2bb06490a7827511972b9d5f371d65a4.ppt
- Количество слайдов: 32
Dynamic Data Driven Finite Element Modeling of Brain Shape Deformation During Neurosurgery A. Majumdar 1, D. Choi 1, P. Krysl 2 , S. K. Warfield 3, N. Archip 3 , K. Baldridge 1, 4 1 San Diego Supercomputer Center & 2 Structural Engineering Dept University of California San Diego Computational Radiology Lab Brigham and Women’s Hospital Harvard Medical School 3 4 Universität Zürich Grants: NSF: ITR 0427183, 0426558; NIH: P 41 RR 13218, P 01 CA 67165, LM 0078651, I 3 grant (IBM) Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 1
Contents of Talk 1. Overview of Image Guided Neurosurgery and Dynamic Data Drive Application System 2. Biomechanical FEM solution 3. Briefly grid scheduling 4. Future : near-continuous DDDAS Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 2
1. Overview of Image Guided Neurosurgery and Dynamic Data Drive Application System Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 3

Neurosurgery Challenge • Challenges : • • Remove as much tumor tissue as possible Minimize the removal of healthy tissue Avoid the disruption of critical anatomical structures Know when to stop the resection process • Pre-op MRI compounded by the intra-operative brain shape deformation as a result of the surgical process • Important to quantify and correct for these deformations while surgery is in progress • Real-time constraints – provide images ~once/hour within few mins during surgery lasting ~6 hours Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 4
