11.2A DNA Replication RNA 1.pptx
- Количество слайдов: 53
 DNA Replication, RNA Structure & Function, and Compare DNA & RNA CIE Biology Jones pp 111 -122 Learning Objective G 11 Biology 2017 -2018 1. Describe the process of DNA replication based on Chargaff rules. 2. Distinguish between types of RNA structure and function. 3. Compare the structure of RNA and DNA.
DNA Replication, RNA Structure & Function, and Compare DNA & RNA CIE Biology Jones pp 111 -122 Learning Objective G 11 Biology 2017 -2018 1. Describe the process of DNA replication based on Chargaff rules. 2. Distinguish between types of RNA structure and function. 3. Compare the structure of RNA and DNA.
 Success Criteria 1. Can apply Chargaff’s rule to calculate a correct number of X bases presented from the information taken from Y bases. Know and explain two rules: 1. Quantity A = Quantity T • Quantity G = Quantity C 2. Relative quantity of DNA varies from one sample to another one particularly in relative quantity of reasons ATGC 2. Know and understand the process of DNA replication. Apply knowledge in completing diagram. Use correctly and explain terms. 3. Analyze structure of RNA and DNA molecules. Find similarities and differences. Complete the table correctly. Meselson and Stahl (watch until 5. 18 min) https: //www. youtube. com/watch? v=4 gd. WOWjio. BE&t=52 s Professor Dave DNA replication (6. 14 min) https: //www. youtube. com/watch? v=9 kp 9 wi. YMQUU Mrs Cooper DNA replication (13 min) https: //www. youtube. com/watch? v=6 Nh. DY 3 IDp 00
Success Criteria 1. Can apply Chargaff’s rule to calculate a correct number of X bases presented from the information taken from Y bases. Know and explain two rules: 1. Quantity A = Quantity T • Quantity G = Quantity C 2. Relative quantity of DNA varies from one sample to another one particularly in relative quantity of reasons ATGC 2. Know and understand the process of DNA replication. Apply knowledge in completing diagram. Use correctly and explain terms. 3. Analyze structure of RNA and DNA molecules. Find similarities and differences. Complete the table correctly. Meselson and Stahl (watch until 5. 18 min) https: //www. youtube. com/watch? v=4 gd. WOWjio. BE&t=52 s Professor Dave DNA replication (6. 14 min) https: //www. youtube. com/watch? v=9 kp 9 wi. YMQUU Mrs Cooper DNA replication (13 min) https: //www. youtube. com/watch? v=6 Nh. DY 3 IDp 00
 Terminology – Replication flash cards https: //quizlet. com/140046226/as-biology-dna-replication-and-the-genetic-code-flash- cards English Google Russian Origin, Origin of Replication fork Chargaff’s Rules Sense, coding, non-template Antisense, non-coding, template Conservative, Semi-conservative Topoisomerases (gyrases) DNA helicase Single Stranded Binding Proteins (SSBP) Nucleases – break down DNA Okazaki fragments DNA polymerase III DNA polymerase I RNA Primase DNA Ligase Replication bubbles Daughter strand Continuous, discontinuous Происхождение, Происхождение репликации Репликационная вилка Прайс-лист Смысл, кодирование, не шаблон Антисмысловой, некодирующий, шаблон Консервативный, полуконсервативный Топоизомеразы (гираны) ДНК-геликаза Однослойные связывающие белки (SSBP) Нуклеазы - разрушить ДНК Окадзаки ДНК-полимераза III ДНК-полимераза I РНК Primase ДНК-лигаза Репликационные пузыри Дочерняя нить Непрерывный, прерывистый
Terminology – Replication flash cards https: //quizlet. com/140046226/as-biology-dna-replication-and-the-genetic-code-flash- cards English Google Russian Origin, Origin of Replication fork Chargaff’s Rules Sense, coding, non-template Antisense, non-coding, template Conservative, Semi-conservative Topoisomerases (gyrases) DNA helicase Single Stranded Binding Proteins (SSBP) Nucleases – break down DNA Okazaki fragments DNA polymerase III DNA polymerase I RNA Primase DNA Ligase Replication bubbles Daughter strand Continuous, discontinuous Происхождение, Происхождение репликации Репликационная вилка Прайс-лист Смысл, кодирование, не шаблон Антисмысловой, некодирующий, шаблон Консервативный, полуконсервативный Топоизомеразы (гираны) ДНК-геликаза Однослойные связывающие белки (SSBP) Нуклеазы - разрушить ДНК Окадзаки ДНК-полимераза III ДНК-полимераза I РНК Primase ДНК-лигаза Репликационные пузыри Дочерняя нить Непрерывный, прерывистый
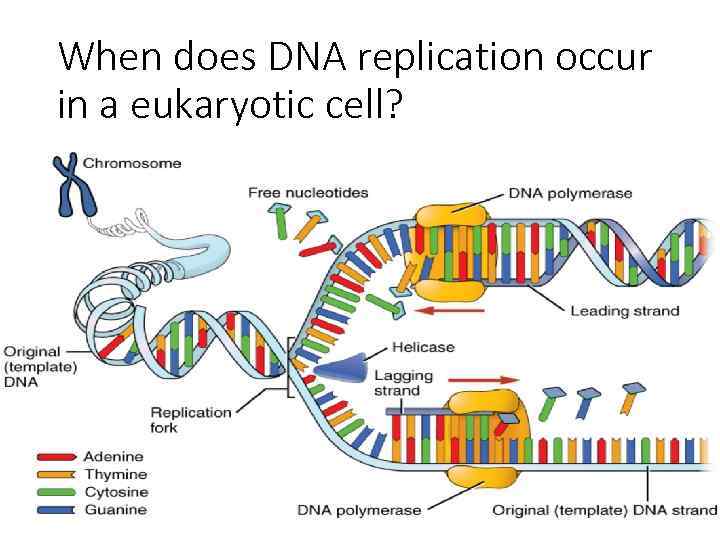 When does DNA replication occur in a eukaryotic cell?
When does DNA replication occur in a eukaryotic cell?
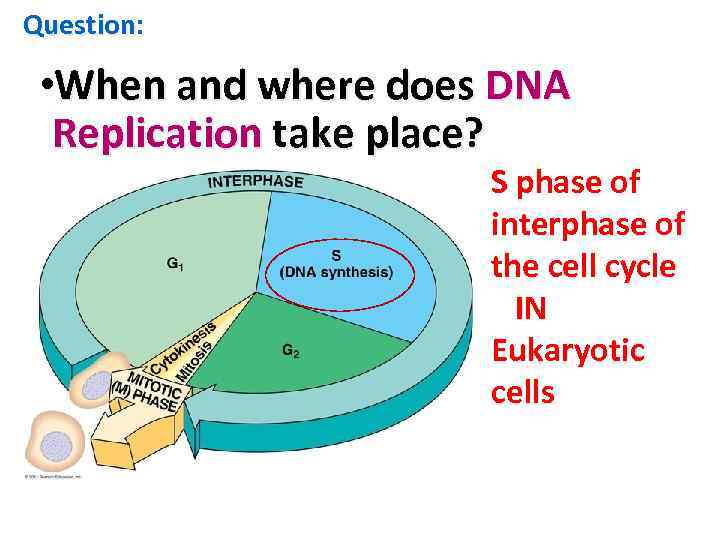 Question: • When and where does DNA Replication take place? S phase of interphase of the cell cycle IN Eukaryotic cells
Question: • When and where does DNA Replication take place? S phase of interphase of the cell cycle IN Eukaryotic cells
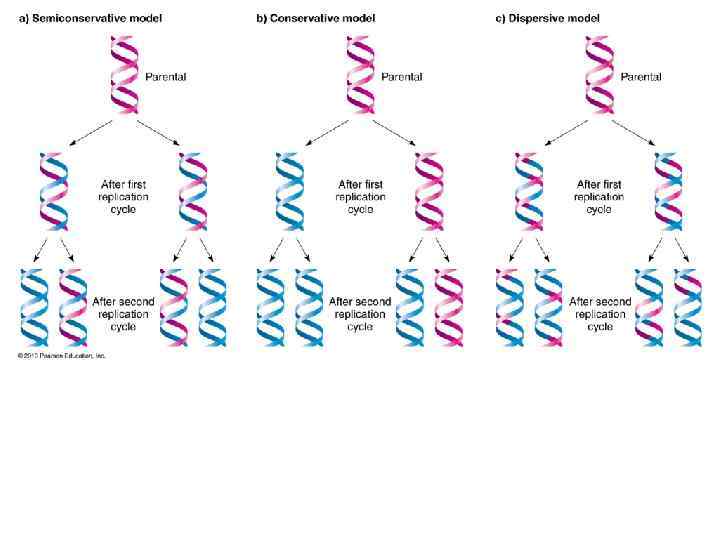
 Mechanisms of DNA Replication Meselsohn and Stahl Experiment There were three theories of how DNA might replication.
Mechanisms of DNA Replication Meselsohn and Stahl Experiment There were three theories of how DNA might replication.
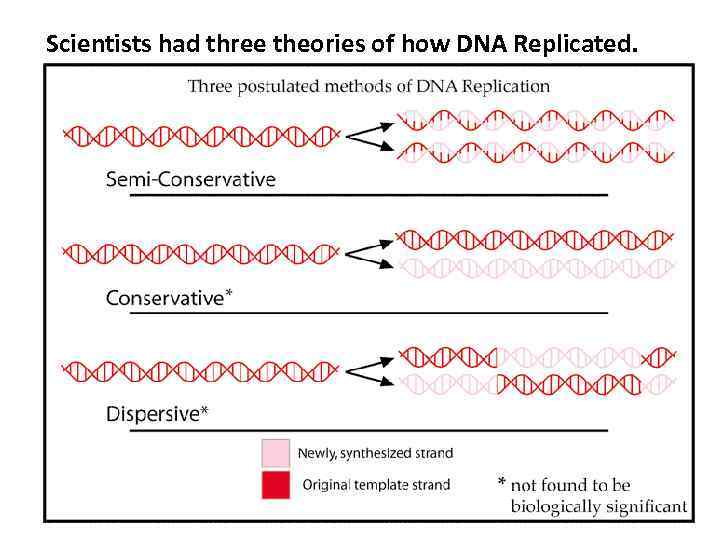 Scientists had three theories of how DNA Replicated.
