eda7cefe231f09525b09f99a5d5bdb4f.ppt
- Количество слайдов: 38

ELIXIR-Pilot Project “BILS-Proteome. Xchange integration using EUDAT resources” Dr. Juan A. Vizcaíno, EMBL-EBI, juan@ebi. ac. uk Dr. Fredrik Levander, BILS, fredrik. levander@bils. se European Life Sciences Infrastructure for Biological Information www. elixir-europe. org

Main people involved directly in this pilot • Andy Jenkinson (Systems group) • Rui Wang (PRIDE) • Juan A. Vizcaíno (PRIDE) • • Fredrik Levander Samuel Lampa Janos Nagy Mikael Borg • Jani Heikkinen Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015

Overview • Short intro to PRIDE & Proteome. Xchange, BILS and EUDAT • Objectives of the pilot • Report on the results • Perspectives for the future and conclusions Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015

Overview • Short intro to PRIDE & Proteome. Xchange, BILS and EUDAT • Objectives of the pilot • Report on the results • Perspectives for the future and conclusions Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015
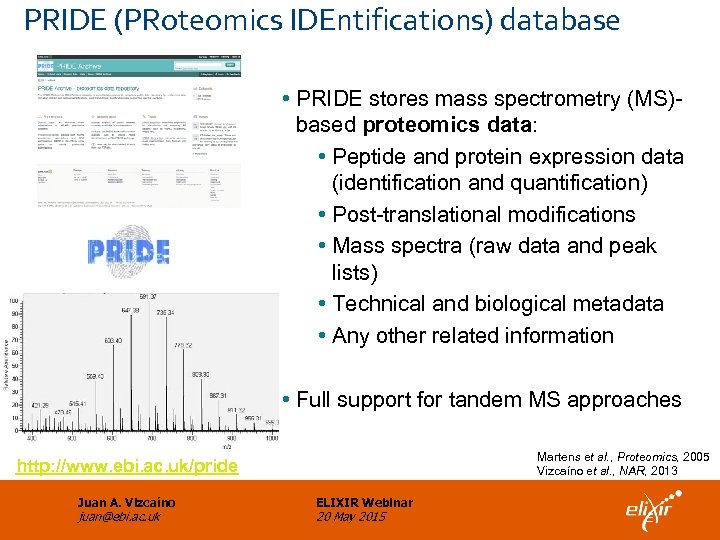
PRIDE (PRoteomics IDEntifications) database • PRIDE stores mass spectrometry (MS)based proteomics data: • Peptide and protein expression data (identification and quantification) • Post-translational modifications • Mass spectra (raw data and peak lists) • Technical and biological metadata • Any other related information • Full support for tandem MS approaches Martens et al. , Proteomics, 2005 Vizcaíno et al. , NAR, 2013 http: //www. ebi. ac. uk/pride Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015
Proteome. Xchange Consortium • Goal: Development of a framework to allow standard data submission and dissemination pipelines between the main existing proteomics repositories. • Includes Peptide. Atlas (ISB, Seattle), PRIDE (Cambridge, UK) and Mass. IVE (UCSD, San Diego). • Tranche and Peptidome initially included but discontinued. • Common identifier space (PXD identifiers) • Two supported data workflows: MS/MS and SRM. • Main objective: Make life easier for researchers Vizcaíno et al. , Nat Biotechnol, 2014 http: //www. proteomexchange. org Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015
