8bb19a8836165af46c5cb466e12ff2d8.ppt
- Количество слайдов: 19
 e-Infrastructure project Virtual Organization Virtual Research Community Structural Biology on the Grid Christophe Schmitz Bijvoet Center for Biomolecular Research Faculty of Science, Utrecht University The Netherlands c. p. f. schmitz@uu. nl e-Bio. Grid Workshop 2011
e-Infrastructure project Virtual Organization Virtual Research Community Structural Biology on the Grid Christophe Schmitz Bijvoet Center for Biomolecular Research Faculty of Science, Utrecht University The Netherlands c. p. f. schmitz@uu. nl e-Bio. Grid Workshop 2011
 The scientific objectives
The scientific objectives
 The molecular machines and network of life
The molecular machines and network of life
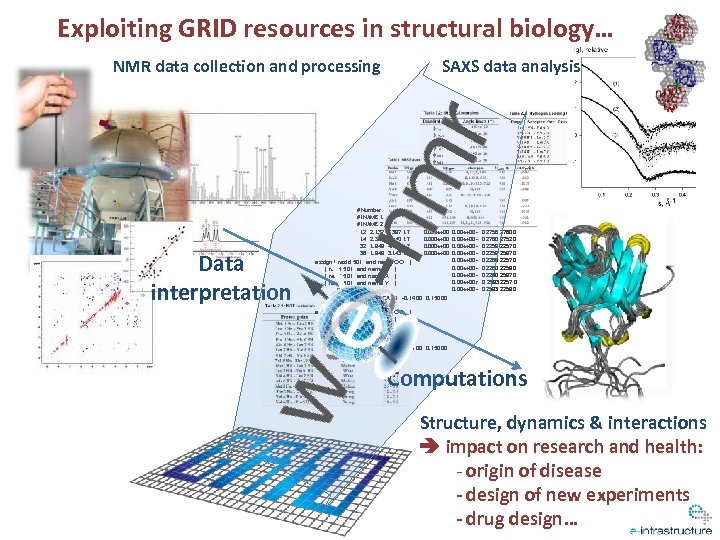 Exploiting GRID resources in structural biology… NMR data collection and processing Data interpretation SAXS data analysis # Number of dimensions 2 # INAME 1 1 H # INAME 2 1 H 12 2. 137 2. 387 1 T 0. 000 e+00 14 2. 387 4. 140 1 T 0. 000 e+00 32 1. 849 4. 432 1 T 0. 000 e+00 36 1. 849 3. 143 1 T 0. 000 e+00 39 1. 760 4. 432 ) 0. 000 e+00 assign ( resid 501 and name OO 1 T 40 1. 760 0. 000 e+00 ( resid 501 and name Z 1. 849 1 T ) 43 1. 760 3. 143 0. 000 e+00 ( resid 501 and name X ) 1 T 46 1. 649 4. 432 1. 035 e+05 ( resid 501 and name Y ) 1 T 47 1. 649 1. 849 1 T 0. 000 e+00 ( resid 2 and name CA ) -0. 1400 0. 15000 0. 00 e+00 0. 00 e+00 r 0. 00 e+00 - 0 2756 2760 0 0 2760 2752 0 0 2259 2257 0 0 2259 2587 0 0 2260 2259 0 0 2260 2587 0 0 2583 2259 0 assign ( resid 501 and name OO ) ( resid 501 and name Z ) ( resid 501 and name X ) ( resid 501 and name Y ) ( resid 3 and name CA ) -0. 0100 0. 15000 Computations Structure, dynamics & interactions impact on research and health: - origin of disease - design of new experiments - drug design …
