9dfc01aca37f9177eb6dc50ef4488f5f.ppt
- Количество слайдов: 23
 Domain-SLi. M mining from High Throughput Protein Interaction Data Hugo Willy August 19, 2010
Domain-SLi. M mining from High Throughput Protein Interaction Data Hugo Willy August 19, 2010
 Introduction to SLi. M It stands for (Protein) Short Linear Motif By its name, it is a short linear stretch of region in a protein sequence that is recognized by another protein for binding It averages at 8 -12 amino acids where some can go as short as three amino acids. It is currently one of the special mechanism of a protein recognizing its interaction partner
Introduction to SLi. M It stands for (Protein) Short Linear Motif By its name, it is a short linear stretch of region in a protein sequence that is recognized by another protein for binding It averages at 8 -12 amino acids where some can go as short as three amino acids. It is currently one of the special mechanism of a protein recognizing its interaction partner
 Protein Interaction in general • Some proteins only function in terms of a complex. They have to be in a certain combination. These are called the obligate complexes. • Their binding surface are usually large to provide strong chemical interactions.
Protein Interaction in general • Some proteins only function in terms of a complex. They have to be in a certain combination. These are called the obligate complexes. • Their binding surface are usually large to provide strong chemical interactions.
 Protein Interaction in general (2) • On the other hand, some complexes are formed only “on-demand”. Once the task is done, they dissociate. These are called the transient complexes. • The interaction surface of this type of interaction is generally smaller. • SLi. M based interaction is one of the transient ones.
Protein Interaction in general (2) • On the other hand, some complexes are formed only “on-demand”. Once the task is done, they dissociate. These are called the transient complexes. • The interaction surface of this type of interaction is generally smaller. • SLi. M based interaction is one of the transient ones.
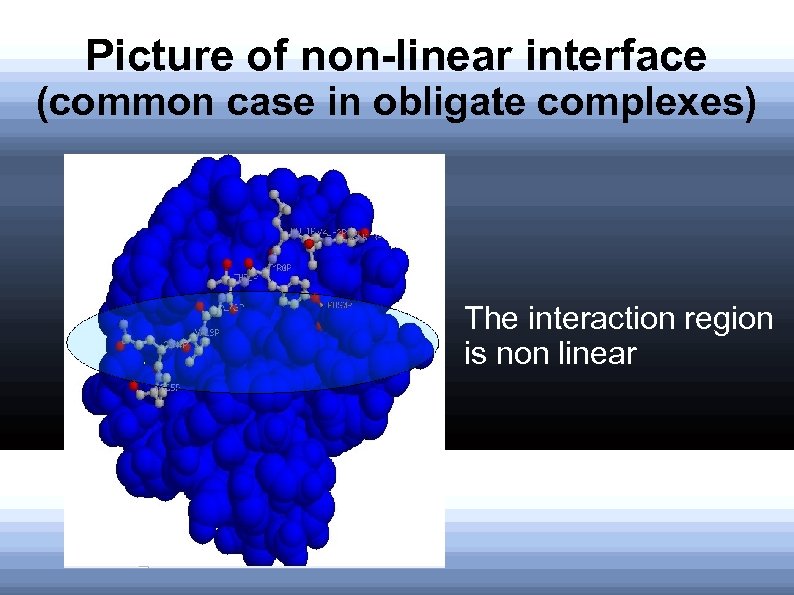 Picture of non-linear interface (common case in obligate complexes) The interaction region is non linear
Picture of non-linear interface (common case in obligate complexes) The interaction region is non linear
 Picture of Linear interface The protein chain bound is linear on the interface of the partner.
Picture of Linear interface The protein chain bound is linear on the interface of the partner.
 Protein domains recognizing SLi. Ms • In reality, the task of recognizing a SLi. M often is performed by specialized protein domain. • Some of the most well known example is the SH 3 domain which recognize P. . P motif where P is a proline amino acid. WW domains recognize PP. Y motif. • These SLi. Ms, along with their functions (or domain association) are listed in databases like Eukaryotic Linear Motif (ELM) [1] and Mini. Motif (Mn. M) [2]
Protein domains recognizing SLi. Ms • In reality, the task of recognizing a SLi. M often is performed by specialized protein domain. • Some of the most well known example is the SH 3 domain which recognize P. . P motif where P is a proline amino acid. WW domains recognize PP. Y motif. • These SLi. Ms, along with their functions (or domain association) are listed in databases like Eukaryotic Linear Motif (ELM) [1] and Mini. Motif (Mn. M) [2]
 Methods of finding SLi. Ms in proteins • The SLi. Ms listed in the two databases are mostly results of experimental procedures like mutagenesis and phage display. • They are laborious and expensive.