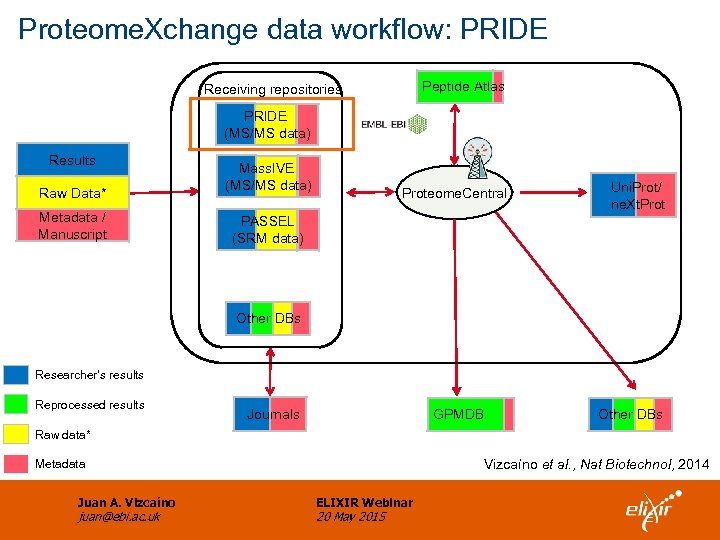
Proteome. Xchange data workflow: PRIDE Peptide Atlas Receiving repositories PRIDE (MS/MS data) Results Raw Data* Metadata / Manuscript Mass. IVE (MS/MS data) Proteome. Central Uni. Prot/ ne. Xt. Prot PASSEL (SRM data) Other DBs Researcher’s results Reprocessed results Journals GPMDB Other DBs Raw data* Vizcaíno et al. , Nat Biotechnol, 2014 Metadata Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015
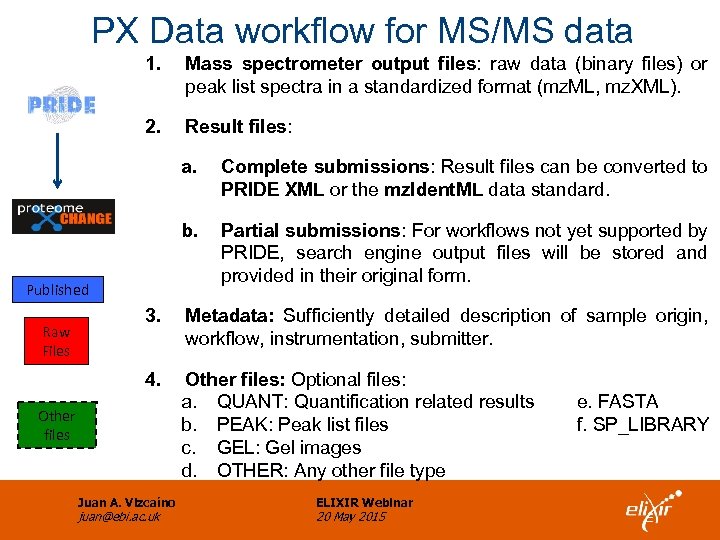
PX Data workflow for MS/MS data 1. Mass spectrometer output files: raw data (binary files) or peak list spectra in a standardized format (mz. ML, mz. XML). 2. Result files: a. b. Published Complete submissions: Result files can be converted to PRIDE XML or the mz. Ident. ML data standard. Partial submissions: For workflows not yet supported by PRIDE, search engine output files will be stored and provided in their original form. Metadata: Sufficiently detailed description of sample origin, workflow, instrumentation, submitter. 4. Raw Files 3. Other files: Optional files: a. QUANT: Quantification related results b. PEAK: Peak list files c. GEL: Gel images d. OTHER: Any other file type Other files Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015 e. FASTA f. SP_LIBRARY
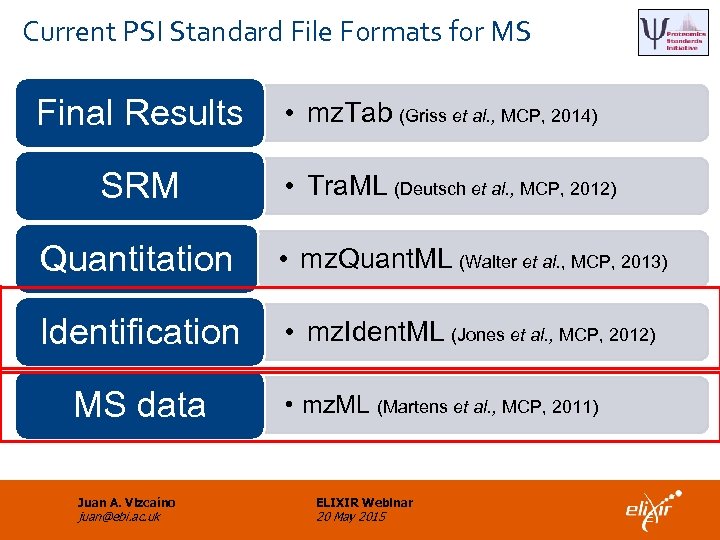
Current PSI Standard File Formats for MS Final Results SRM • mz. Tab (Griss et al. , MCP, 2014) • Tra. ML (Deutsch et al. , MCP, 2012) Quantitation • mz. Quant. ML (Walter et al. , MCP, 2013) Identification • mz. Ident. ML (Jones et al. , MCP, 2012) MS data Juan A. Vizcaíno juan@ebi. ac. uk • mz. ML (Martens et al. , MCP, 2011) ELIXIR Webinar 20 May 2015
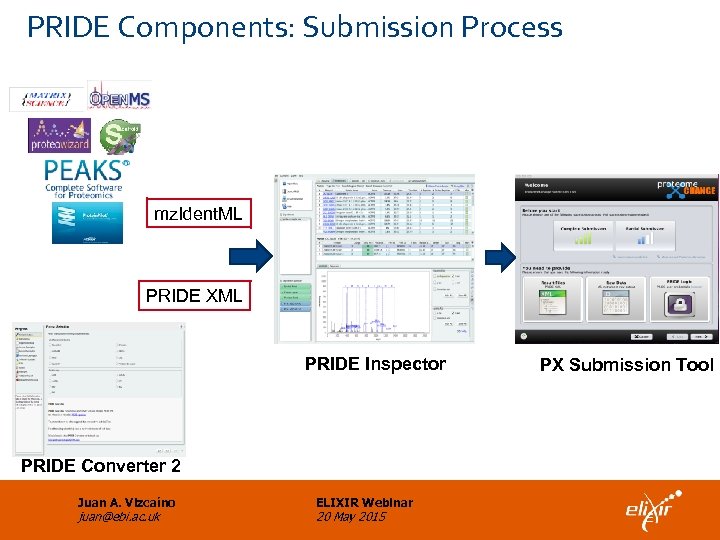
PRIDE Components: Submission Process mz. Ident. ML PRIDE XML PRIDE Inspector PRIDE Converter 2 Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015 PX Submission Tool
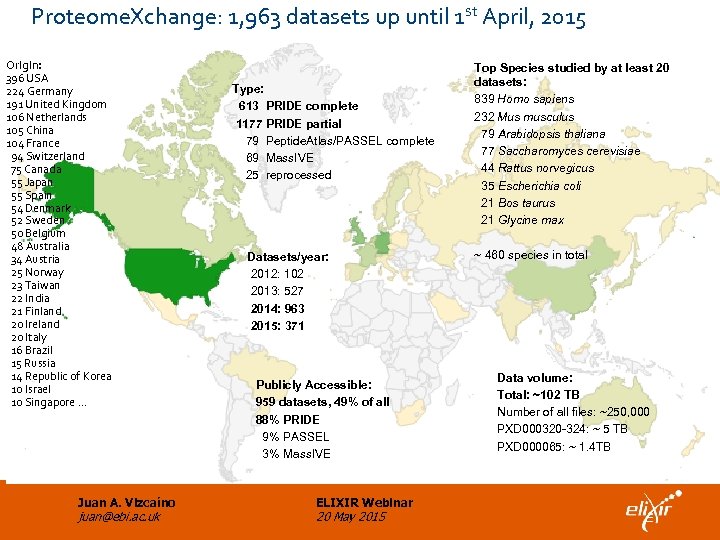
