fe234ac1d2061e8c9ac307be89e425dd.ppt
- Количество слайдов: 52
 Distance functions and IE – 4? William W. Cohen CALD
Distance functions and IE – 4? William W. Cohen CALD
 Announcements • Current statistics: – days with unscheduled student talks: 6 – students with unscheduled student talks: 4 – Projects are due: 4/28 (last day of class) – Additional requirement: draft (for comments) no later than 4/21
Announcements • Current statistics: – days with unscheduled student talks: 6 – students with unscheduled student talks: 4 – Projects are due: 4/28 (last day of class) – Additional requirement: draft (for comments) no later than 4/21
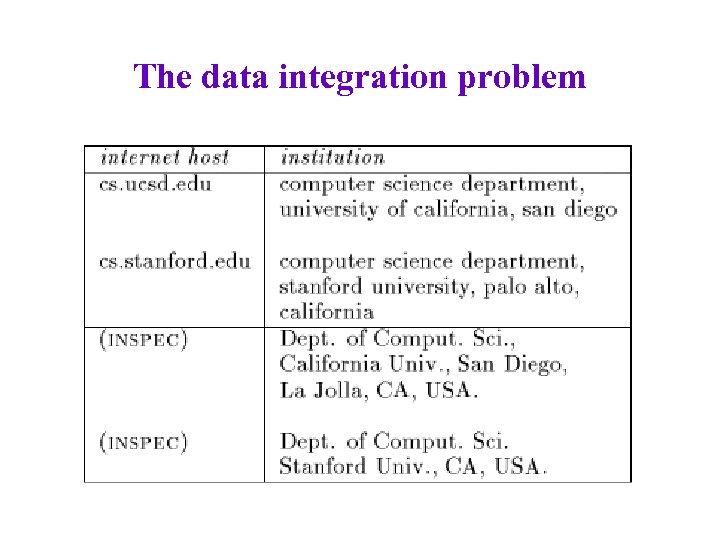 The data integration problem
The data integration problem
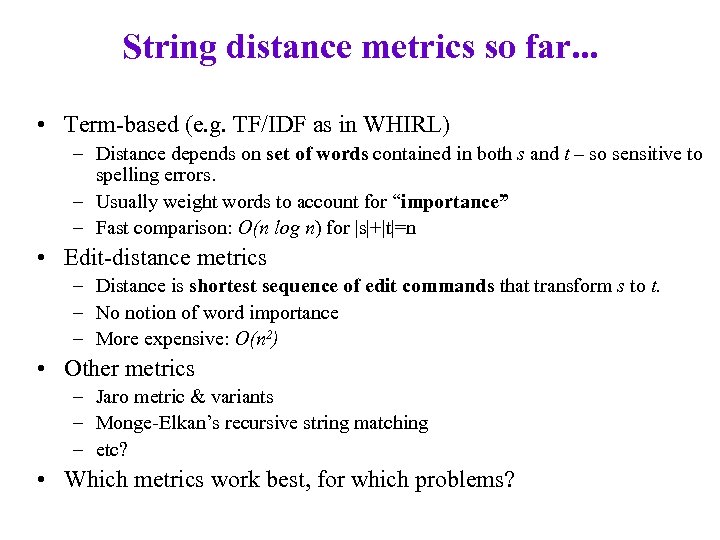 String distance metrics so far. . . • Term-based (e. g. TF/IDF as in WHIRL) – Distance depends on set of words contained in both s and t – so sensitive to spelling errors. – Usually weight words to account for “importance” – Fast comparison: O(n log n) for |s|+|t|=n • Edit-distance metrics – Distance is shortest sequence of edit commands that transform s to t. – No notion of word importance – More expensive: O(n 2) • Other metrics – Jaro metric & variants – Monge-Elkan’s recursive string matching – etc? • Which metrics work best, for which problems?
String distance metrics so far. . . • Term-based (e. g. TF/IDF as in WHIRL) – Distance depends on set of words contained in both s and t – so sensitive to spelling errors. – Usually weight words to account for “importance” – Fast comparison: O(n log n) for |s|+|t|=n • Edit-distance metrics – Distance is shortest sequence of edit commands that transform s to t. – No notion of word importance – More expensive: O(n 2) • Other metrics – Jaro metric & variants – Monge-Elkan’s recursive string matching – etc? • Which metrics work best, for which problems?