Exploiting GRID resources in structural biology… NMR data collection and processing Data interpretation SAXS data analysis # Number of dimensions 2 # INAME 1 1 H # INAME 2 1 H 12 2. 137 2. 387 1 T 0. 000 e+00 14 2. 387 4. 140 1 T 0. 000 e+00 32 1. 849 4. 432 1 T 0. 000 e+00 36 1. 849 3. 143 1 T 0. 000 e+00 39 1. 760 4. 432 ) 0. 000 e+00 assign ( resid 501 and name OO 1 T 40 1. 760 0. 000 e+00 ( resid 501 and name Z 1. 849 1 T ) 43 1. 760 3. 143 0. 000 e+00 ( resid 501 and name X ) 1 T 46 1. 649 4. 432 1. 035 e+05 ( resid 501 and name Y ) 1 T 47 1. 649 1. 849 1 T 0. 000 e+00 ( resid 2 and name CA ) -0. 1400 0. 15000 0. 00 e+00 0. 00 e+00 r 0. 00 e+00 - 0 2756 2760 0 0 2760 2752 0 0 2259 2257 0 0 2259 2587 0 0 2260 2259 0 0 2260 2587 0 0 2583 2259 0 assign ( resid 501 and name OO ) ( resid 501 and name Z ) ( resid 501 and name X ) ( resid 501 and name Y ) ( resid 3 and name CA ) -0. 0100 0. 15000 Computations Structure, dynamics & interactions impact on research and health: - origin of disease - design of new experiments - drug design …
 The institute and the users
The institute and the users
 The Institute Partner Institute BCBR Utrecht, The Netherlands BMRZ Frankfurt, Germany CIRMMP Florence, Italy INFN Padova, Italy RUN Nijmegen, The Netherlands UCAM Cambridge, United Kingdom EMBL Hamburg, Germany Alexandre Bonvin, We. NMR coordinator
The Institute Partner Institute BCBR Utrecht, The Netherlands BMRZ Frankfurt, Germany CIRMMP Florence, Italy INFN Padova, Italy RUN Nijmegen, The Netherlands UCAM Cambridge, United Kingdom EMBL Hamburg, Germany Alexandre Bonvin, We. NMR coordinator