Proteome. Xchange: 1, 963 datasets up until 1 st April, 2015 Origin: 396 USA 224 Germany 191 United Kingdom 106 Netherlands 105 China 104 France 94 Switzerland 75 Canada 55 Japan 55 Spain 54 Denmark 52 Sweden 50 Belgium 48 Australia 34 Austria 25 Norway 23 Taiwan 22 India 21 Finland 20 Ireland 20 Italy 16 Brazil 15 Russia 14 Republic of Korea 10 Israel 10 Singapore … Juan A. Vizcaíno juan@ebi. ac. uk Type: 613 PRIDE complete 1177 PRIDE partial 79 Peptide. Atlas/PASSEL complete 69 Mass. IVE 25 reprocessed Datasets/year: 2012: 102 2013: 527 2014: 963 2015: 371 Publicly Accessible: 959 datasets, 49% of all 88% PRIDE 9% PASSEL 3% Mass. IVE ELIXIR Webinar 20 May 2015 Top Species studied by at least 20 datasets: 839 Homo sapiens 232 Mus musculus 79 Arabidopsis thaliana 77 Saccharomyces cerevisiae 44 Rattus norvegicus 35 Escherichia coli 21 Bos taurus 21 Glycine max ~ 460 species in total Data volume: Total: ~102 TB Number of all files: ~250, 000 PXD 000320 -324: ~ 5 TB PXD 000065: ~ 1. 4 TB
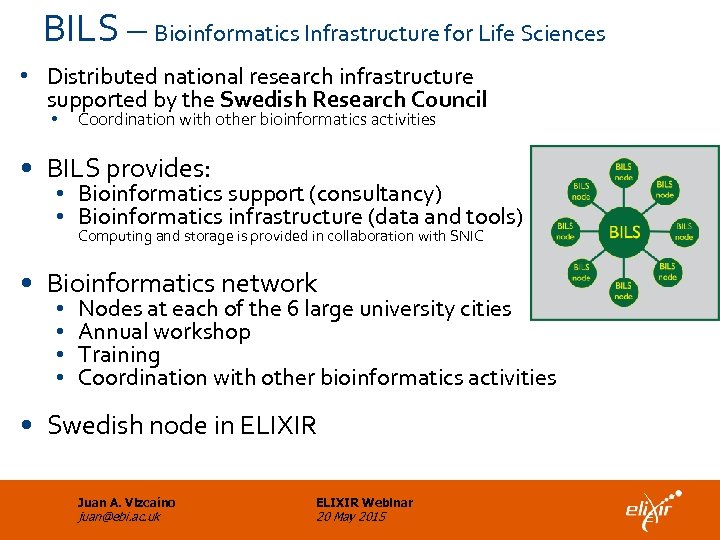
BILS – Bioinformatics Infrastructure for Life Sciences • Distributed national research infrastructure supported by the Swedish Research Council • Coordination with other bioinformatics activities • BILS provides: • Bioinformatics support (consultancy) • Bioinformatics infrastructure (data and tools) Computing and storage is provided in collaboration with SNIC • Bioinformatics network • • Nodes at each of the 6 large university cities Annual workshop Training Coordination with other bioinformatics activities • Swedish node in ELIXIR Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015
Main BILS proteomics support aims • Data storage: • • • Secure Long-time Metadata Automated Publishing Standardised formats • Data processing: • Accessible data processing workflows Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015
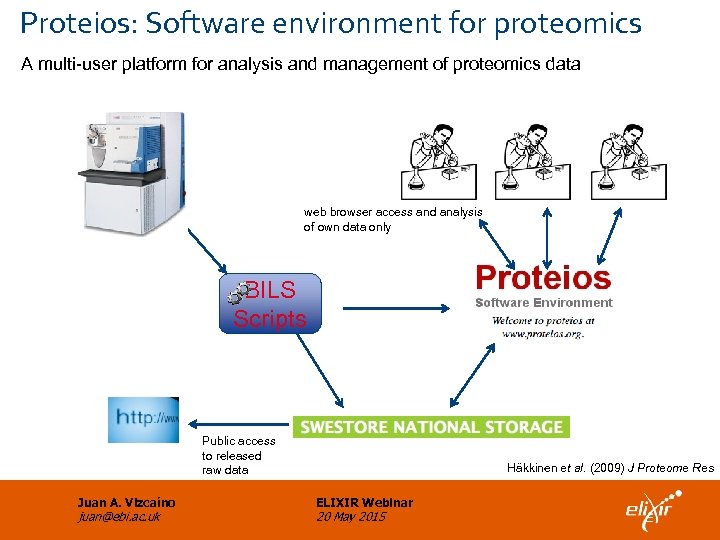
Proteios: Software environment for proteomics A multi-user platform for analysis and management of proteomics data web browser access and analysis of own data only BILS Scripts Public access to released raw data Juan A. Vizcaíno juan@ebi. ac. uk Häkkinen et al. (2009) J Proteome Res ELIXIR Webinar 20 May 2015

