given as +A etc regardless of modification ØNo distinction between different topological variants (12 different variants can be found for HIS in PDB)
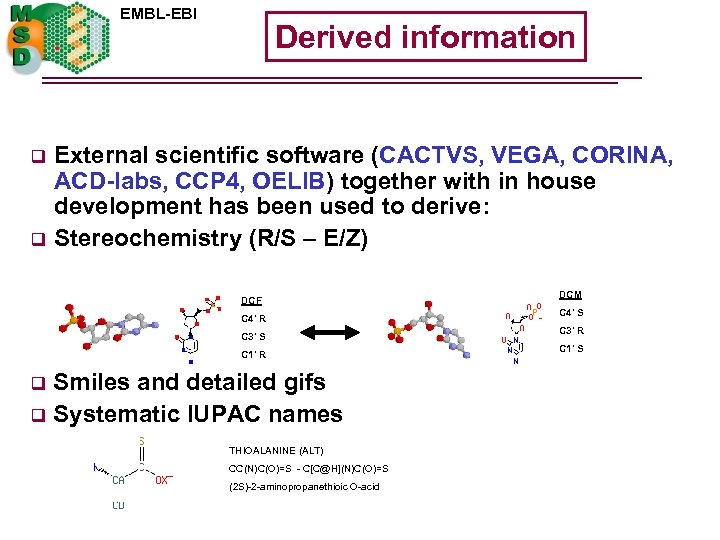
EMBL-EBI q q Derived information External scientific software (CACTVS, VEGA, CORINA, ACD-labs, CCP 4, OELIB) together with in house development has been used to derive: Stereochemistry (R/S – E/Z) DCF C 4' R C 3' S C 1' R Smiles and detailed gifs q Systematic IUPAC names q THIOALANINE (ALT) CC(N)C(O)=S - C[C@H](N)C(O)=S (2 S)-2 -aminopropanethioic O-acid DCM C 4' S C 3' R C 1' S
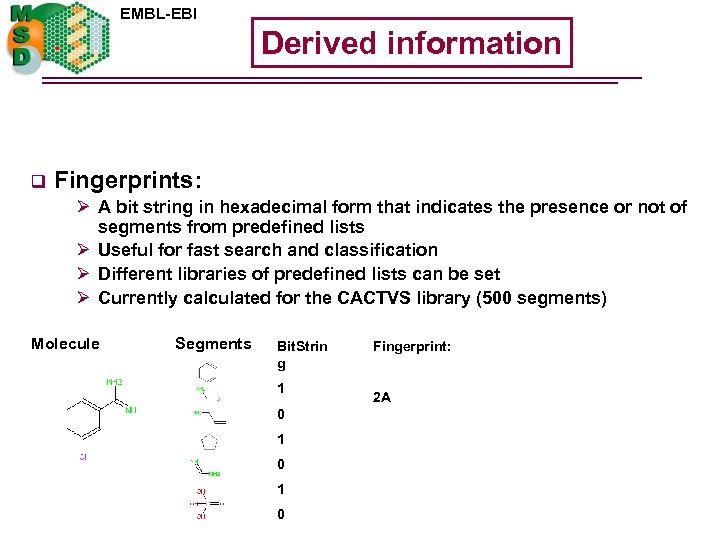
EMBL-EBI Derived information q Fingerprints: Ø A bit string in hexadecimal form that indicates the presence or not of segments from predefined lists Ø Useful for fast search and classification Ø Different libraries of predefined lists can be set Ø Currently calculated for the CACTVS library (500 segments) Molecule Segments Bit. Strin g 1 0 1 0 Fingerprint: 2 A