Methods of finding SLi. Ms in proteins • The SLi. Ms listed in the two databases are mostly results of experimental procedures like mutagenesis and phage display. • They are laborious and expensive.
 Computational Method to detect SLi. Ms • From Sequence-based data (the focus of this talk) • From Structural data – earlier this year, we published SLi. MDiet [3], which is currently the most comprehensive SLi. M listing from the PDB.
Computational Method to detect SLi. Ms • From Sequence-based data (the focus of this talk) • From Structural data – earlier this year, we published SLi. MDiet [3], which is currently the most comprehensive SLi. M listing from the PDB.
 Sequence-based SLi. M detection • Protein sequence based – – • Given a set of grouped sequence, find motif that occurs in unrelated sequences. Example: DILIMOT [4], Sli. MDisc [5], SLi. MFinder [6] Protein interaction based – Find correlated motifs that is over-represented in interacting proteins – Example: D-STAR [7], Motif. Cluster [8], SLIDER [9]
Sequence-based SLi. M detection • Protein sequence based – – • Given a set of grouped sequence, find motif that occurs in unrelated sequences. Example: DILIMOT [4], Sli. MDisc [5], SLi. MFinder [6] Protein interaction based – Find correlated motifs that is over-represented in interacting proteins – Example: D-STAR [7], Motif. Cluster [8], SLIDER [9]
 Protein Sequence Based Methods • Rely on occurrences on unrelated sequences. • May need to remove protein domains from the motif search space because of their similarity • The grouping of the sequences can be manual – by manually selecting known sequences with a certain property. For example, proteins that are exported outside the cells can be grouped to find the motif that is responsible for the export mechanism. • Automated grouping – using the protein domain information or GO ontology annotation
Protein Sequence Based Methods • Rely on occurrences on unrelated sequences. • May need to remove protein domains from the motif search space because of their similarity • The grouping of the sequences can be manual – by manually selecting known sequences with a certain property. For example, proteins that are exported outside the cells can be grouped to find the motif that is responsible for the export mechanism. • Automated grouping – using the protein domain information or GO ontology annotation
 Protein Sequence Based Methods (2) • Once the grouping is done, the motif is mined using standard motif searching like MEME or TEIRESIAS. • Because of the speed and rigid requirement of motif length of MEME, usually TEIRESIAS is the program of choice (it can start with a motif length and try to combine the motifs into longer ones). • Teiresias uses L, W motif – motif of length L over window of length W.
Protein Sequence Based Methods (2) • Once the grouping is done, the motif is mined using standard motif searching like MEME or TEIRESIAS. • Because of the speed and rigid requirement of motif length of MEME, usually TEIRESIAS is the program of choice (it can start with a motif length and try to combine the motifs into longer ones). • Teiresias uses L, W motif – motif of length L over window of length W.
 Protein Sequence Based Methods (3) • The problem of this method is that it relies too much on the initial grouping. • The grouping must have the motif really overrepresented. • All paper in this line have been comparing their performances in the ELM set (a dataset of curated sequences which are known to contain the ELM motif).
Protein Sequence Based Methods (3) • The problem of this method is that it relies too much on the initial grouping. • The grouping must have the motif really overrepresented. • All paper in this line have been comparing their performances in the ELM set (a dataset of curated sequences which are known to contain the ELM motif).
 Protein Sequence Based Methods (4) • They also found some significant motifs from the group of protein known to interact with a certain protein domain which is known to have such SLi. M interaction. • DILIMOT got published in PLo. S Biology as they managed some biological validations.
Protein Sequence Based Methods (4) • They also found some significant motifs from the group of protein known to interact with a certain protein domain which is known to have such SLi. M interaction. • DILIMOT got published in PLo. S Biology as they managed some biological validations.