EUDAT • EUDAT aims to contribute to building and operating a Collaborative Data Infrastructure for European science. • This involves a suite of co-ordinated and interoperable services for preserving scientific data, and for making them accessible to researchers. • EUDAT collaborates with research communities across a range of disciplines, from social sciences to environmental science and including molecular biology (as represented by ELIXIR). • These communities have diverse structures, cultures and scales but also share some common requirements regarding the management of data. http: //www. eudat. eu Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015
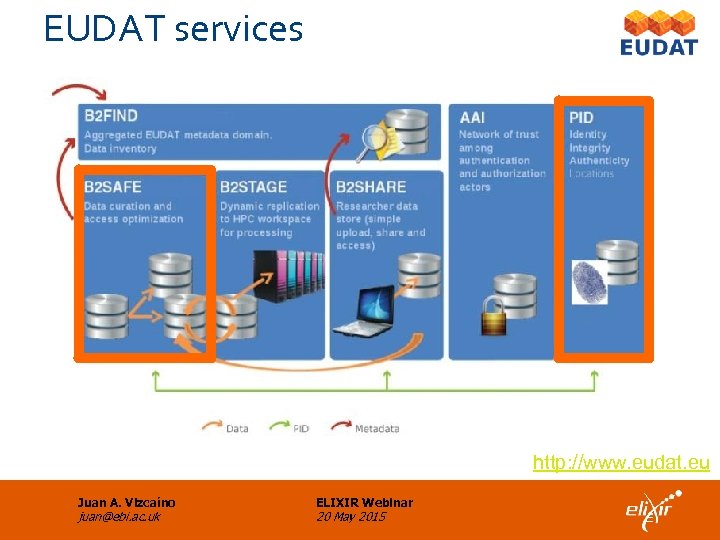
EUDAT services http: //www. eudat. eu Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015