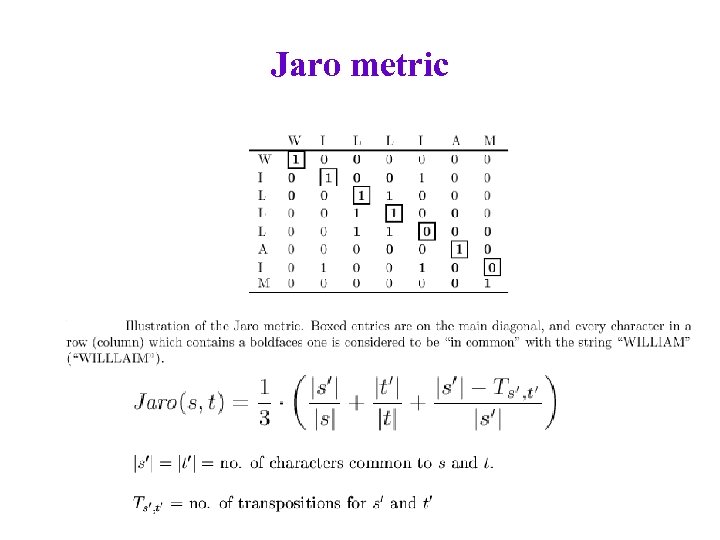 Jaro metric
Jaro metric
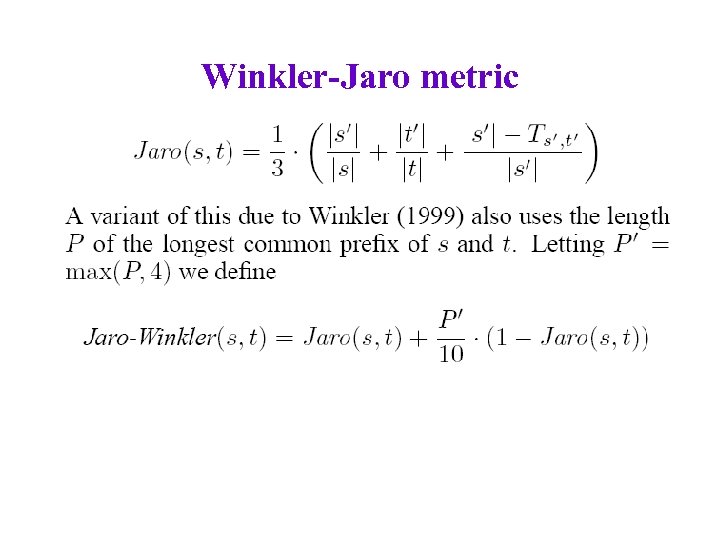 Winkler-Jaro metric
Winkler-Jaro metric
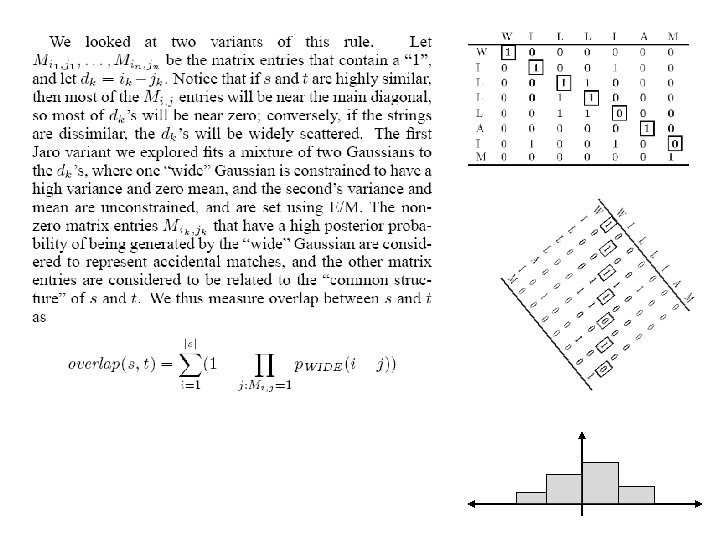
 String distance metrics so far. . . • Term-based (e. g. TF/IDF as in WHIRL) – Distance depends on set of words contained in both s and t – so sensitive to spelling errors. – Usually weight words to account for “importance” – Fast comparison: O(n log n) for |s|+|t|=n • Edit-distance metrics – Distance is shortest sequence of edit commands that transform s to t. – No notion of word importance – More expensive: O(n 2) • Other metrics – Jaro metric & variants – Monge-Elkan’s recursive string matching – etc? • Which metrics work best, for which problems?
String distance metrics so far. . . • Term-based (e. g. TF/IDF as in WHIRL) – Distance depends on set of words contained in both s and t – so sensitive to spelling errors. – Usually weight words to account for “importance” – Fast comparison: O(n log n) for |s|+|t|=n • Edit-distance metrics – Distance is shortest sequence of edit commands that transform s to t. – No notion of word importance – More expensive: O(n 2) • Other metrics – Jaro metric & variants – Monge-Elkan’s recursive string matching – etc? • Which metrics work best, for which problems?
 So which metric should you use? Second. String (Cohen, Ravikumar, Fienberg): • Java toolkit of string-matching methods from AI, Statistics, IR and DB communities • Tools for evaluating performance on test data • Exploratory tool for adding, testing, combining string distances – e. g. Second. String implements a generic “Winkler rescorer” which can rescale any distance function with range of [0, 1] • URL – http: //secondstring. sourceforge. net • Distribution also includes several sample matching problems.
So which metric should you use? Second. String (Cohen, Ravikumar, Fienberg): • Java toolkit of string-matching methods from AI, Statistics, IR and DB communities • Tools for evaluating performance on test data • Exploratory tool for adding, testing, combining string distances – e. g. Second. String implements a generic “Winkler rescorer” which can rescale any distance function with range of [0, 1] • URL – http: //secondstring. sourceforge. net • Distribution also includes several sample matching problems.