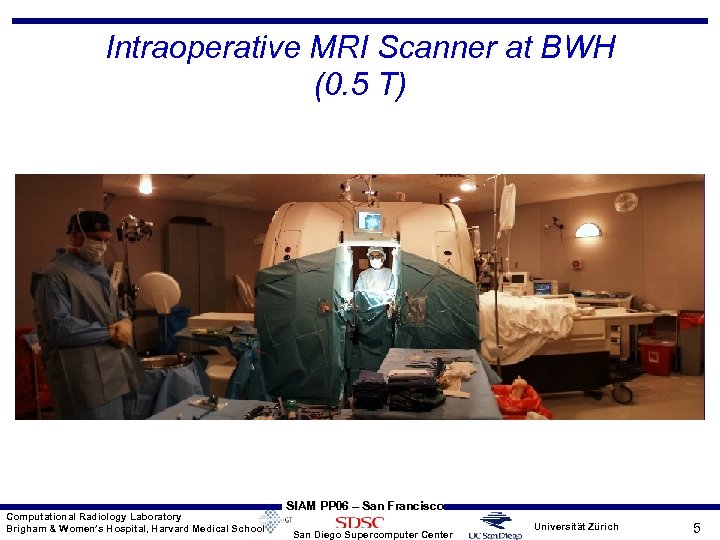
Intraoperative MRI Scanner at BWH (0. 5 T) Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 5
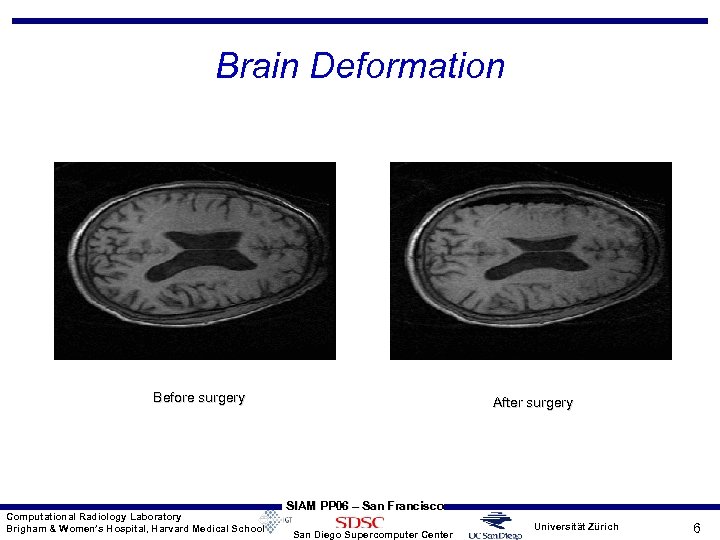
Brain Deformation Before surgery Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School After surgery SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 6
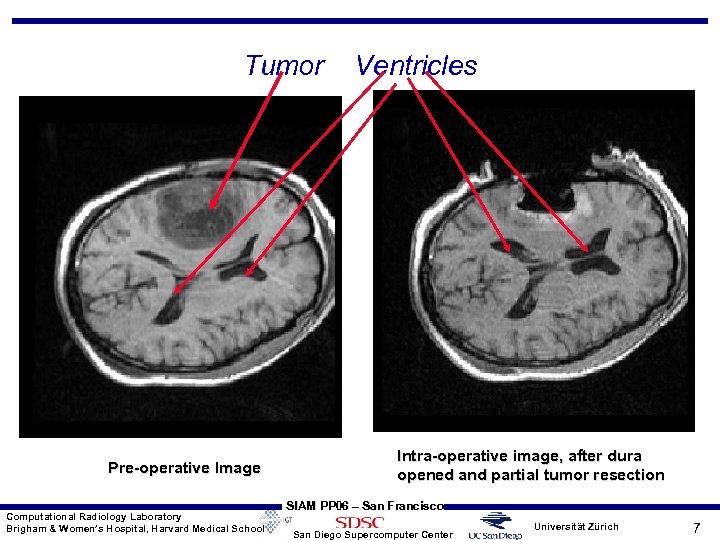
Tumor Pre-operative Image Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School Ventricles Intra-operative image, after dura opened and partial tumor resection SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 7
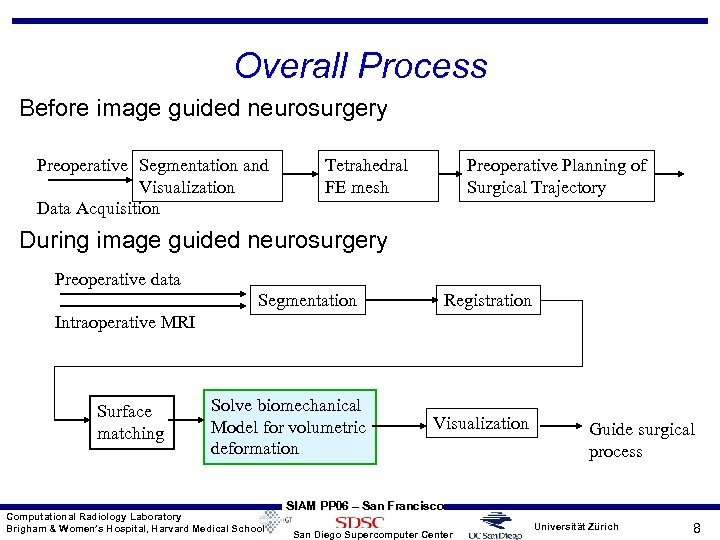
Overall Process Before image guided neurosurgery Preoperative Segmentation and Visualization Data Acquisition Tetrahedral FE mesh Preoperative Planning of Surgical Trajectory During image guided neurosurgery Preoperative data Segmentation Registration Intraoperative MRI Surface matching Solve biomechanical Model for volumetric deformation Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School Visualization Guide surgical process SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 8
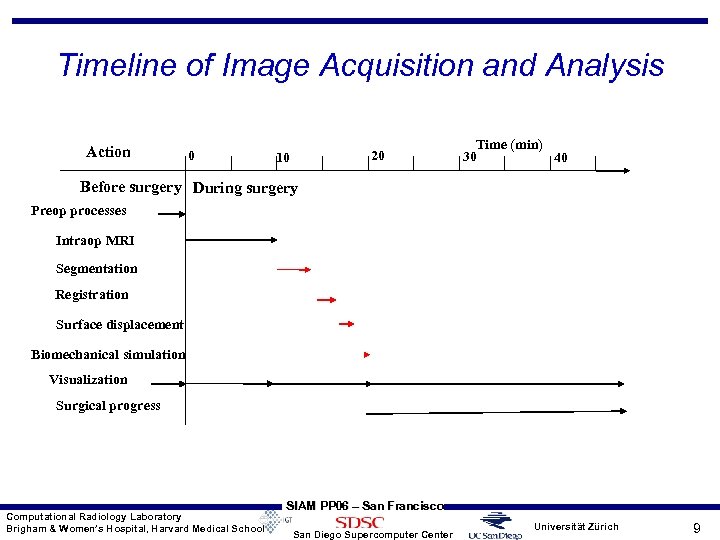
Timeline of Image Acquisition and Analysis Action 0 20 10 Time (min) 30 40 Before surgery During surgery Preop processes Intraop MRI Segmentation Registration Surface displacement Biomechanical simulation Visualization Surgical progress Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 9