Scientists had three theories of how DNA Replicated.
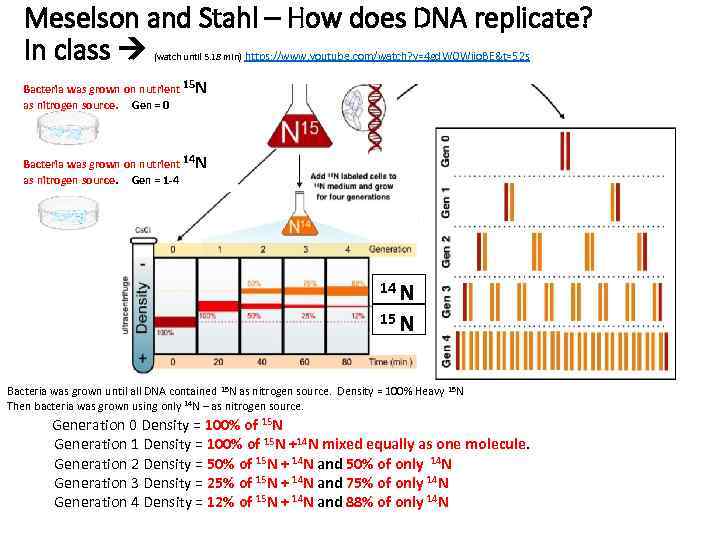 Meselson and Stahl – How does DNA replicate? In class (watch until 5. 18 min) https: //www. youtube. com/watch? v=4 gd. WOWjio. BE&t=52 s Bacteria was grown on nutrient 15 N as nitrogen source. Gen = 0 Bacteria was grown on nutrient 14 N as nitrogen source. Gen = 1 -4 14 N 15 N Bacteria was grown until all DNA contained 15 N as nitrogen source. Density = 100% Heavy 15 N Then bacteria was grown using only 14 N – as nitrogen source. Generation 0 Density = 100% of 15 N Generation 1 Density = 100% of 15 N +14 N mixed equally as one molecule. Generation 2 Density = 50% of 15 N + 14 N and 50% of only 14 N Generation 3 Density = 25% of 15 N + 14 N and 75% of only 14 N Generation 4 Density = 12% of 15 N + 14 N and 88% of only 14 N
Meselson and Stahl – How does DNA replicate? In class (watch until 5. 18 min) https: //www. youtube. com/watch? v=4 gd. WOWjio. BE&t=52 s Bacteria was grown on nutrient 15 N as nitrogen source. Gen = 0 Bacteria was grown on nutrient 14 N as nitrogen source. Gen = 1 -4 14 N 15 N Bacteria was grown until all DNA contained 15 N as nitrogen source. Density = 100% Heavy 15 N Then bacteria was grown using only 14 N – as nitrogen source. Generation 0 Density = 100% of 15 N Generation 1 Density = 100% of 15 N +14 N mixed equally as one molecule. Generation 2 Density = 50% of 15 N + 14 N and 50% of only 14 N Generation 3 Density = 25% of 15 N + 14 N and 75% of only 14 N Generation 4 Density = 12% of 15 N + 14 N and 88% of only 14 N
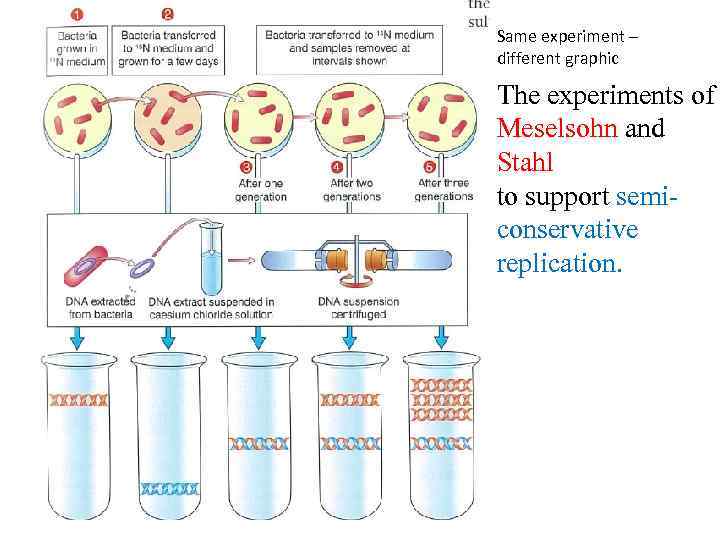 Same experiment – different graphic The experiments of Meselsohn and Stahl to support semiconservative replication.
Same experiment – different graphic The experiments of Meselsohn and Stahl to support semiconservative replication.
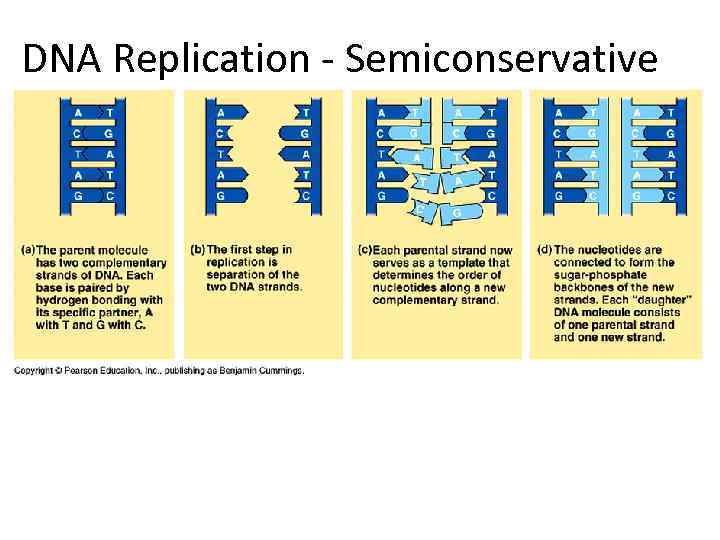 DNA Replication - Semiconservative
DNA Replication - Semiconservative
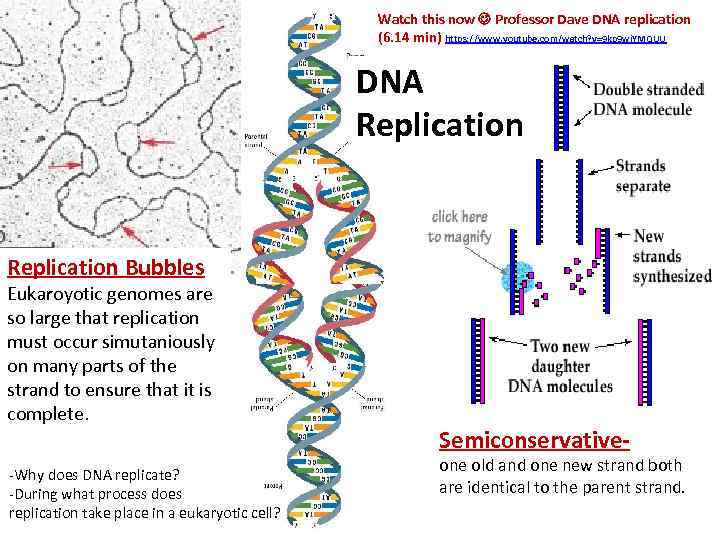 Watch this now Professor Dave DNA replication (6. 14 min) https: //www. youtube. com/watch? v=9 kp 9 wi. YMQUU DNA Replication Bubbles Eukaroyotic genomes are so large that replication must occur simutaniously on many parts of the strand to ensure that it is complete. -Why does DNA replicate? -During what process does replication take place in a eukaryotic cell? Semiconservative- one old and one new strand both are identical to the parent strand.
Watch this now Professor Dave DNA replication (6. 14 min) https: //www. youtube. com/watch? v=9 kp 9 wi. YMQUU DNA Replication Bubbles Eukaroyotic genomes are so large that replication must occur simutaniously on many parts of the strand to ensure that it is complete. -Why does DNA replicate? -During what process does replication take place in a eukaryotic cell? Semiconservative- one old and one new strand both are identical to the parent strand.
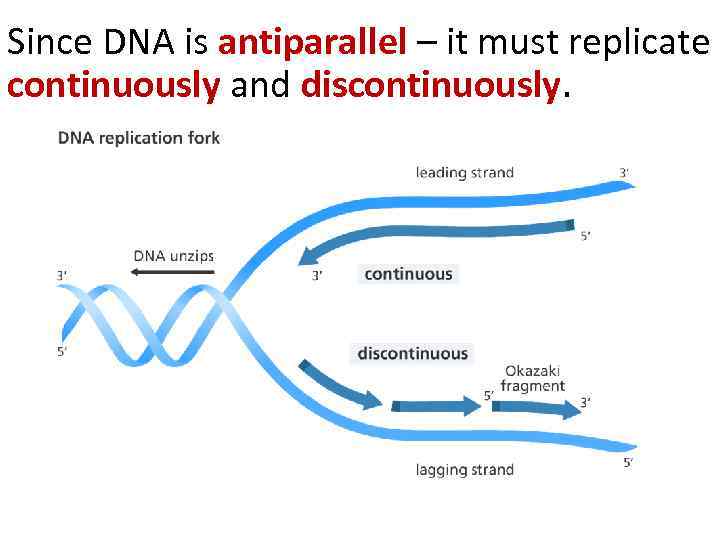 Since DNA is antiparallel – it must replicate continuously and discontinuously.