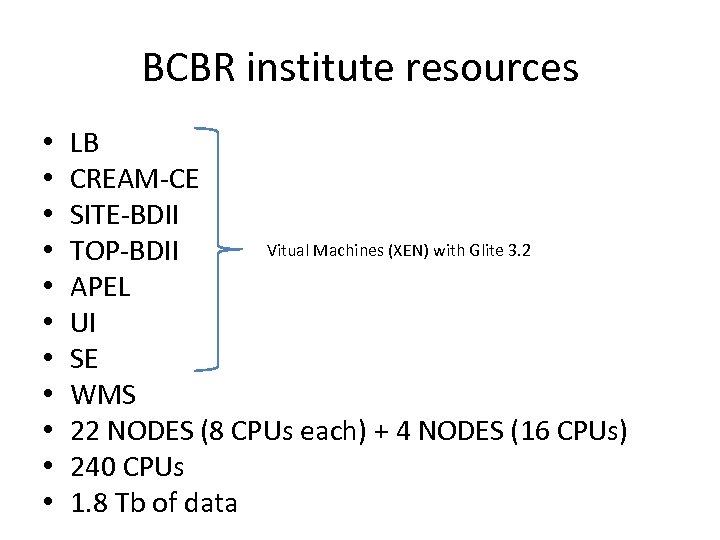 BCBR institute resources • • • LB CREAM-CE SITE-BDII Vitual Machines (XEN) with Glite 3. 2 TOP-BDII APEL UI SE WMS 22 NODES (8 CPUs each) + 4 NODES (16 CPUs) 240 CPUs 1. 8 Tb of data
BCBR institute resources • • • LB CREAM-CE SITE-BDII Vitual Machines (XEN) with Glite 3. 2 TOP-BDII APEL UI SE WMS 22 NODES (8 CPUs each) + 4 NODES (16 CPUs) 240 CPUs 1. 8 Tb of data
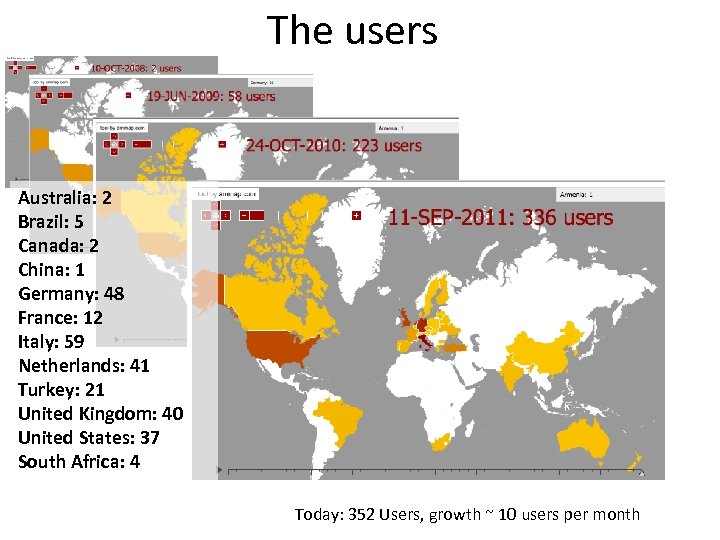 The users Australia: 2 Brazil: 5 Canada: 2 China: 1 Germany: 48 France: 12 Italy: 59 Netherlands: 41 Turkey: 21 United Kingdom: 40 United States: 37 South Africa: 4 Today: 352 Users, growth ~ 10 users per month
The users Australia: 2 Brazil: 5 Canada: 2 China: 1 Germany: 48 France: 12 Italy: 59 Netherlands: 41 Turkey: 21 United Kingdom: 40 United States: 37 South Africa: 4 Today: 352 Users, growth ~ 10 users per month
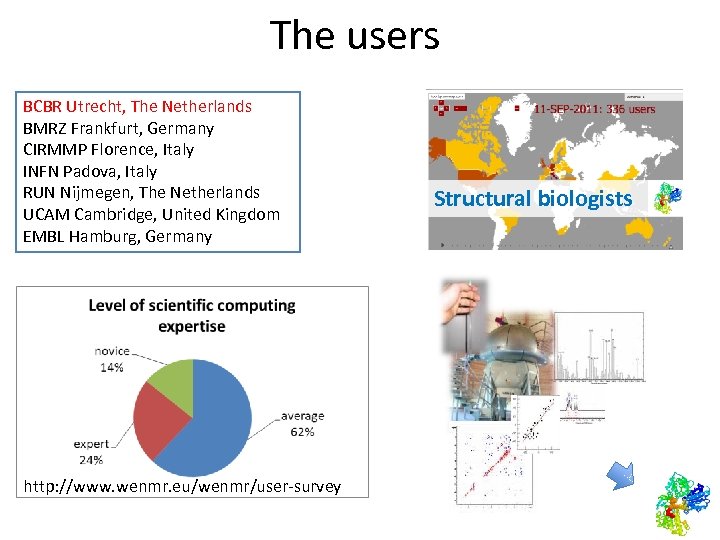 The users BCBR Utrecht, The Netherlands BMRZ Frankfurt, Germany CIRMMP Florence, Italy INFN Padova, Italy RUN Nijmegen, The Netherlands UCAM Cambridge, United Kingdom EMBL Hamburg, Germany http: //www. wenmr. eu/wenmr/user-survey Structural biologists
The users BCBR Utrecht, The Netherlands BMRZ Frankfurt, Germany CIRMMP Florence, Italy INFN Padova, Italy RUN Nijmegen, The Netherlands UCAM Cambridge, United Kingdom EMBL Hamburg, Germany http: //www. wenmr. eu/wenmr/user-survey Structural biologists
 The software
The software
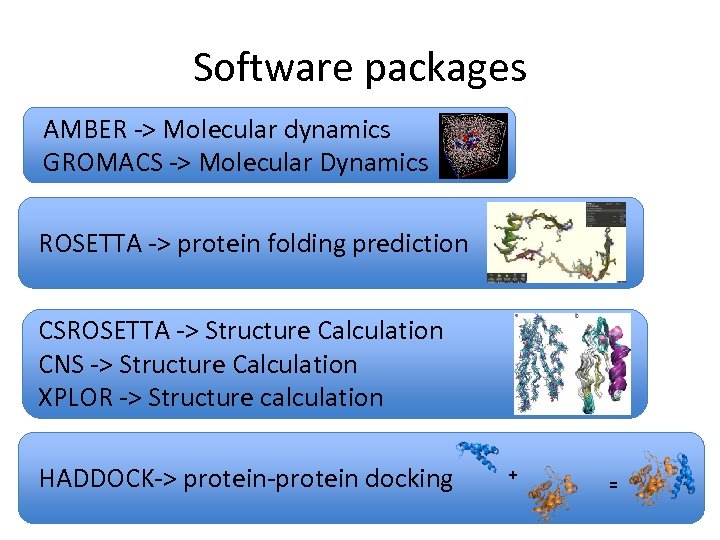 Software packages AMBER -> Molecular dynamics GROMACS -> Molecular Dynamics ROSETTA -> protein folding prediction CSROSETTA -> Structure Calculation CNS -> Structure Calculation XPLOR -> Structure calculation HADDOCK-> protein-protein docking + =