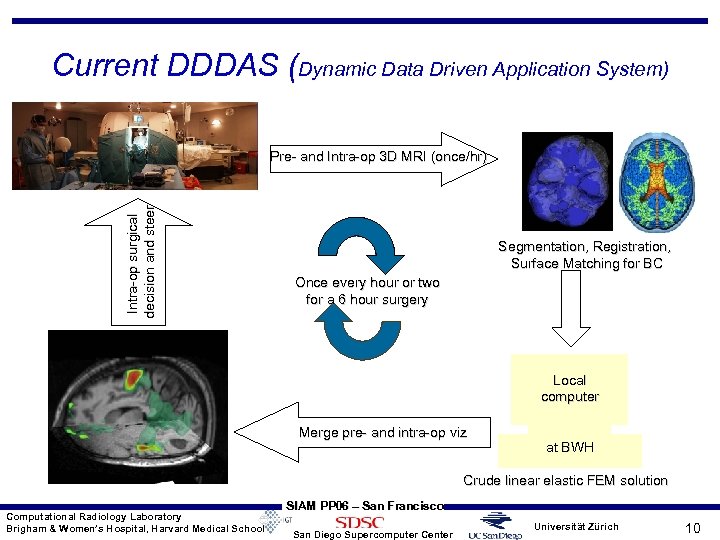
Current DDDAS (Dynamic Data Driven Application System) Intra-op surgical decision and steer Pre- and Intra-op 3 D MRI (once/hr) Segmentation, Registration, Surface Matching for BC Once every hour or two for a 6 hour surgery Local computer Merge pre- and intra-op viz at BWH Crude linear elastic FEM solution Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 10
Two Research Aspects • Parallel solution of the linear elastic biomechanical model for brain shape deformation during surgery • Grid Architecture – grid scheduling, on demand remote access to multi-teraflop machines, data transfer/sharing Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 11
2. Biomechanical FEM solution Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 12
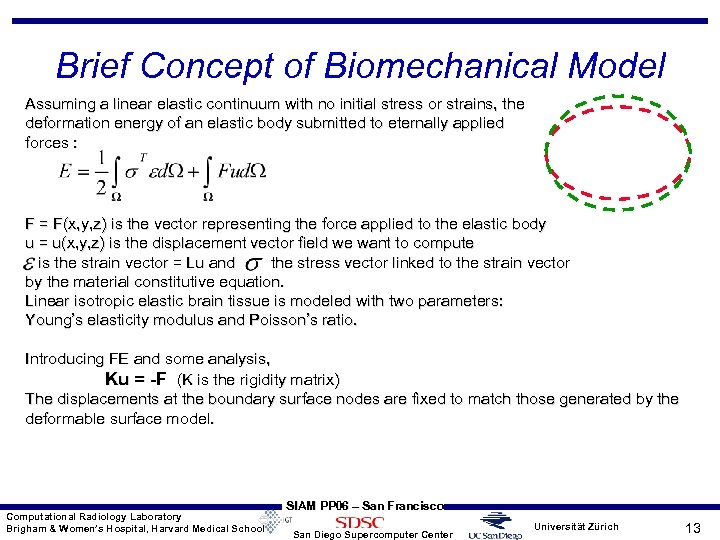
Brief Concept of Biomechanical Model Assuming a linear elastic continuum with no initial stress or strains, the deformation energy of an elastic body submitted to eternally applied forces : F = F(x, y, z) is the vector representing the force applied to the elastic body u = u(x, y, z) is the displacement vector field we want to compute is the strain vector = Lu and the stress vector linked to the strain vector by the material constitutive equation. Linear isotropic elastic brain tissue is modeled with two parameters: Young’s elasticity modulus and Poisson’s ratio. Introducing FE and some analysis, Ku = -F (K is the rigidity matrix) The displacements at the boundary surface nodes are fixed to match those generated by the deformable surface model. Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 13