 Interaction based methods • To be precise, none of the interaction based methods designed up to date were specifically designed to find SLi. Ms. • Most of them are finding “correlated motif pair”. • These are a pair of motif which occur consistently more frequently in interacting proteins as opposed to some background model. • Examples: D-STAR, Motif. Cluster and SLIDER
Interaction based methods • To be precise, none of the interaction based methods designed up to date were specifically designed to find SLi. Ms. • Most of them are finding “correlated motif pair”. • These are a pair of motif which occur consistently more frequently in interacting proteins as opposed to some background model. • Examples: D-STAR, Motif. Cluster and SLIDER
 Interaction based methods (2) • These methods rely solely on the density of interactions between the two set of proteins that contain the motif pair respectively. • D-STAR and SLIDER uses a Chi-Square scoring while Motif. Cluster uses hypergeometric scoring. • As I shall show later, they may not be suitable in finding SLi. Ms – they are by design finding interaction motif which may not be the binding motif themselves.
Interaction based methods (2) • These methods rely solely on the density of interactions between the two set of proteins that contain the motif pair respectively. • D-STAR and SLIDER uses a Chi-Square scoring while Motif. Cluster uses hypergeometric scoring. • As I shall show later, they may not be suitable in finding SLi. Ms – they are by design finding interaction motif which may not be the binding motif themselves.
 My current attempt - SLIMMER • I learnt that most of the time SLi. Ms are bound by a non-linear interface. • Thus, it is not very feasible to hope that both side of the interface contain linear motifs. • This was mentioned by one of D-STAR’s reviewer. • So, I try to find correlated motifs where one of them is a protein domain – which is by definition non-linear (they are distinct protein folds in 3 D)
My current attempt - SLIMMER • I learnt that most of the time SLi. Ms are bound by a non-linear interface. • Thus, it is not very feasible to hope that both side of the interface contain linear motifs. • This was mentioned by one of D-STAR’s reviewer. • So, I try to find correlated motifs where one of them is a protein domain – which is by definition non-linear (they are distinct protein folds in 3 D)
 SLIMMER (2) • I basically combine the good ideas from many programs to accomplish this. • The strength of correlated motifs is that they can find seemingly insignificant motifs (by virtue of their sequence occurrence) by using the fact that once they occur, they interact intensively with the partner motif. • The correlated motif uses over-representation of the interaction occurrence, as opposed to sequence occurrence.
SLIMMER (2) • I basically combine the good ideas from many programs to accomplish this. • The strength of correlated motifs is that they can find seemingly insignificant motifs (by virtue of their sequence occurrence) by using the fact that once they occur, they interact intensively with the partner motif. • The correlated motif uses over-representation of the interaction occurrence, as opposed to sequence occurrence.
 SLIMMER (3) • However, the tricks of sequence based method can also be applied. • They requires occurrence of the SLi. Ms in nonhomologous sequences (which can be considered as independent occurrences). • This non homology is never considered in DSTAR, Motif. Cluster and SLIDER. • We should consider only non-homologous interactions when we count the occurrence of the motif pair.
SLIMMER (3) • However, the tricks of sequence based method can also be applied. • They requires occurrence of the SLi. Ms in nonhomologous sequences (which can be considered as independent occurrences). • This non homology is never considered in DSTAR, Motif. Cluster and SLIDER. • We should consider only non-homologous interactions when we count the occurrence of the motif pair.
 SLIMMER (4) • The SLi. M itself must have an occurrence probability better than random. Motif. Cluster uses the binomial distribution to compute the probability of seeing a motif M, k times in the sequence set (this is threshold approach). • I also tried another approach where I combine the binomial p-value of the motif occurrence and the hypergeometric p-value of the interaction occurrence.
SLIMMER (4) • The SLi. M itself must have an occurrence probability better than random. Motif. Cluster uses the binomial distribution to compute the probability of seeing a motif M, k times in the sequence set (this is threshold approach). • I also tried another approach where I combine the binomial p-value of the motif occurrence and the hypergeometric p-value of the interaction occurrence.