B 2 SAFE Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015
EUDAT: B 2 SAFE AND i. RODS • B 2 SAFE aims to provide a software ecosystem for persistently available data, including persistent identification, abstracted data storage, and reliable automated replication via auditable rules. • It is built on top of the i. RODS data management software (http: //irods. org) and integrates a PID system such as the European Persistent Identification Consortium (EPIC - (http: //www. pidconsortium. eu) Handle API). Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015

Overview • PRIDE, Proteome. Xchange, BILS and EUDAT • Objectives of the pilot • Report on the results • Perspectives for the future and conclusions Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015
Objective • To integrate the data repositories for MS proteomics data run by BILS (Sweden) and Proteome. Xchange (via the PRIDE database, EMBL-EBI, UK), using EUDAT’s B 2 SAFE software. Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015
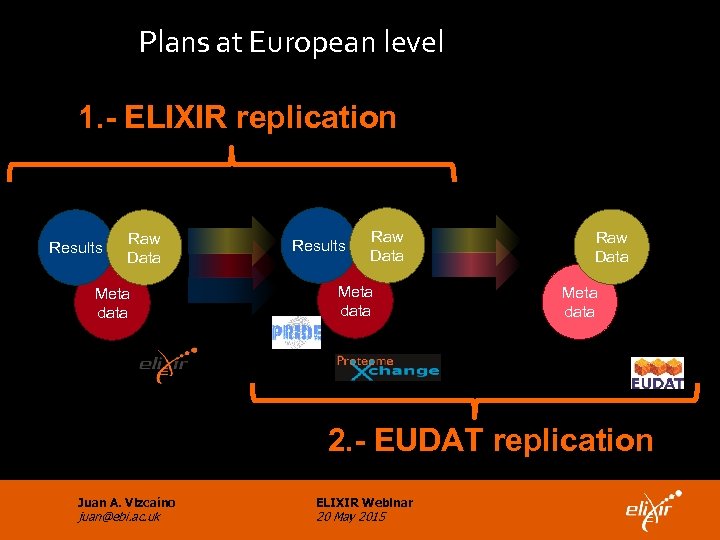
Plans at European level 1. - ELIXIR replication National proteomics centers Results Raw Data Meta data Central repository Results Raw Data Meta data Data storage centers Raw Data Meta data 2. - EUDAT replication Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015
Objective • To integrate the data repositories for MS proteomics data run by BILS (Sweden) and Proteome. Xchange (via the PRIDE database, EMBL-EBI, UK), using EUDAT’s B 2 SAFE software. • This project will also show the potential of collaboration among research infrastructures and e-infrastructures to better manage the data deluge. It will help to evaluate the requirements of such federated systems. Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015

Overview • Short intro to PRIDE & Proteome. Xchange, BILS and EUDAT • Objectives of the pilot • Report on the results • Perspectives for the future and conclusions Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015