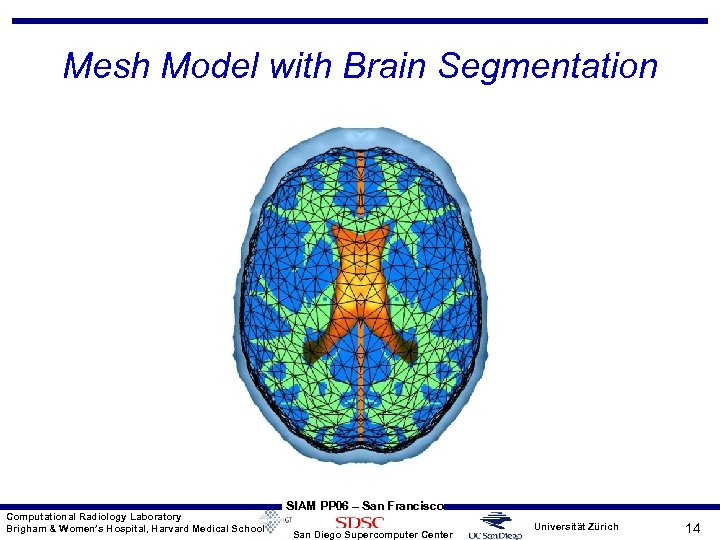
Mesh Model with Brain Segmentation Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 14
Current and New Biomechanical Models • Current linear elastic material model RTBM • Advanced biomechanical model FAMULS (AMR) • Advanced model is based on conforming adaptive refinement method • Inspired by theory of wavelets this refinement produces globally compatible meshes by construction • Replicate the linear elastic result produced by RTBM using FAMULS Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 15
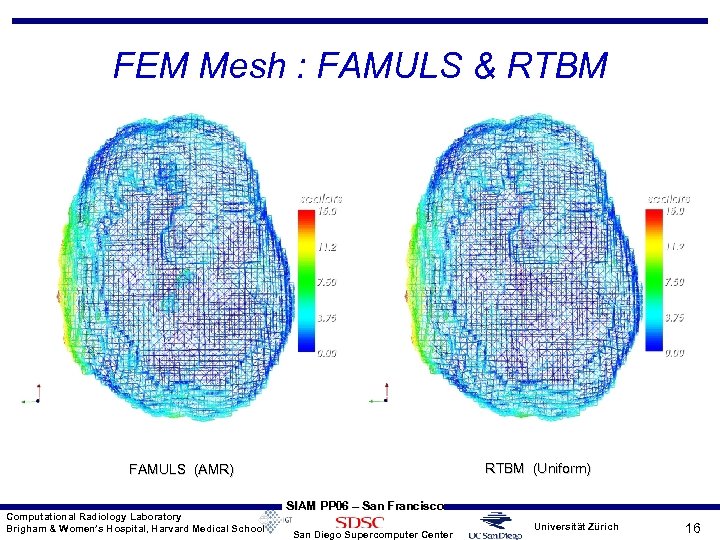
FEM Mesh : FAMULS & RTBM (Uniform) FAMULS (AMR) Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 16
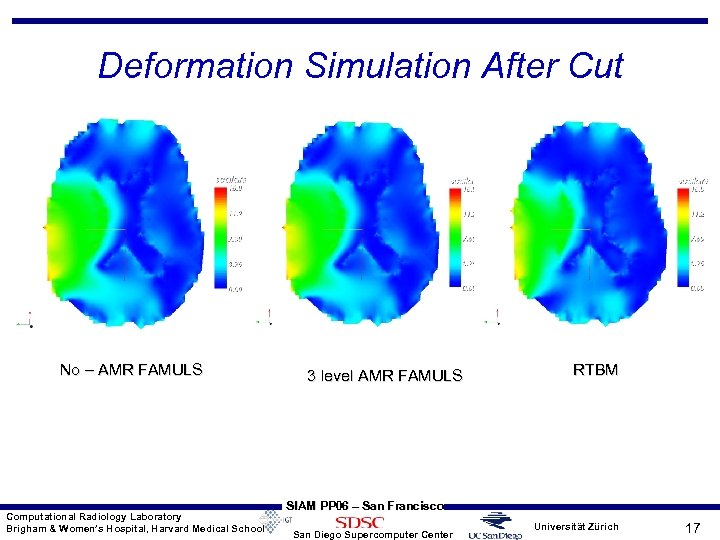
Deformation Simulation After Cut No – AMR FAMULS Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School 3 level AMR FAMULS RTBM SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 17

Petsc setup • Petsc. Map. Create. MPI(PETSC_COMM_WORLD, PE TSC_DECIDE, n, &map) ; • Mat. Create. MPIAIJ(PETSC_COMM_WORLD, . . &K_g lobal) ; Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 18
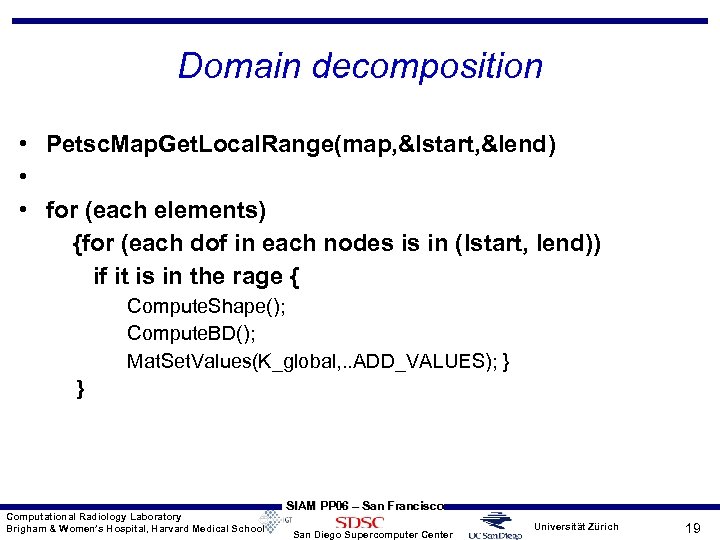
Domain decomposition • Petsc. Map. Get. Local. Range(map, &Istart, &Iend) • • for (each elements) {for (each dof in each nodes is in (lstart, lend)) if it is in the rage { Compute. Shape(); Compute. BD(); Mat. Set. Values(K_global, . . ADD_VALUES); } } Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 19