Since DNA is antiparallel – it must replicate continuously and discontinuously.
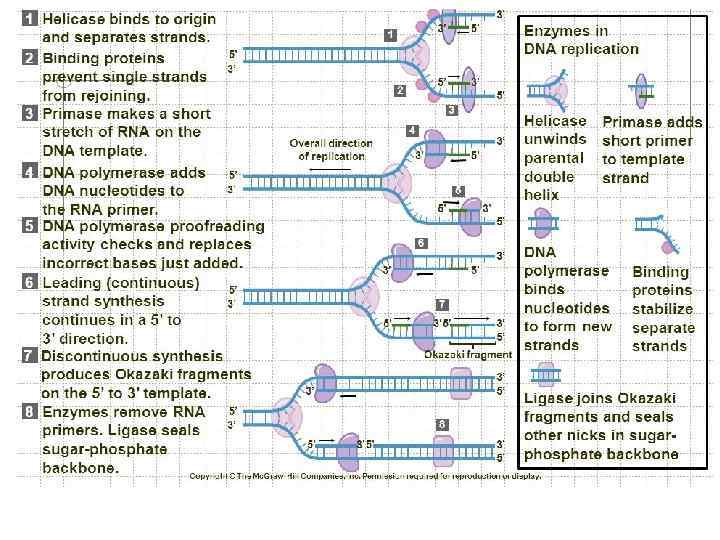
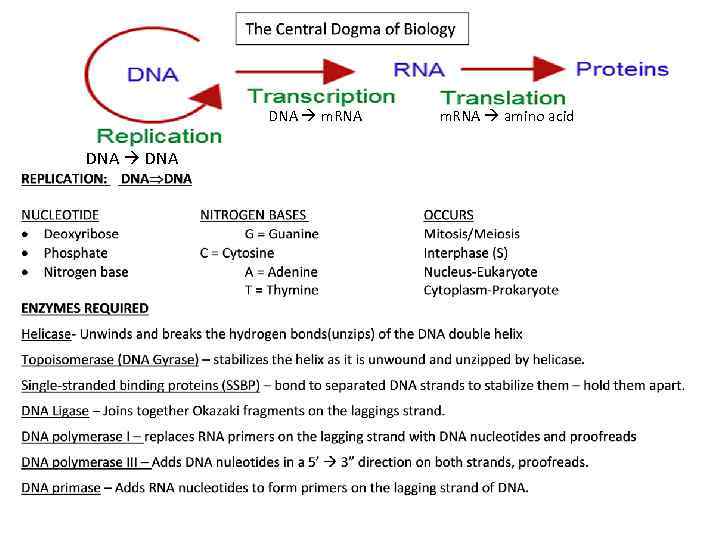
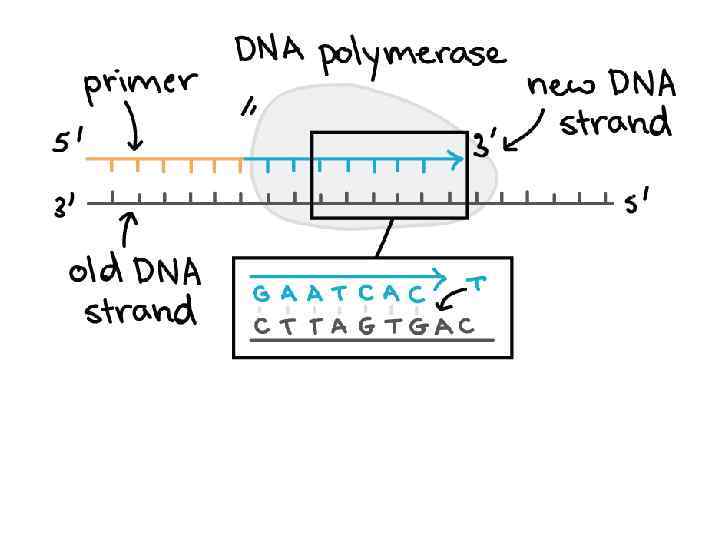
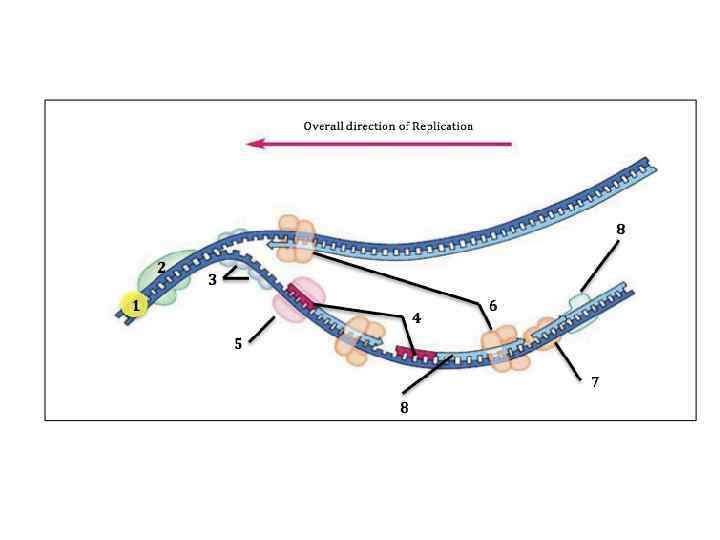
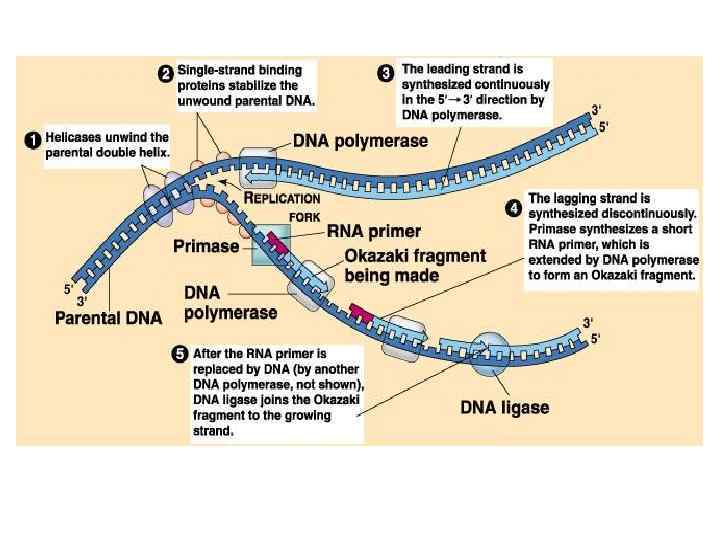
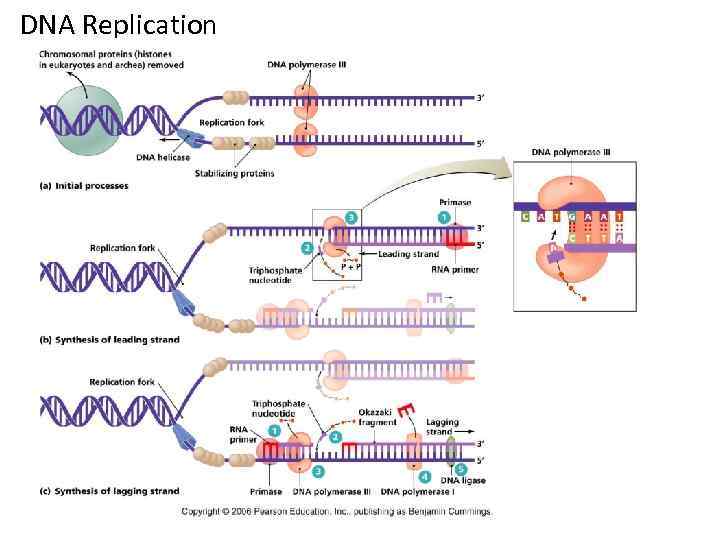
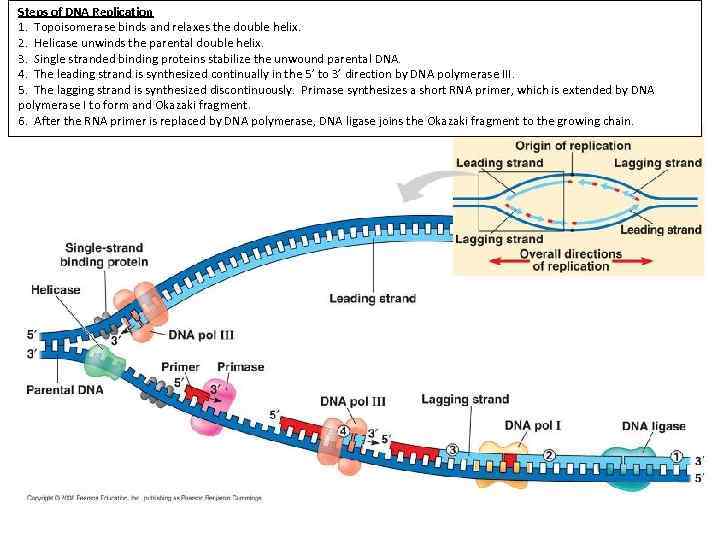 Steps of DNA Replication 1. Topoisomerase binds and relaxes the double helix. 2. Helicase unwinds the parental double helix. 3. Single stranded binding proteins stabilize the unwound parental DNA. 4. The leading strand is synthesized continually in the 5’ to 3’ direction by DNA polymerase III. 5. The lagging strand is synthesized discontinuously. Primase synthesizes a short RNA primer, which is extended by DNA polymerase I to form and Okazaki fragment. 6. After the RNA primer is replaced by DNA polymerase, DNA ligase joins the Okazaki fragment to the growing chain.