Software packages AMBER -> Molecular dynamics GROMACS -> Molecular Dynamics ROSETTA -> protein folding prediction CSROSETTA -> Structure Calculation CNS -> Structure Calculation XPLOR -> Structure calculation HADDOCK-> protein-protein docking + =
 The Web Portals
The Web Portals
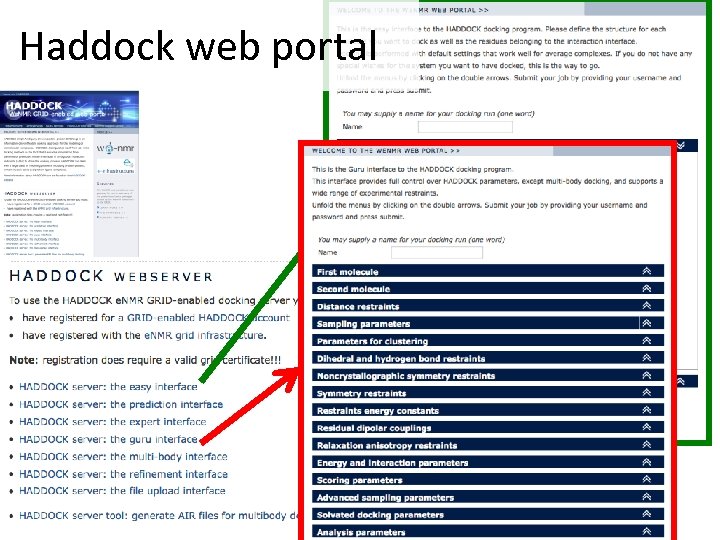 Haddock web portal
Haddock web portal
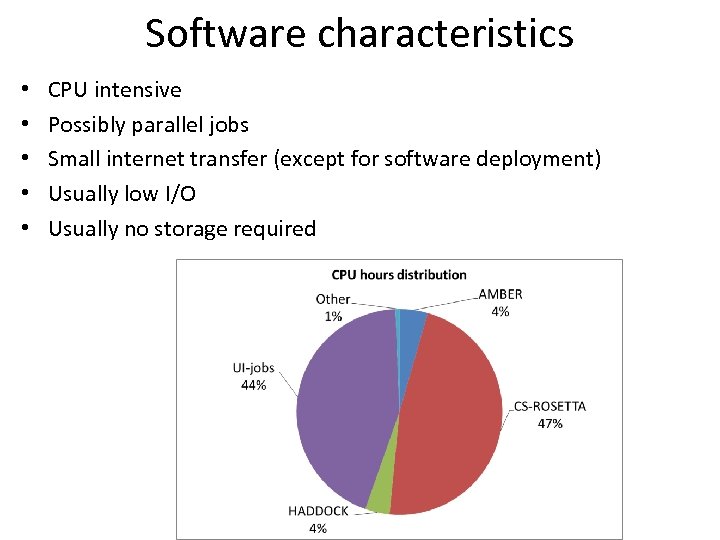 Software characteristics • • • CPU intensive Possibly parallel jobs Small internet transfer (except for software deployment) Usually low I/O Usually no storage required
Software characteristics • • • CPU intensive Possibly parallel jobs Small internet transfer (except for software deployment) Usually low I/O Usually no storage required
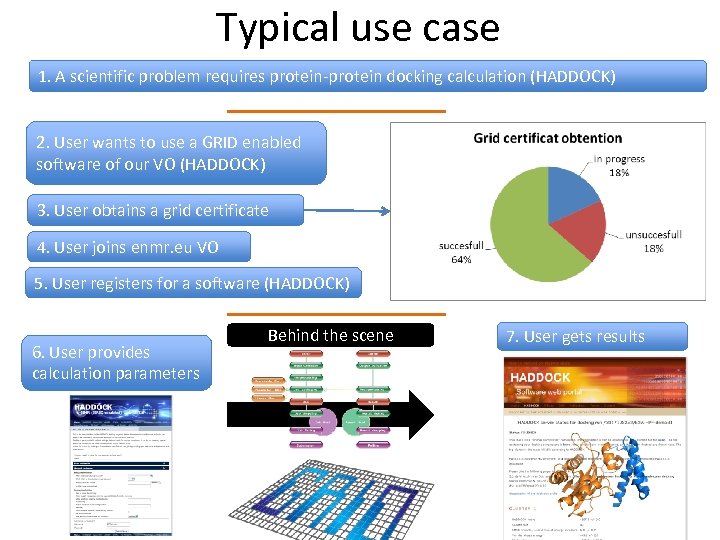 Typical use case 1. A scientific problem requires protein-protein docking calculation (HADDOCK) 2. User wants to use a GRID enabled software of our VO (HADDOCK) 3. User obtains a grid certificate 4. User joins enmr. eu VO 5. User registers for a software (HADDOCK) 6. User provides calculation parameters Behind the scene 7. User gets results
Typical use case 1. A scientific problem requires protein-protein docking calculation (HADDOCK) 2. User wants to use a GRID enabled software of our VO (HADDOCK) 3. User obtains a grid certificate 4. User joins enmr. eu VO 5. User registers for a software (HADDOCK) 6. User provides calculation parameters Behind the scene 7. User gets results
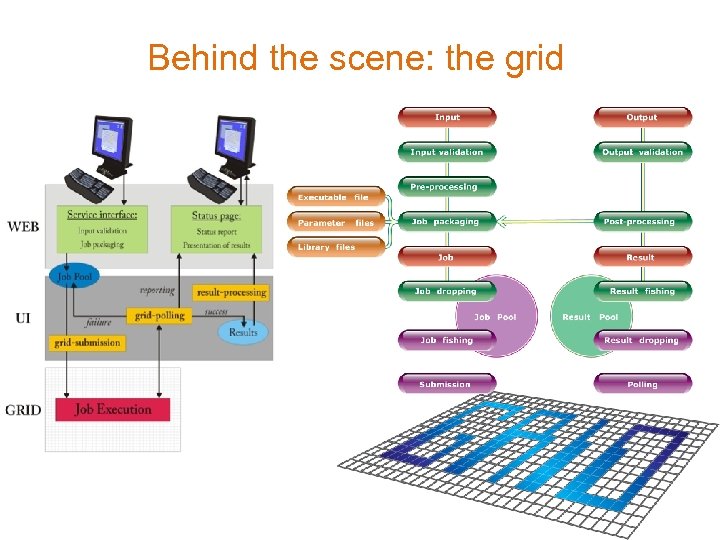 Behind the scene: the grid
Behind the scene: the grid
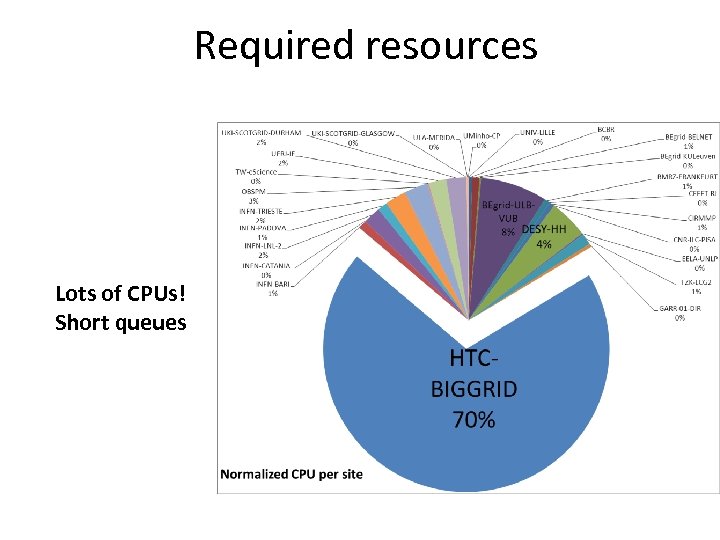 Required resources Lots of CPUs! Short queues
Required resources Lots of CPUs! Short queues