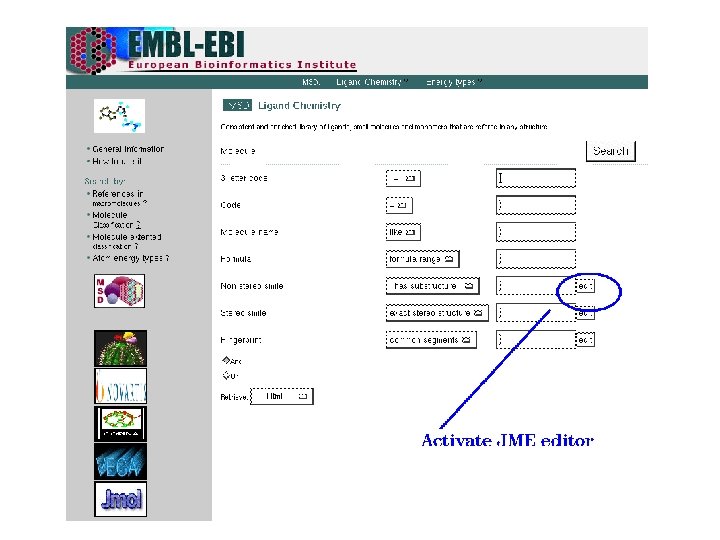
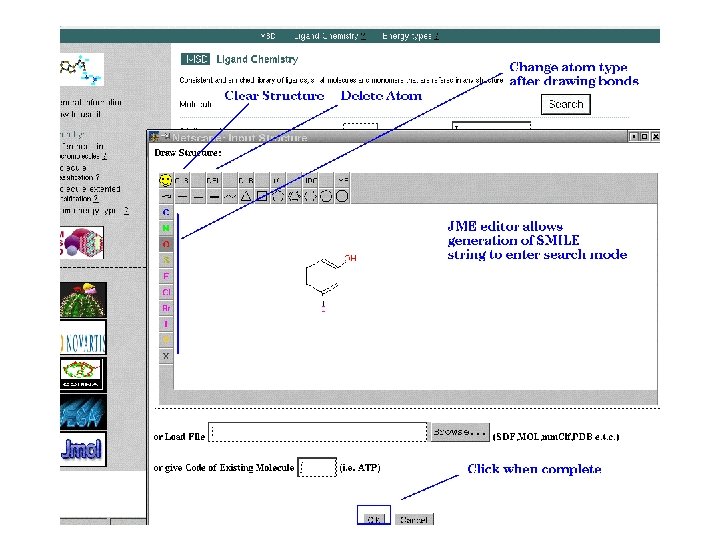
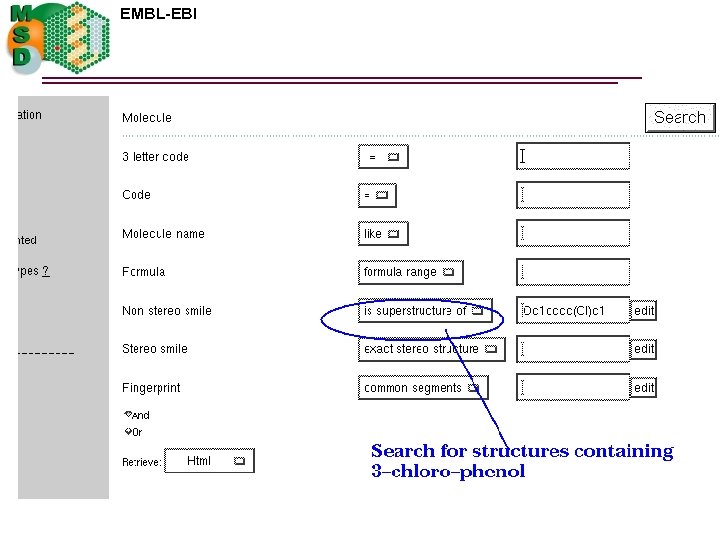
EMBL-EBI Search options q By ligand code q By ligand name or synonym q By formula or formula range q By non stereo substructure q By non stereo superstructure q By exact stereo or non stereo structure q By fingerprint similarity
EMBL-EBI
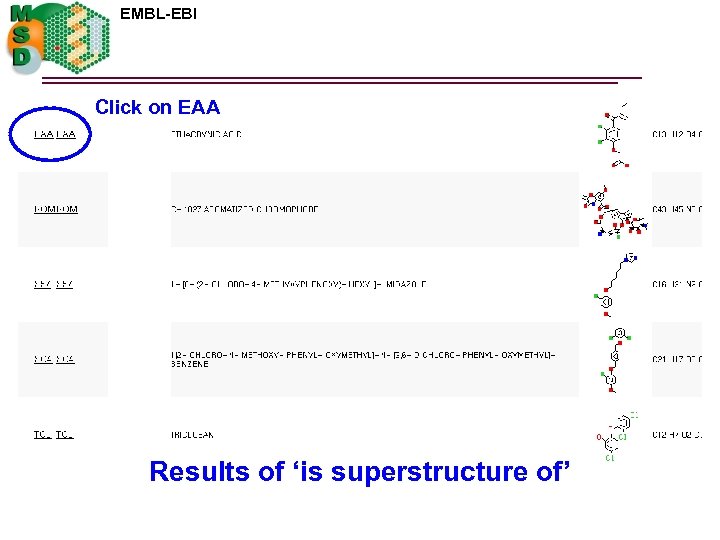
EMBL-EBI Click on EAA Results of ‘is superstructure of’

EAA details 3 -chloro-phenol
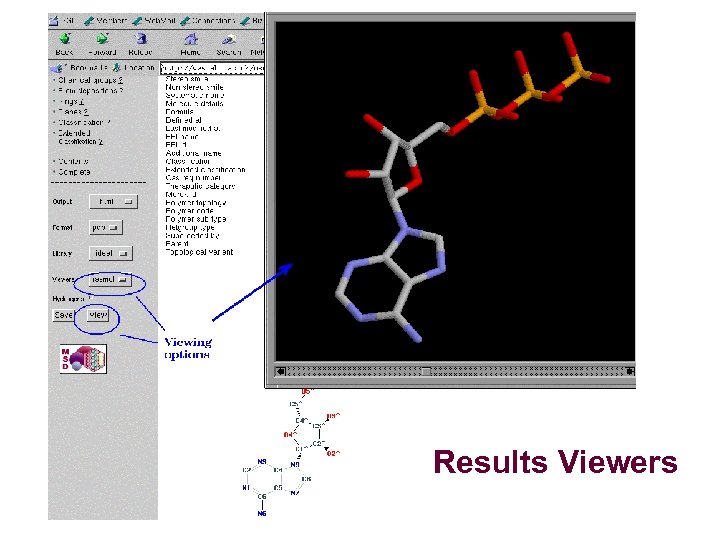
Results Viewers
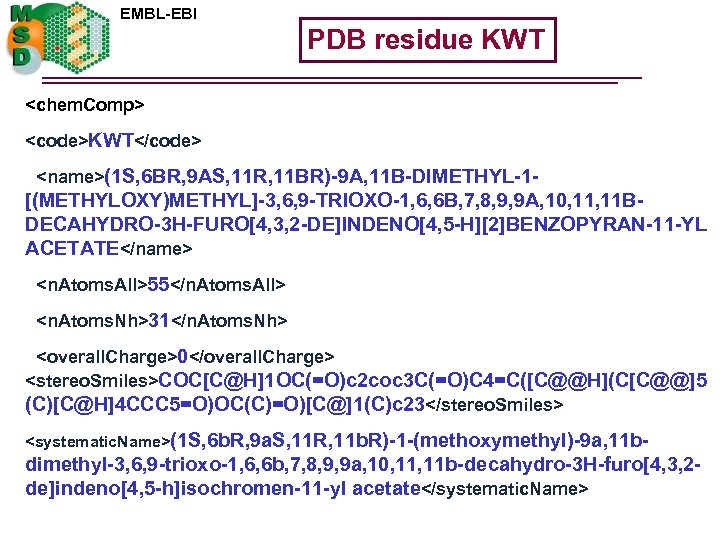
EMBL-EBI PDB residue KWT <chem. Comp> <code>KWT</code> <name>(1 S, 6 BR, 9 AS, 11 R, 11 BR)-9 A, 11 B-DIMETHYL-1 - [(METHYLOXY)METHYL]-3, 6, 9 -TRIOXO-1, 6, 6 B, 7, 8, 9, 9 A, 10, 11 