Steps of DNA Replication 1. Topoisomerase binds and relaxes the double helix. 2. Helicase unwinds the parental double helix. 3. Single stranded binding proteins stabilize the unwound parental DNA. 4. The leading strand is synthesized continually in the 5’ to 3’ direction by DNA polymerase III. 5. The lagging strand is synthesized discontinuously. Primase synthesizes a short RNA primer, which is extended by DNA polymerase I to form and Okazaki fragment. 6. After the RNA primer is replaced by DNA polymerase, DNA ligase joins the Okazaki fragment to the growing chain.
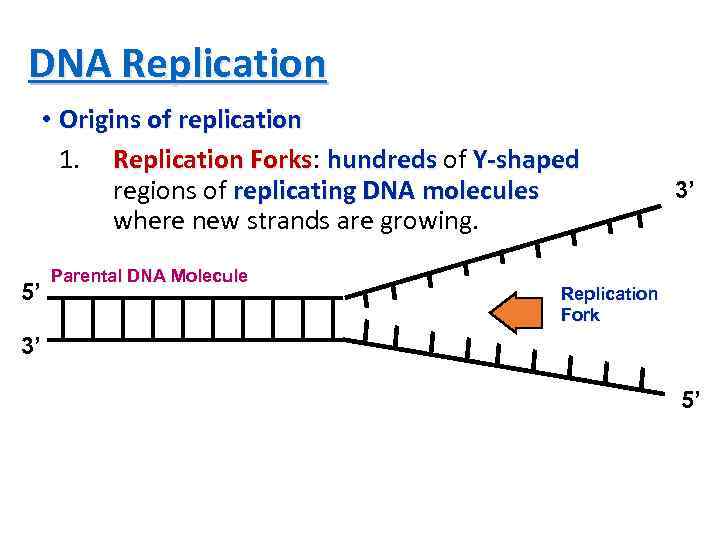 DNA Replication • Origins of replication 1. Replication Forks: hundreds of Y-shaped Forks regions of replicating DNA molecules where new strands are growing. 5’ Parental DNA Molecule 3’ Replication Fork 3’ 5’
DNA Replication • Origins of replication 1. Replication Forks: hundreds of Y-shaped Forks regions of replicating DNA molecules where new strands are growing. 5’ Parental DNA Molecule 3’ Replication Fork 3’ 5’
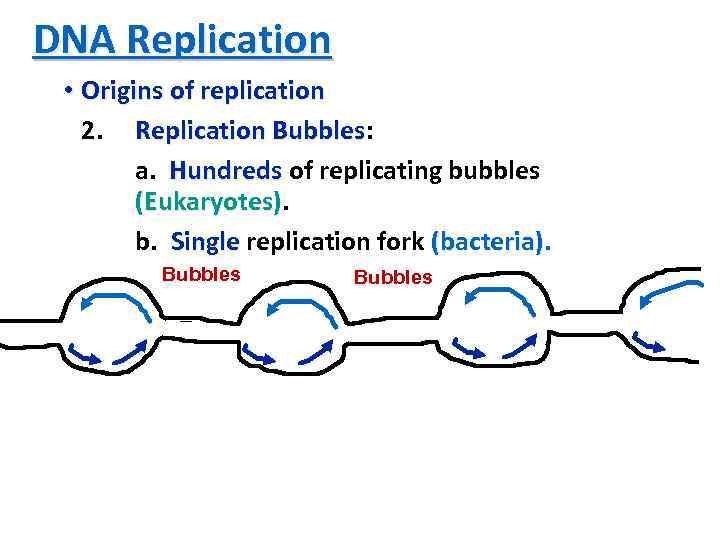 DNA Replication • Origins of replication 2. Replication Bubbles: Bubbles a. Hundreds of replicating bubbles (Eukaryotes) b. Single replication fork (bacteria). Bubbles
DNA Replication • Origins of replication 2. Replication Bubbles: Bubbles a. Hundreds of replicating bubbles (Eukaryotes) b. Single replication fork (bacteria). Bubbles
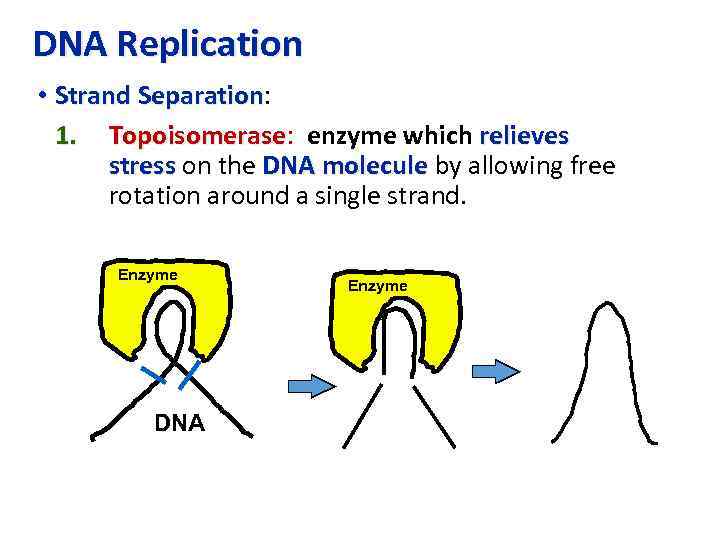 DNA Replication • Strand Separation: Separation 1. Topoisomerase: enzyme which relieves Topoisomerase stress on the DNA molecule by allowing free rotation around a single strand. Enzyme DNA Enzyme
DNA Replication • Strand Separation: Separation 1. Topoisomerase: enzyme which relieves Topoisomerase stress on the DNA molecule by allowing free rotation around a single strand. Enzyme DNA Enzyme
 DNA Replication • Strand Separation: Separation 2. Helicase: enzyme which catalyze the unwinding and Helicase separation (breaking H-Bonds) of the parental double helix. 3. Single-Strand Binding Proteins (SSBP): proteins (SSBP) which attach and help keep the separated strands apart.
DNA Replication • Strand Separation: Separation 2. Helicase: enzyme which catalyze the unwinding and Helicase separation (breaking H-Bonds) of the parental double helix. 3. Single-Strand Binding Proteins (SSBP): proteins (SSBP) which attach and help keep the separated strands apart.
 DNA Replication • Priming: 2. RNA primers: before new DNA strands can form, primers there must be small pre-existing primers (RNA) present to start the addition of new nucleotides (DNA Polymerase) 3. Primase: enzyme that polymerizes (synthesizes) Primase the RNA Primer.
DNA Replication • Priming: 2. RNA primers: before new DNA strands can form, primers there must be small pre-existing primers (RNA) present to start the addition of new nucleotides (DNA Polymerase) 3. Primase: enzyme that polymerizes (synthesizes) Primase the RNA Primer.