 SLIMMER (5) • Current results, SLIMMER is better than all methods available and it is also fast. • I am still implementing a better background model to deal with low complexity regions – using a simple 3 rd or 4 th order markov. • I also in the middle of trying a motif model that allows choices like [LIVM], [FWY] or [KRH] • The program allowing these currently is only SLi. MFinder and it is very slow and inaccurate for now.
SLIMMER (5) • Current results, SLIMMER is better than all methods available and it is also fast. • I am still implementing a better background model to deal with low complexity regions – using a simple 3 rd or 4 th order markov. • I also in the middle of trying a motif model that allows choices like [LIVM], [FWY] or [KRH] • The program allowing these currently is only SLi. MFinder and it is very slow and inaccurate for now.
![References [1] P Puntervoll et al. ELM server: A new resource for investigating short References [1] P Puntervoll et al. ELM server: A new resource for investigating short](https://present5.com/presentation/9dfc01aca37f9177eb6dc50ef4488f5f/image-22.jpg) References [1] P Puntervoll et al. ELM server: A new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. , 31(13): 3625– 3630, 2003. [2] S Rajasekaran et al. Minimotif miner 2 nd release: a database and web system for motif search. Nucleic Acids Res. , 37(Database issue): D 185– 190, 2009. [3] W Hugo et al. SLi. M on Diet: finding short linear motifs on domain interaction interfaces in Protein Data Bank. Bioinformatics 2010 26(8): 1036 -1042 [4] V Neduva et al. Systematic discovery of new recognition peptides mediating protein interaction networks. PLo. S Biol. , 3(12): e 405, 2005. [5] N E Davey et al. SLi. MDisc: short, linear motif discovery, correcting for common evolutionary descent. Nucleic Acids Res. , 34(12): 3546– 3554, 2006. [6] R J Edwards et al. Slim. Finder: a probabilistic method for identifying overrepresented, convergently evolved, short linear motifs in proteins. PLo. S ONE, 2(10): e(967), 2007.
References [1] P Puntervoll et al. ELM server: A new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. , 31(13): 3625– 3630, 2003. [2] S Rajasekaran et al. Minimotif miner 2 nd release: a database and web system for motif search. Nucleic Acids Res. , 37(Database issue): D 185– 190, 2009. [3] W Hugo et al. SLi. M on Diet: finding short linear motifs on domain interaction interfaces in Protein Data Bank. Bioinformatics 2010 26(8): 1036 -1042 [4] V Neduva et al. Systematic discovery of new recognition peptides mediating protein interaction networks. PLo. S Biol. , 3(12): e 405, 2005. [5] N E Davey et al. SLi. MDisc: short, linear motif discovery, correcting for common evolutionary descent. Nucleic Acids Res. , 34(12): 3546– 3554, 2006. [6] R J Edwards et al. Slim. Finder: a probabilistic method for identifying overrepresented, convergently evolved, short linear motifs in proteins. PLo. S ONE, 2(10): e(967), 2007.
![References (2) [7] S H Tan et al. A correlated motif approach for finding References (2) [7] S H Tan et al. A correlated motif approach for finding](https://present5.com/presentation/9dfc01aca37f9177eb6dc50ef4488f5f/image-23.jpg) References (2) [7] S H Tan et al. A correlated motif approach for finding short linear motifs from protein interaction networks. BMC Bioinformatics, 7: 502, 2006. [8] H C Leung et al. Clustering-based approach for predicting motif pairs from protein interaction data. J Bioinform Comput Biol. 2009 Aug; 7(4): 701 -16. [9] P Boyen et al. SLIDER: Mining correlated motifs in protein-protein interaction networks. In Proceedings of the 2009 Ninth IEEE International Conference on Data Mining, pages 716– 721, 2009.
References (2) [7] S H Tan et al. A correlated motif approach for finding short linear motifs from protein interaction networks. BMC Bioinformatics, 7: 502, 2006. [8] H C Leung et al. Clustering-based approach for predicting motif pairs from protein interaction data. J Bioinform Comput Biol. 2009 Aug; 7(4): 701 -16. [9] P Boyen et al. SLIDER: Mining correlated motifs in protein-protein interaction networks. In Proceedings of the 2009 Ninth IEEE International Conference on Data Mining, pages 716– 721, 2009.