BDECAHYDRO-3 H-FURO[4, 3, 2 -DE]INDENO[4, 5 -H][2]BENZOPYRAN-11 -YL ACETATE</name> <n. Atoms. All>55</n. Atoms. All> <n. Atoms. Nh>31</n. Atoms. Nh> <overall. Charge>0</overall. Charge> <stereo. Smiles>COC[C@H]1 OC(=O)c 2 coc 3 C(=O)C 4=C([C@@H](C[C@@]5 (C)[C@H]4 CCC 5=O)OC(C)=O)[C@]1(C)c 23</stereo. Smiles> <systematic. Name>(1 S, 6 b. R, 9 a. S, 11 R, 11 b. R)-1 -(methoxymethyl)-9 a, 11 b- dimethyl-3, 6, 9 -trioxo-1, 6, 6 b, 7, 8, 9, 9 a, 10, 11 b-decahydro-3 H-furo[4, 3, 2 de]indeno[4, 5 -h]isochromen-11 -yl acetate</systematic. Name>
EMBL-EBI Future targets q Identify and model protein inhibitors as ligands q Pre-classify functional groups for ligands and ligand atoms based on substructure fragments. q Optimise and boost the performance of substructure searches q Enhance visualisation and integration with other MSD tools
df034565863cda260cd5c2554dc80172.ppt