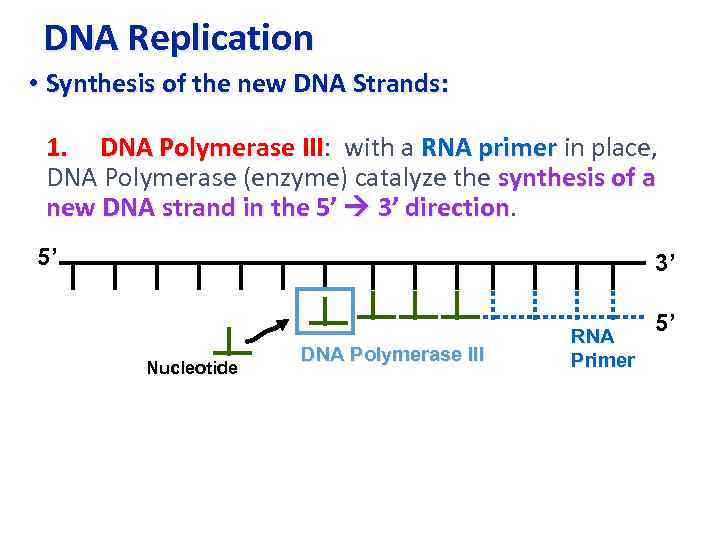 DNA Replication • Synthesis of the new DNA Strands: 1. DNA Polymerase III: with a RNA primer in place, III DNA Polymerase (enzyme) catalyze the synthesis of a new DNA strand in the 5’ 3’ direction 5’ 3’ Nucleotide DNA Polymerase III RNA Primer 5’
DNA Replication • Synthesis of the new DNA Strands: 1. DNA Polymerase III: with a RNA primer in place, III DNA Polymerase (enzyme) catalyze the synthesis of a new DNA strand in the 5’ 3’ direction 5’ 3’ Nucleotide DNA Polymerase III RNA Primer 5’
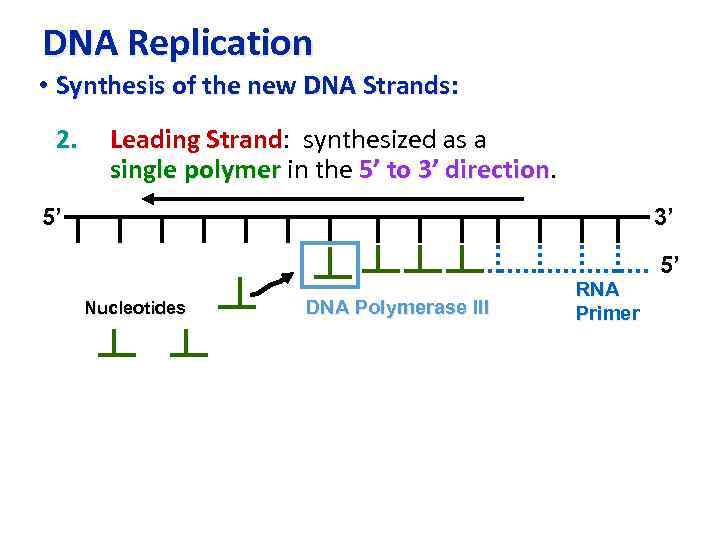 DNA Replication • Synthesis of the new DNA Strands: 2. Leading Strand: synthesized as a Strand single polymer in the 5’ to 3’ direction 5’ 3’ 5’ Nucleotides DNA Polymerase III RNA Primer
DNA Replication • Synthesis of the new DNA Strands: 2. Leading Strand: synthesized as a Strand single polymer in the 5’ to 3’ direction 5’ 3’ 5’ Nucleotides DNA Polymerase III RNA Primer
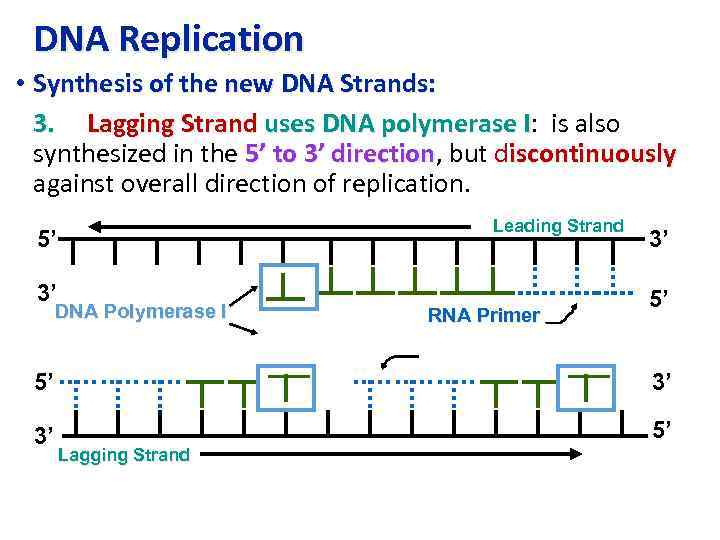 DNA Replication • Synthesis of the new DNA Strands: 3. Lagging Strand uses DNA polymerase I: is also I synthesized in the 5’ to 3’ direction, but discontinuously direction against overall direction of replication. Leading Strand 5’ 3’ DNA Polymerase I RNA Primer 3’ 5’ 5’ 3’ 3’ 5’ Lagging Strand
DNA Replication • Synthesis of the new DNA Strands: 3. Lagging Strand uses DNA polymerase I: is also I synthesized in the 5’ to 3’ direction, but discontinuously direction against overall direction of replication. Leading Strand 5’ 3’ DNA Polymerase I RNA Primer 3’ 5’ 5’ 3’ 3’ 5’ Lagging Strand
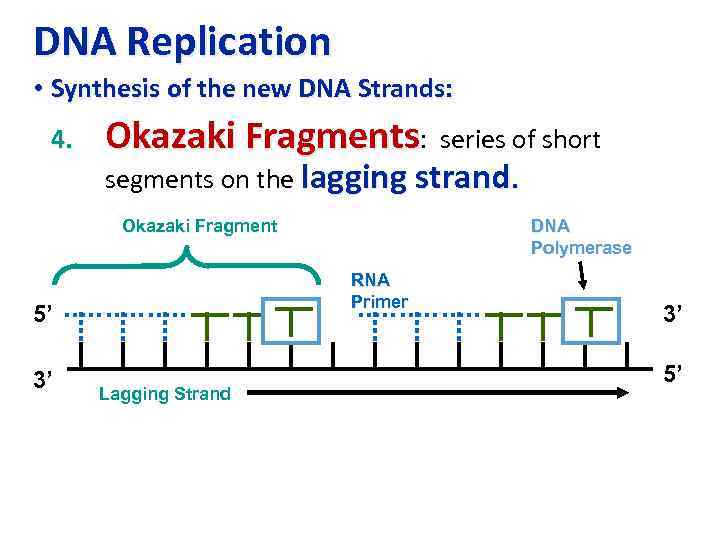 DNA Replication • Synthesis of the new DNA Strands: 4. Okazaki Fragments: series of short segments on the lagging strand. Okazaki Fragment RNA Primer 5’ 3’ DNA Polymerase Lagging Strand 3’ 5’
DNA Replication • Synthesis of the new DNA Strands: 4. Okazaki Fragments: series of short segments on the lagging strand. Okazaki Fragment RNA Primer 5’ 3’ DNA Polymerase Lagging Strand 3’ 5’