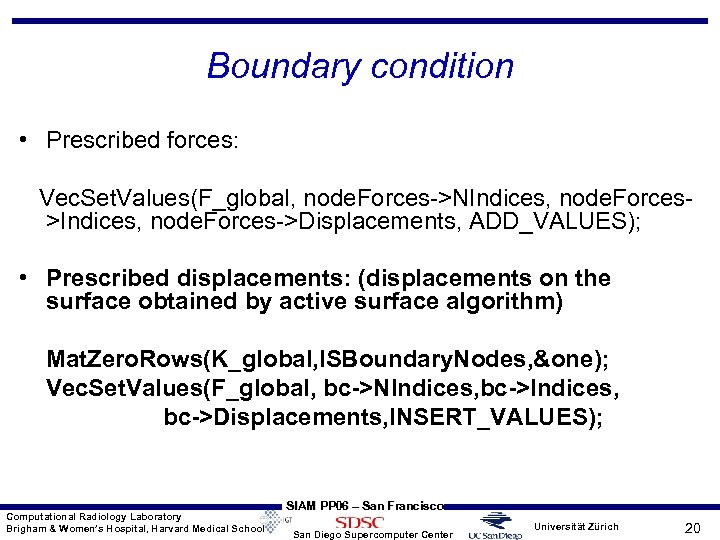
Boundary condition • Prescribed forces: Vec. Set. Values(F_global, node. Forces->NIndices, node. Forces>Indices, node. Forces->Displacements, ADD_VALUES); • Prescribed displacements: (displacements on the surface obtained by active surface algorithm) Mat. Zero. Rows(K_global, ISBoundary. Nodes, &one); Vec. Set. Values(F_global, bc->NIndices, bc->Displacements, INSERT_VALUES); Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 20

Solver setup • • KSPCreate(PETSC_COMM_WORLD, &ksp) KSPSet. Operators(ksp, K_global. . ) KSPGet. PC(ksp, &pc) PCSet. Type(pc, PCBJACOBI) KSPSet. Tolerances(ksp, 1. e-7. . ) KSPSet. From. Options(ksp) KSPSolve(ksp, F_global, u_displ, &its) Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 21
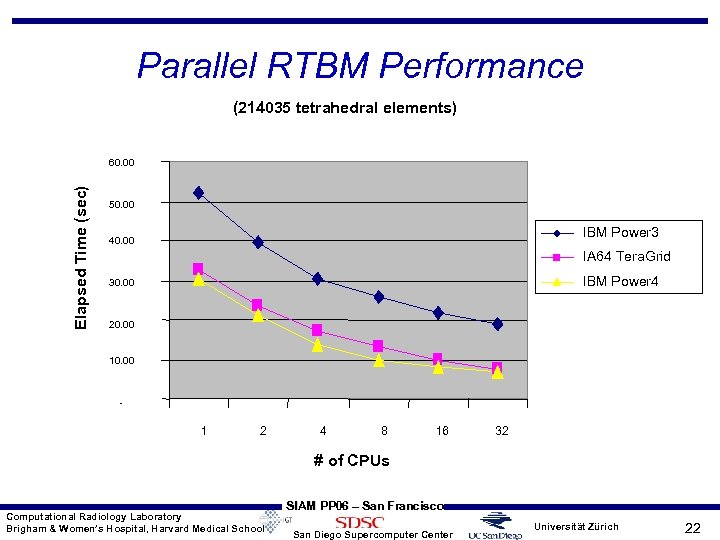
Parallel RTBM Performance (214035 tetrahedral elements) Elapsed Time (sec) 60. 00 50. 00 IBM Power 3 40. 00 IA 64 Tera. Grid IBM Power 4 30. 00 20. 00 10. 00 - 1 2 4 8 16 32 # of CPUs Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 22
Advanced Biomechanical Model • The current solver is based on small strain isotropic elastic principle • New biomechanical model • Inhomogeneous scalable non-linear hyper-elastic or visco-elastic model with AMR • Increase resolution close to the level of MRI voxels i. e. millions of FEM meshes • New high resolution complex model still has to meet the real time constraint of neurosurgery • Requires fast access to remote multi-teraflop systems Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 23