Timeline • The pilot started when Jani Heikkinen (EUDAT) installed B 2 SAFE at EMBL-EBI (July 2014). • Data workflow was defined on September/ October 2014. • Implementation work happened in parallel, with regular weekly calls from January 2015. • The pilot is now finishing (May 2015). Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015
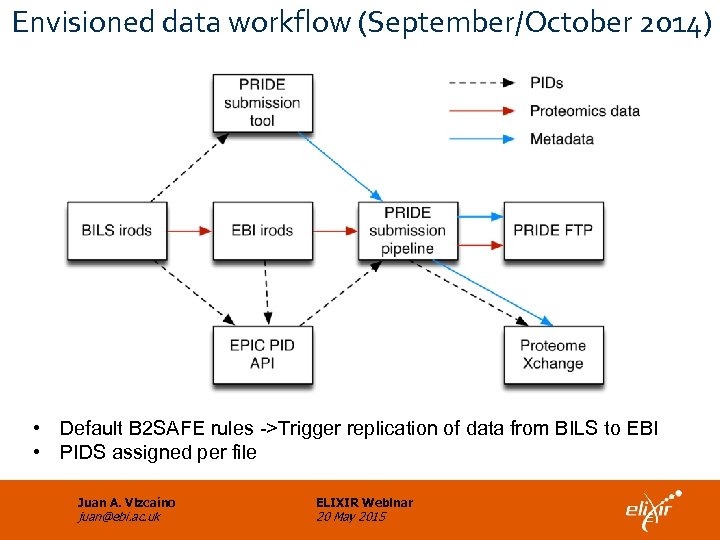
Envisioned data workflow (September/October 2014) • Default B 2 SAFE rules ->Trigger replication of data from BILS to EBI • PIDS assigned per file Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015
Implementation process (1) • B 2 SAFE 3. 0. 0 (including i. RODS 3. 3. 1) was initially installed at EMBL-EBI. • However, BILS had moved already to i. RODS v 4. • Incompatibility problems were found. • It was decided to install i. RODS 4. 0 at the EBI, to solve the incompatibility issue. • At the time i. RODS v 4 was not officially supported with i. RODS version 4. 0. 3, so changes were necessary to the original install procedure to accommodate 4. 0. 3. Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015
Implementation process (2) • EBI and BILS obtained Handle prefixes and made them available within EPIC. The integration with i. RODS was successfully tested. • The next step was to configure B 2 SAFE and achieve a test replication of a file from BILS to EBI using the B 2 SAFE PID creation and file transfer rules. • Unexpected delays: • EBI experienced some network issues that affected communications between the EBI and BILS i. RODS. • Two successive bugs were discovered. Both centered on the rule execution engine and prevented B 2 SAFE from functioning. • These bugs were solved by EUDAT & i. RODs developers. Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015
Implementation process (3) • With workarounds now in place it was possible to manually trigger a successful replication of a file from BILS to EBI. • However it became apparent that the authorisation mechanism employed by i. RODS in a federation would make the proposed submission workflow difficult to manage in a production environment. • This means every BILS researcher able to submit data must have a user created for them on the EBI server first. Alternative customised solutions could solve this issue by decoupling the actions of researchers from the replication itself. However this would inevitably add complexity. Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015
Implementation process (4) • At this point (March 2015) the pilot had overrun (it was expected to last 6 months), with more work required to integrate the B 2 SAFE replication process with the PRIDE submission pipeline. • It was decided to halt the process and find an alternative way to achieve the same goals using existing resources. • A detailed report has been written and has been sent to all the parties involved. Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015