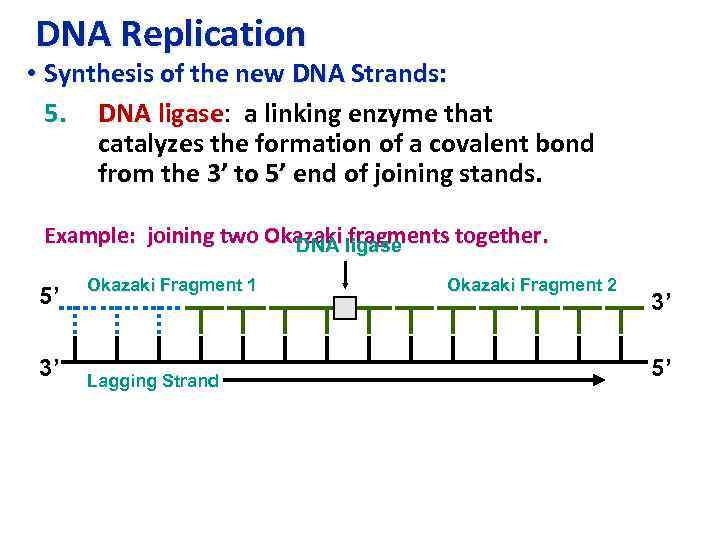 DNA Replication • Synthesis of the new DNA Strands: 5. DNA ligase: a linking enzyme that ligase catalyzes the formation of a covalent bond from the 3’ to 5’ end of joining stands. Example: joining two Okazaki ligase DNA fragments together. 5’ 3’ Okazaki Fragment 1 Lagging Strand Okazaki Fragment 2 3’ 5’
DNA Replication • Synthesis of the new DNA Strands: 5. DNA ligase: a linking enzyme that ligase catalyzes the formation of a covalent bond from the 3’ to 5’ end of joining stands. Example: joining two Okazaki ligase DNA fragments together. 5’ 3’ Okazaki Fragment 1 Lagging Strand Okazaki Fragment 2 3’ 5’
 DNA Replication • Synthesis of the new DNA Strands: 6. Proofreading: initial base-pairing Proofreading errors may be corrected by DNA polymerase I or III
DNA Replication • Synthesis of the new DNA Strands: 6. Proofreading: initial base-pairing Proofreading errors may be corrected by DNA polymerase I or III
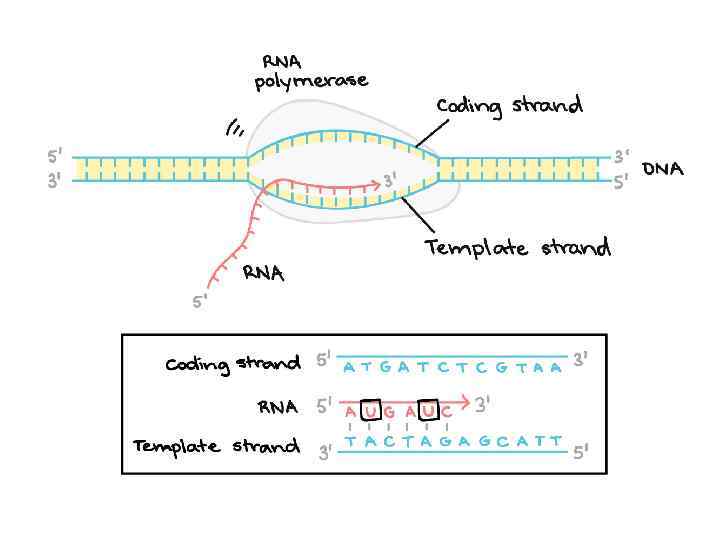
 DNA m. RNA DNA m. RNA amino acid
DNA m. RNA DNA m. RNA amino acid
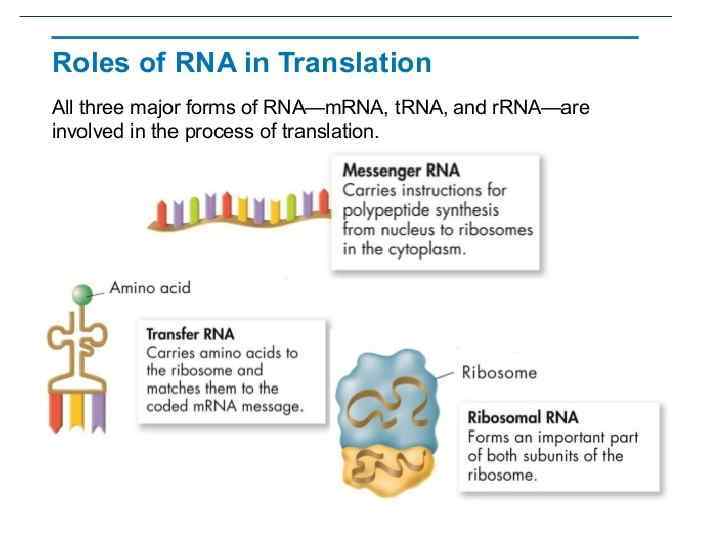
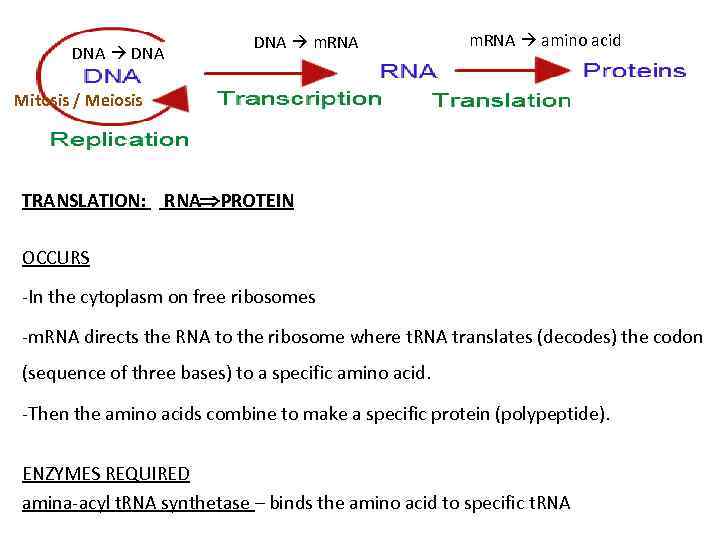 DNA m. RNA amino acid Mitosis / Meiosis TRANSLATION: RNA PROTEIN OCCURS -In the cytoplasm on free ribosomes -m. RNA directs the RNA to the ribosome where t. RNA translates (decodes) the codon (sequence of three bases) to a specific amino acid. -Then the amino acids combine to make a specific protein (polypeptide). ENZYMES REQUIRED amina-acyl t. RNA synthetase – binds the amino acid to specific t. RNA
DNA m. RNA amino acid Mitosis / Meiosis TRANSLATION: RNA PROTEIN OCCURS -In the cytoplasm on free ribosomes -m. RNA directs the RNA to the ribosome where t. RNA translates (decodes) the codon (sequence of three bases) to a specific amino acid. -Then the amino acids combine to make a specific protein (polypeptide). ENZYMES REQUIRED amina-acyl t. RNA synthetase – binds the amino acid to specific t. RNA
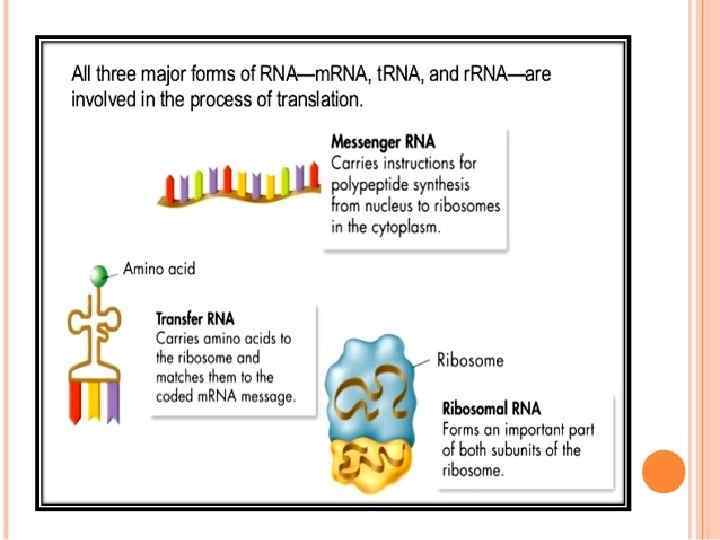
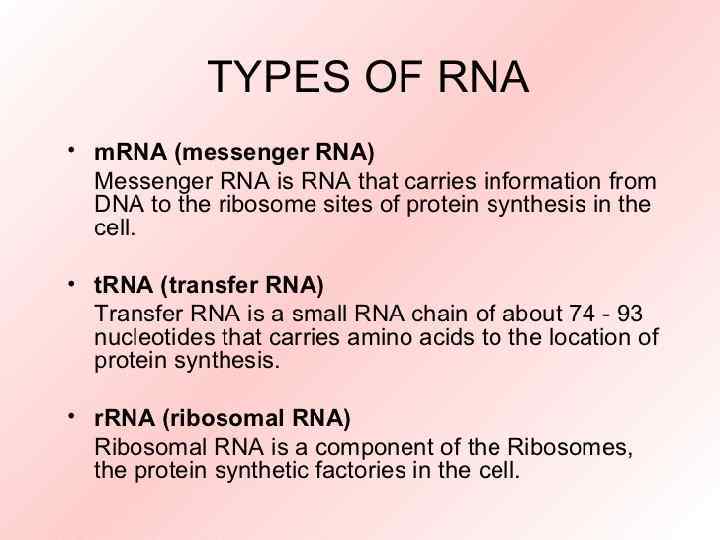
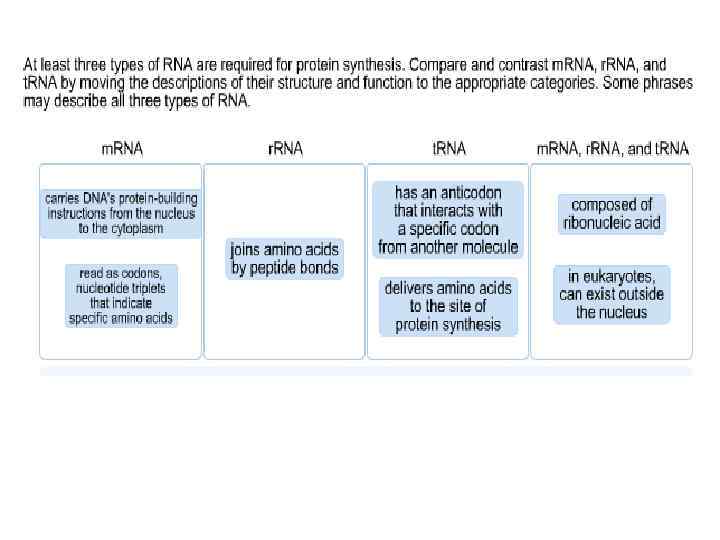
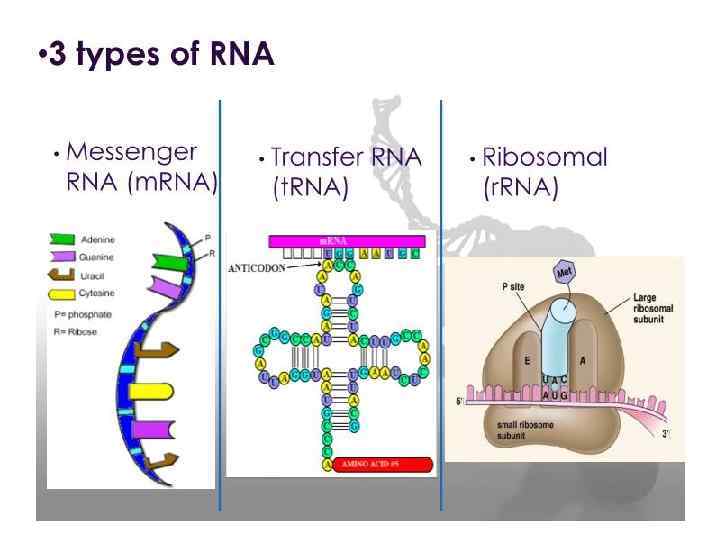