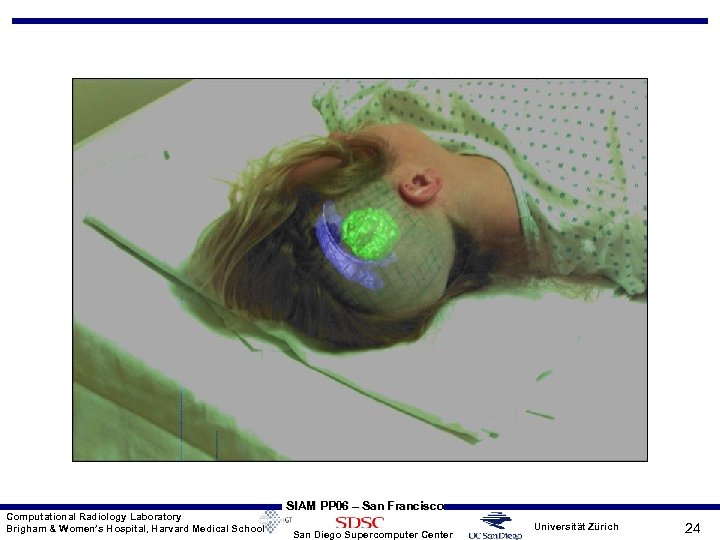
Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 24
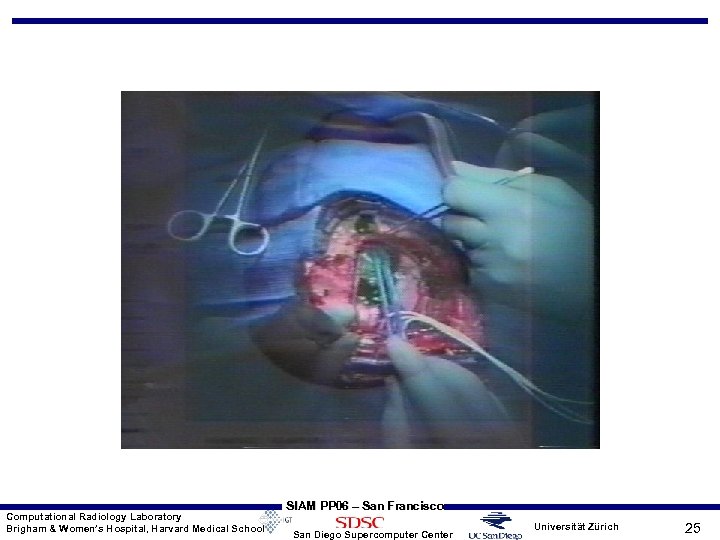
Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 25
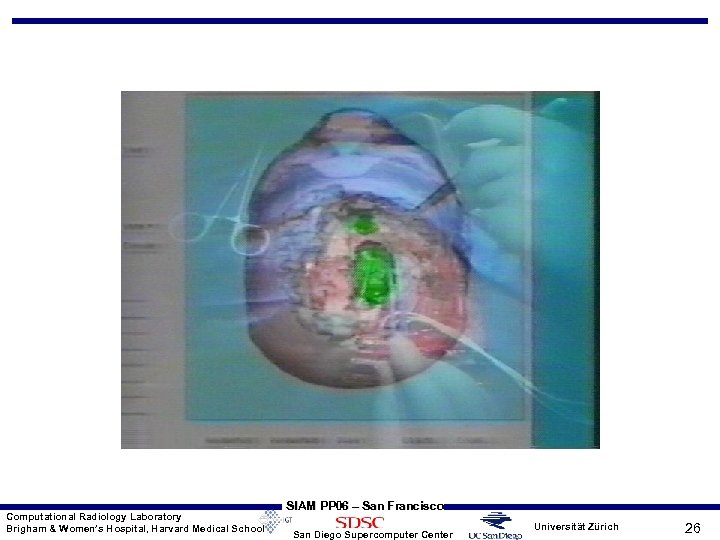
Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 26
3. Briefly Grid Scheduling Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 27
On-demand Scheduling Experiment on 5 Tera. Grid Clusters • The real-time constraint of this application requires that data transfer and simulation altogether take about 10 mins, otherwise these results are not of use to surgeons • Assume simulation and data transfer (both ways) together takes 10 mins and data transfer takes 4 mins • Leaves 6 mins for biomechanical simulation on remote HPC machines • Assume biomechanical model is scalable i. e. better results achieved on higher number of processors • Objective : • Get simulation done in 6 mins • Get maximum number of processors available within 6 mins • Allow 4 mins to wait in the queue; this leaves 2 mins for actual simulation Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 28
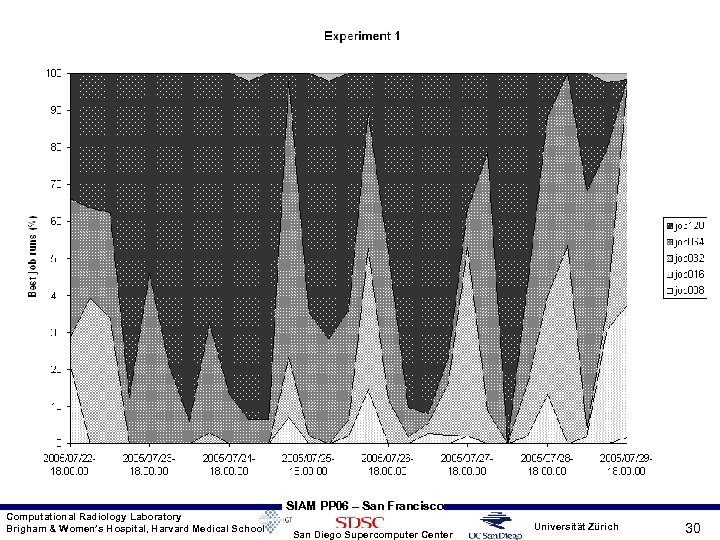