 We. NMR platform operational and well used! • • Largest global VO in the life sciences Over 352 registered users from 38 nationalities, and growing >33 000 CPUs >700 (normalized) CPU years over the last 12 months 1. 4 million jobs over the last 12 months 150 Tb of storage space ~20% of Life Sciences on the Grid User-friendly access to e-Infrastructure via web portals
We. NMR platform operational and well used! • • Largest global VO in the life sciences Over 352 registered users from 38 nationalities, and growing >33 000 CPUs >700 (normalized) CPU years over the last 12 months 1. 4 million jobs over the last 12 months 150 Tb of storage space ~20% of Life Sciences on the Grid User-friendly access to e-Infrastructure via web portals
 Acknowledgments Prof. Alexandre Bonvin (We. NMR coordinator) All the Bijvoet Center in Utrecht University, Bijvoet Center for Biomolecular Research, NL Johann Wolfgang Goethe Universität Frankfurt a. M. , Center for Biomolecular Magnetic Resonance DE University of Florence, Magnetic Resonance Center, IT Istituto Nazionale di Fisica Nucleare , Padova, IT Raboud University, Nijmegen, NL University of Cambridge UK European Molecular Biology Laboratory, Hamburg, DE Spronk NMR Consultancy, LT
Acknowledgments Prof. Alexandre Bonvin (We. NMR coordinator) All the Bijvoet Center in Utrecht University, Bijvoet Center for Biomolecular Research, NL Johann Wolfgang Goethe Universität Frankfurt a. M. , Center for Biomolecular Magnetic Resonance DE University of Florence, Magnetic Resonance Center, IT Istituto Nazionale di Fisica Nucleare , Padova, IT Raboud University, Nijmegen, NL University of Cambridge UK European Molecular Biology Laboratory, Hamburg, DE Spronk NMR Consultancy, LT


