3e4cfe2acb0eeca99950a30a291b7199.ppt
- Количество слайдов: 26

Early studies on the Eco. B restriction enzyme using filamentous phage DNA Kensuke Horiuchi The Rockefeller University

Restriction Endonuclease Binds Does not bind Me Recognition site Cleaved Recognition site Intact

What we discovered about Eco. B • The cleavage site is different from the recognition site. • Cleavage does not occur at a defined site but occurs after the enzyme translocates along the DNA.

Norton raised the possibility that the cleavage site and the recognition site are distinct.

Phage f 1 is restricted by Eco. B but not by Eco. K e. o. p. on E. coli B f 1. K 1. 0 7 x 10 -4 f 1. B 1. 0

F 1 has two E. coli B sensitive sites Phage Genotype No. of SB e. o. p. on B Wild type SB 1+ SB 2+ SB = 2 7 X 10 -4 One step mutant SB 1+ SB 20 SB = 1 3 X 10 -2 One step mutant SB 10 SB 2+ SB = 1 3 X 10 -2 Two step mutant SB 10 SB 20 SB = 0 1. 0 Arber & Kuehnlein (1969) Path. Microbiol. Boon & Zinder (1971) JMB

Genetic Map of f 1 Lyons & Zinder (1972) Virology

Cleavage of f 1 RFI by Eco. B enzyme I supercoiled DNA II nicked circular DNA III linear DNA Horiuchi & Zinder (1972) PNAS

Eco. B does not cleave DNA at defined sites Mutant with a single SB site 1) If Eco. B cleaves f 1 RF DNA at a single specific site, annealing after denaturation should yield only linear molecules. 2) If cleavage sites are not specific, reannealing should yield circular DNA and multimers. Horiuchi & Zinder (1972) PNAS

ATP hydrolysis continues after DNA cleavage Horiuchi, Vovis & Zinder (1974) JBC
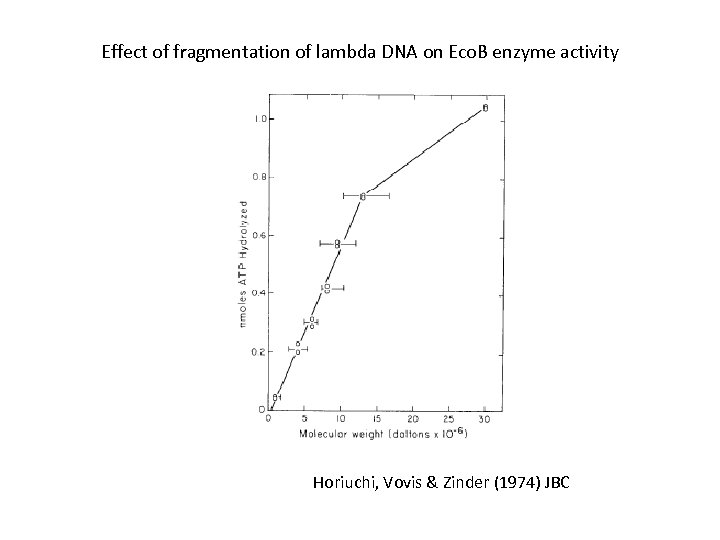
Effect of fragmentation of lambda DNA on Eco. B enzyme activity Horiuchi, Vovis & Zinder (1974) JBC

Steps in Eco. B endonuclease action 1) Eco. B recognizes DNA at SB sites. Recognition is independent of DNA length. 2) The probability that linear DNA is cleaved by bound enzyme depends on DNA length. 3) Circular DNA has an increased probability of cleavage. 4) Thus the enzyme likely needs to translocate along DNA before cleavage. 5) After DNA cleavage, the enzyme (or its components) remains on DNA and causes massive ATP hydrolysis. Horiuchi, Vovis & Zinder (1974) JBC
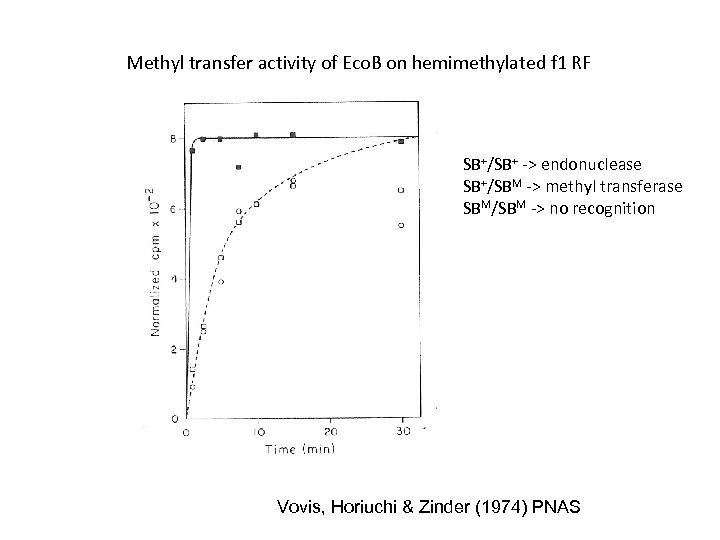
Methyl transfer activity of Eco. B on hemimethylated f 1 RF SB+/SB+ -> endonuclease SB+/SBM -> methyl transferase SBM/SBM -> no recognition Vovis, Horiuchi & Zinder (1974) PNAS