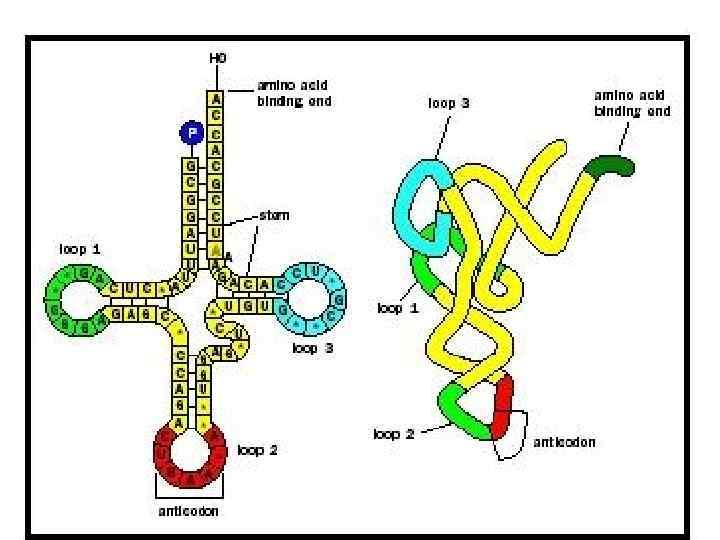
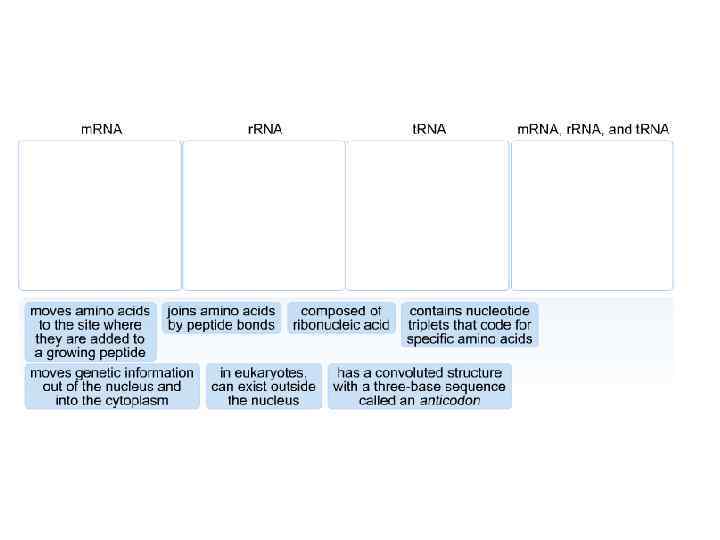
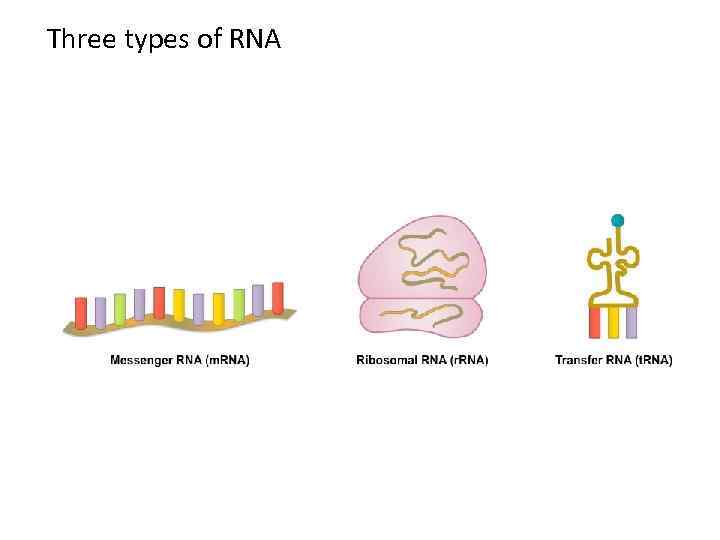
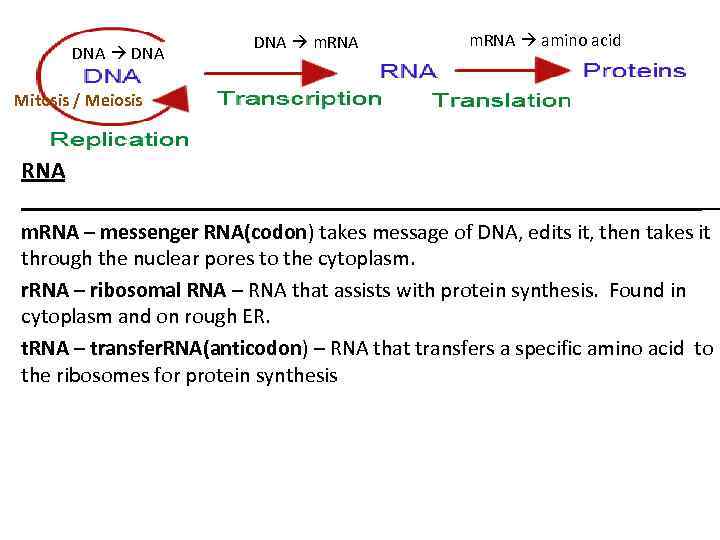 DNA m. RNA amino acid Mitosis / Meiosis RNA _____________________________ m. RNA – messenger RNA(codon) takes message of DNA, edits it, then takes it through the nuclear pores to the cytoplasm. r. RNA – ribosomal RNA – RNA that assists with protein synthesis. Found in cytoplasm and on rough ER. t. RNA – transfer. RNA(anticodon) – RNA that transfers a specific amino acid to the ribosomes for protein synthesis
DNA m. RNA amino acid Mitosis / Meiosis RNA _____________________________ m. RNA – messenger RNA(codon) takes message of DNA, edits it, then takes it through the nuclear pores to the cytoplasm. r. RNA – ribosomal RNA – RNA that assists with protein synthesis. Found in cytoplasm and on rough ER. t. RNA – transfer. RNA(anticodon) – RNA that transfers a specific amino acid to the ribosomes for protein synthesis
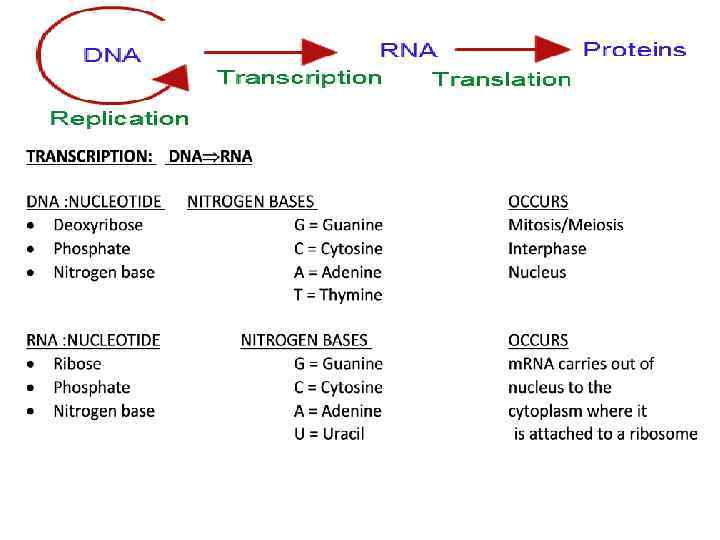
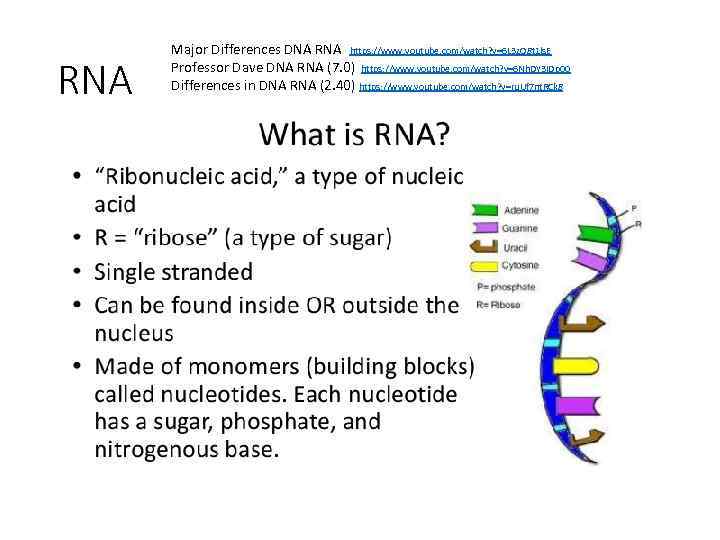 RNA Major Differences DNA RNA https: //www. youtube. com/watch? v=6 L 3 z. O 8 t 1 ls. E Professor Dave DNA RNA (7. 0) https: //www. youtube. com/watch? v=6 Nh. DY 3 IDp 00 Differences in DNA RNA (2. 40) https: //www. youtube. com/watch? v=ru. Uf 7 nt. RCk 8
RNA Major Differences DNA RNA https: //www. youtube. com/watch? v=6 L 3 z. O 8 t 1 ls. E Professor Dave DNA RNA (7. 0) https: //www. youtube. com/watch? v=6 Nh. DY 3 IDp 00 Differences in DNA RNA (2. 40) https: //www. youtube. com/watch? v=ru. Uf 7 nt. RCk 8
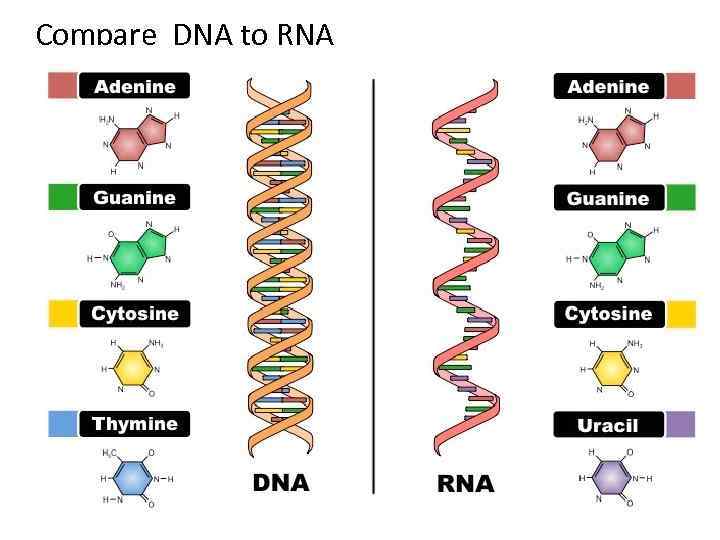 Compare DNA to RNA
Compare DNA to RNA
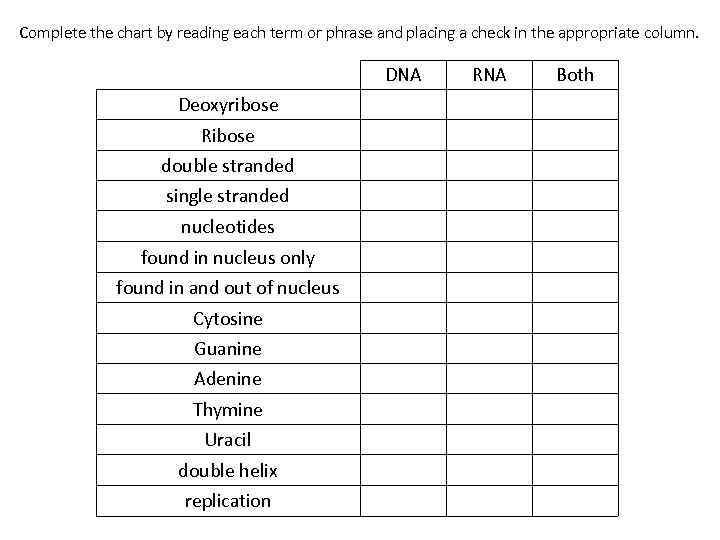 Complete the chart by reading each term or phrase and placing a check in the appropriate column. DNA Deoxyribose single stranded nucleotides found in nucleus only Both Ribose double stranded RNA found in and out of nucleus Cytosine Guanine Adenine Thymine Uracil double helix replication