Experiment Characteristics • Flooding scheduler approach – experiment 1: • Simultaneously submit 8, 16, 32, 64, 128 procs jobs to multiple clusters - SDSC Data. Star, SDSC TG, NCSA TG, ANL TG, PSC TG • When a higher count job starts (at any center) kill all the lower CPU count jobs at all the other centers • Results : out of 1464 job submissions over ~7 days, only 6 failed giving success of 99. 59%; 128 CPU jobs ran greater than 50% of time; at least 64 CPU jobs ran more than 80% of time • Next slide gives time varying behavior with 6 hour intervals for this experiment • 4 other experiments were performed by taking out some of the successful clusters as well as taking scheduler cycle time into account on Data. Star • As number of clusters were reduced, success rate goes down Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 29
Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 30
4. Future : Near-continuous DDDAS Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 31
Current DDDAS vs. (future) near-continuous DDDAS • Problem of current DDDAS: • Using current DDDAS procedure, surgeon does not have near-continuous brain deformation info • It takes more than 20 minutes to have whole 3 d scan, segmentation, surface matching and FEM solution • Solution is to extend to near continuous DDDAS: • DDDAS approach to provide near-continuous closed loop registration updates using near-continuous 2 D MRI slice scans Computational Radiology Laboratory Brigham & Women’s Hospital, Harvard Medical School SIAM PP 06 – San Francisco San Diego Supercomputer Center Universität Zürich 32
2bb06490a7827511972b9d5f371d65a4.ppt