Physical map of f 1 by type II restriction enzymes Hae III Hpa II Hha I Genes

Ravetch, Horiuchi & Zinder (1978) PNAS

Origin and direction of f 1 DNA replication in vivo Horiuchi & Zinder (1976) PNAS

A Zinder lab at a party at Peter Model’s house in 1989

At the 50 th CSH Phage Meeting (1995)


Four point cross: genetic mapping of f 1 Lyons & Zinder (1972) Virology

ATP hydrolysis continues without new DNA-protein interaction Horiuchi, Vovis & Zinder (1974) JBC
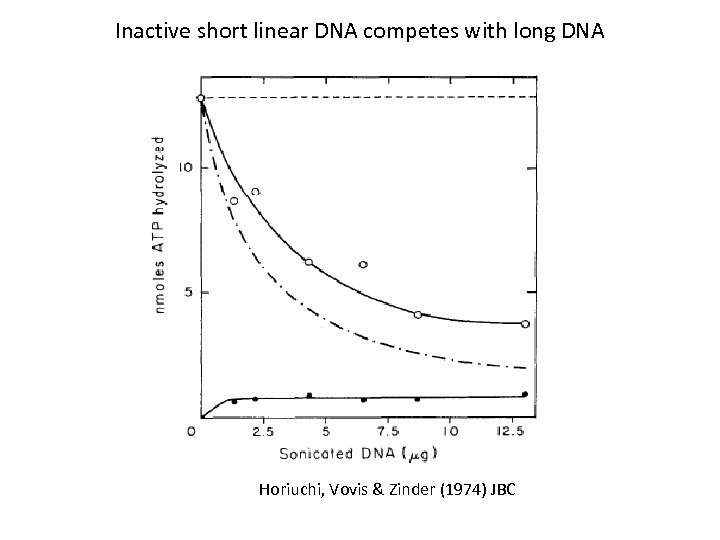
Inactive short linear DNA competes with long DNA Horiuchi, Vovis & Zinder (1974) JBC
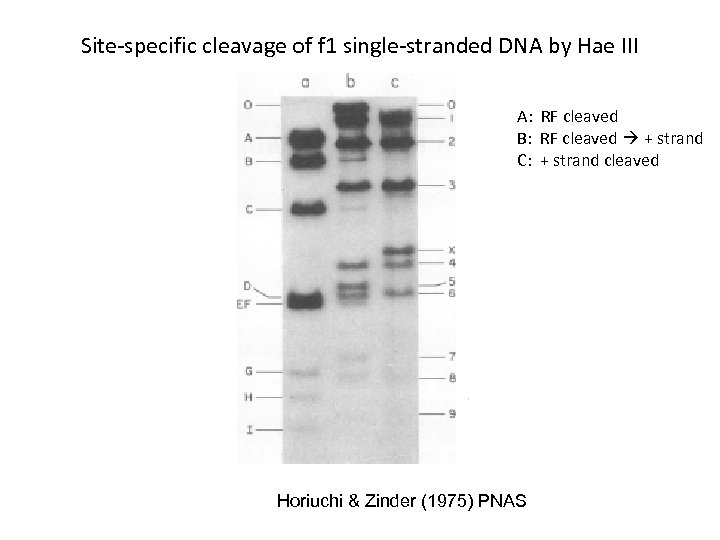
Site-specific cleavage of f 1 single-stranded DNA by Hae III A: RF cleaved B: RF cleaved + strand C: + strand cleaved Horiuchi & Zinder (1975) PNAS


Genetic assay for DNA breaks

Sites of f 1 DNA scission by Eco. RI star mutant endonucleases Heitman & Model (1990) EMBO J.
3e4cfe2acb0eeca99950a30a291b7199.ppt