Implemented alternative solution • Proteios is able to generate the metadata file needed for the submission to Proteome. Xchange via PRIDE. • The PX submission tool was extended to support loading of files not available locally at the moment of submission (URLs are specified). • As a proof of concept, dataset PXD 002037 was submitted to PRIDE. Now it is publicly available. Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015
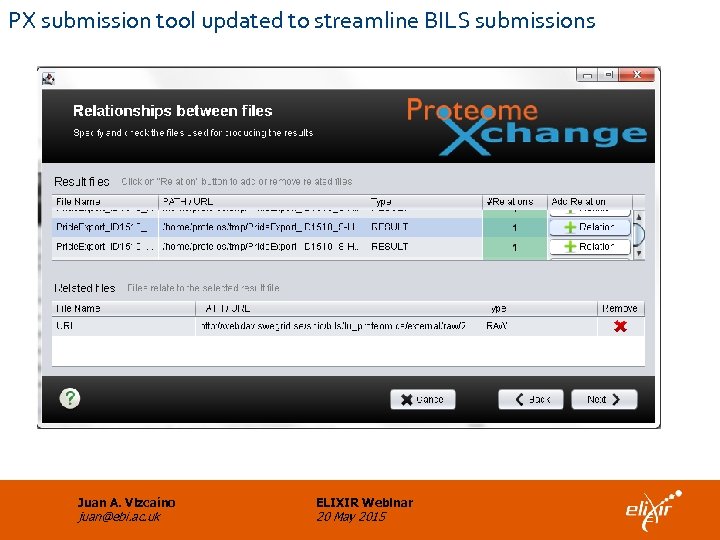
PX submission tool updated to streamline BILS submissions Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015
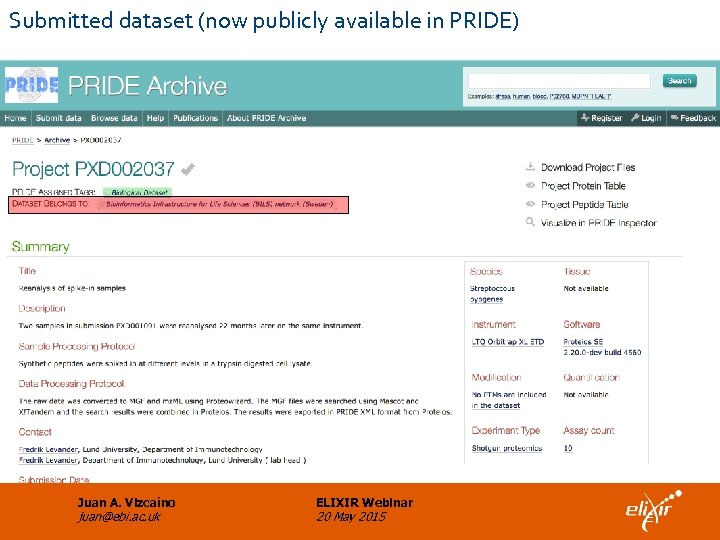
Submitted dataset (now publicly available in PRIDE) Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015

Dataset tags in PRIDE Archive - Datasets can be tags with different attributes. - Functionality available in the submission process. - Stable URLs can be generated. http: //www. ebi. ac. uk/pride/archive/simple. Search? q=&project. Tag. Filters=Bioinformatics%20 Infrastructure%20 for%20 Life%20 Scienc es%20(BILS)%20 network%20(Sweden) Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015

Overview • Short intro to PRIDE & Proteome. Xchange, BILS and EUDAT • Objectives of the pilot • Report on the results • Perspectives for the future and conclusions Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015
At present and in the near future… • EMBL-EBI is involved in the EUDAT 2020 project (PI is Steven Newhouse). • EMBL-EBI will then continue to collaborate with EUDAT, for gaining experience in the use of this software. • PRIDE will evaluate the situation in the future to decide if the originally envisioned submission pipeline (based on B 2 SAFE and IRODS) is implemented. Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015
Conclusions • The pilot establishes that the original use case is not the best application of B 2 SAFE at the present time. However, the situation will be kept under review by PRIDE. • This conclusion is not a reflection on B 2 SAFE per se, indeed B 2 SAFE and i. RODS have been found to be very flexible and are likely to be interesting candidates for other use cases outside of PRIDE elsewhere in EMBL-EBI or ELIXIR. • In particular, use cases focused on data management within or between data centres (i. e. bipartite collaborations) or environments where mature data submission, curation and archiving solutions do not already exist. • In addition, we recommend ELIXIR continues to explore EUDAT services and their relevance in ELIXIR use cases. Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015
Conclusions: Technical recommendations • Incorporate a fully-functional RESTful interface for i. RODS into B 2 SAFE, that can be used by a client to avoid installing i. Commands on the client machine. • The security model should be adapted to allow anonymous RW to a specified URL. • If widespread deployment of EUDAT software is expected, effort must be committed by EUDAT 2020 to make the software more easily and quickly deployable by ‘ordinary’ system administrators. Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015

Acknowledgements • Henning Hermjakob • Steven Newhouse • Rafael Jimenez • Bengt Persson • EUDAT management & developers Juan A. Vizcaíno juan@ebi. ac. uk ELIXIR Webinar 20 May 2015
eda7cefe231f09525b09f99a5d5bdb4f.ppt