Complete the chart by reading each term or phrase and placing a check in the appropriate column. DNA Deoxyribose single stranded nucleotides found in nucleus only Both Ribose double stranded RNA found in and out of nucleus Cytosine Guanine Adenine Thymine Uracil double helix replication
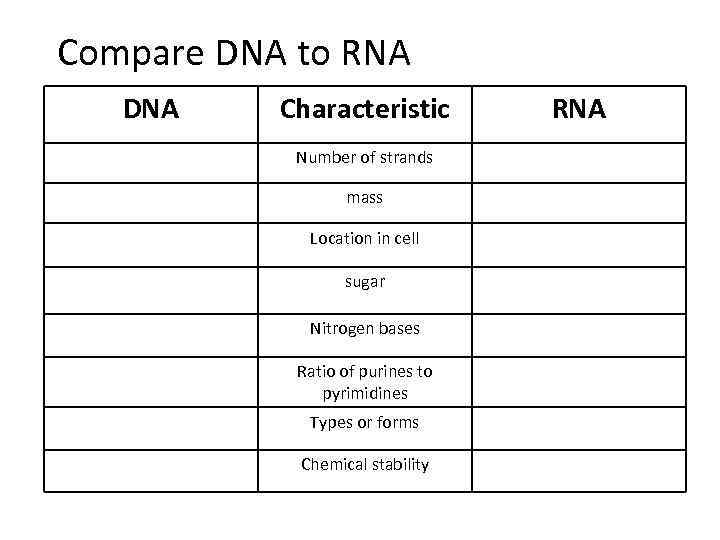 Compare DNA to RNA DNA Characteristic Number of strands mass Location in cell sugar Nitrogen bases Ratio of purines to pyrimidines Types or forms Chemical stability RNA
Compare DNA to RNA DNA Characteristic Number of strands mass Location in cell sugar Nitrogen bases Ratio of purines to pyrimidines Types or forms Chemical stability RNA
 Quiz Formative
Quiz Formative
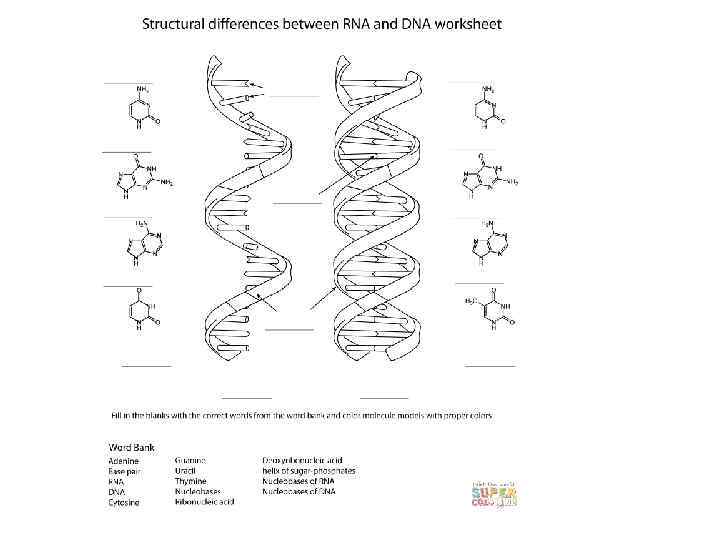
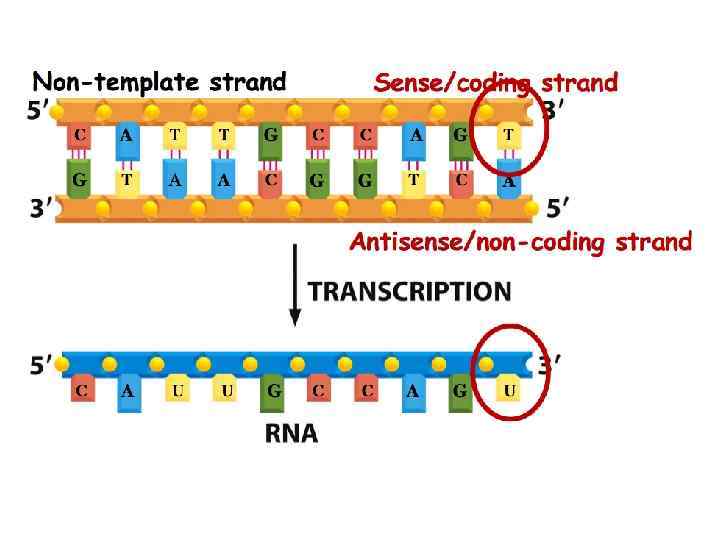
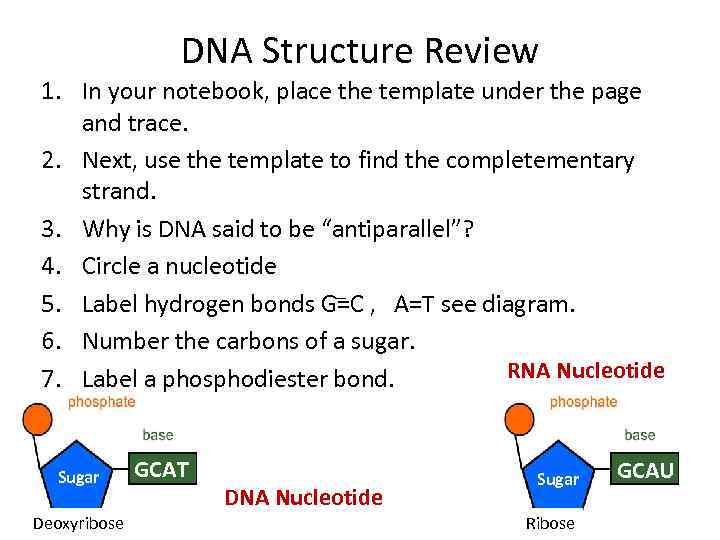 DNA Structure Review 1. In your notebook, place the template under the page and trace. 2. Next, use the template to find the completementary strand. 3. Why is DNA said to be “antiparallel”? 4. Circle a nucleotide _ 5. Label hydrogen bonds G=C , A=T see diagram. 6. Number the carbons of a sugar. RNA Nucleotide 7. Label a phosphodiester bond. Sugar Deoxyribose GCAT DNA Nucleotide Sugar Ribose GCAU
DNA Structure Review 1. In your notebook, place the template under the page and trace. 2. Next, use the template to find the completementary strand. 3. Why is DNA said to be “antiparallel”? 4. Circle a nucleotide _ 5. Label hydrogen bonds G=C , A=T see diagram. 6. Number the carbons of a sugar. RNA Nucleotide 7. Label a phosphodiester bond. Sugar Deoxyribose GCAT DNA Nucleotide Sugar Ribose GCAU
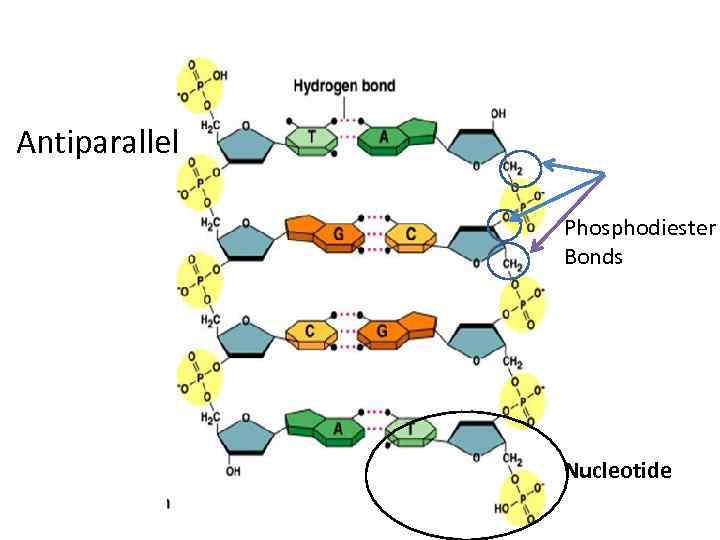 Antiparallel Phosphodiester Bonds Nucleotide
Antiparallel Phosphodiester Bonds Nucleotide
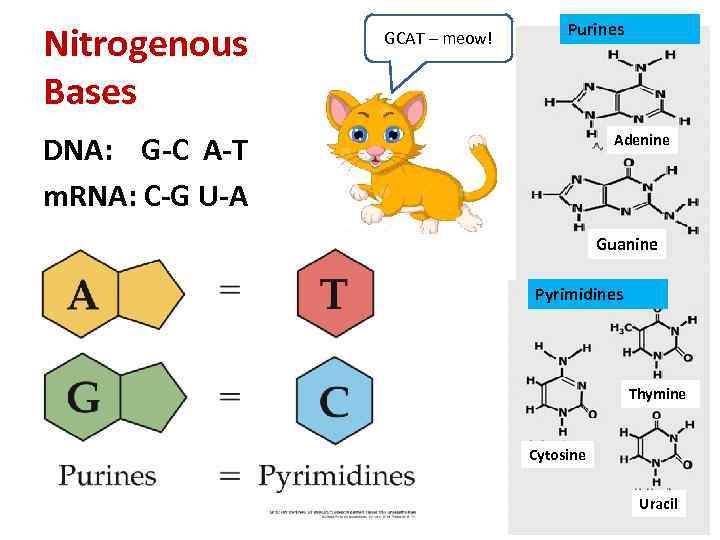 Nitrogenous Bases GCAT – meow! Purines DNA: G-C A-T m. RNA: C-G U-A Adenine Guanine Pyrimidines Thymine Cytosine Uracil
Nitrogenous Bases GCAT – meow! Purines DNA: G-C A-T m. RNA: C-G U-A Adenine Guanine Pyrimidines Thymine Cytosine Uracil
 DNA Replication
DNA Replication
 Three types of RNA
Three types of RNA