 Second. String distance functions • Edit-distance like: – Levenshtein – unit costs – untuned Smith-Waterman – Monge-Elkan (tuned Smith-Waterman) – Jaro and Jaro-Winkler
Second. String distance functions • Edit-distance like: – Levenshtein – unit costs – untuned Smith-Waterman – Monge-Elkan (tuned Smith-Waterman) – Jaro and Jaro-Winkler
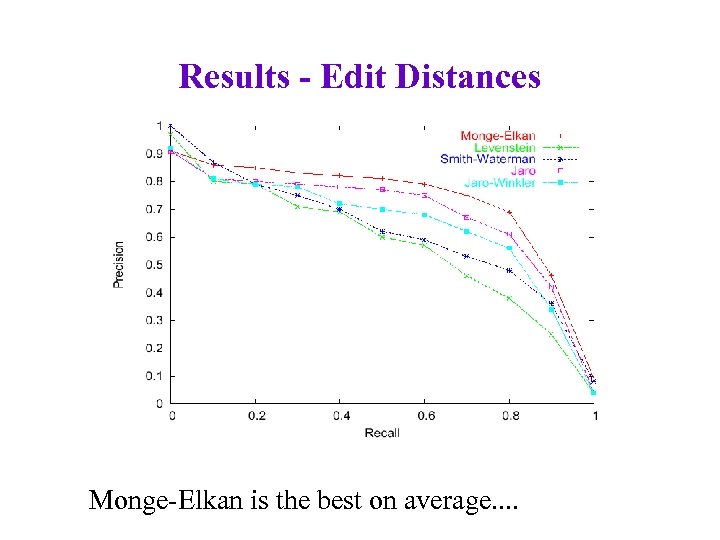 Results - Edit Distances Monge-Elkan is the best on average. .
Results - Edit Distances Monge-Elkan is the best on average. .
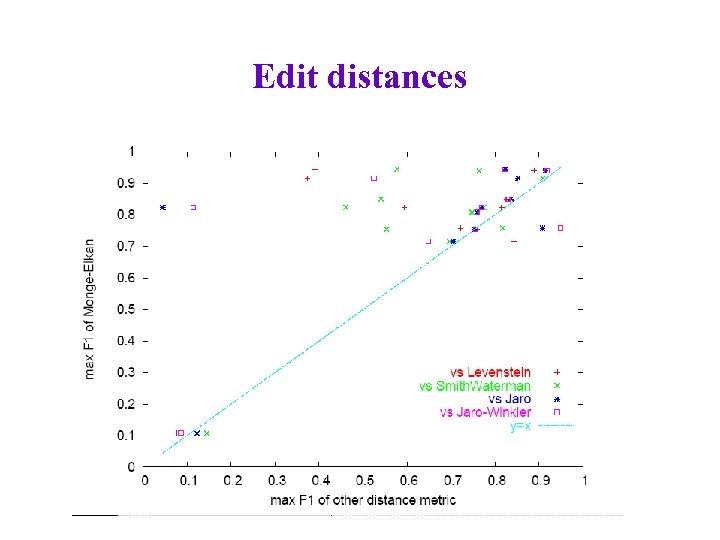 Edit distances
Edit distances
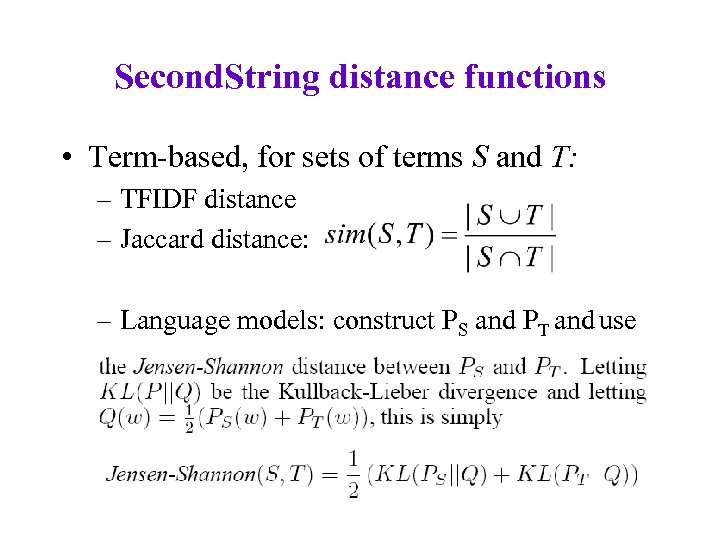 Second. String distance functions • Term-based, for sets of terms S and T: – TFIDF distance – Jaccard distance: – Language models: construct PS and PT and use
Second. String distance functions • Term-based, for sets of terms S and T: – TFIDF distance – Jaccard distance: – Language models: construct PS and PT and use
 Second. String distance functions • Term-based, for sets of terms S and T: – TFIDF distance – Jaccard distance – Jensen-Shannon distance • smoothing toward union of S, T reduces cost of disagreeing on common terms • unsmoothed PS, Dirichlet smoothing, Jelenik-Mercer – “Simplified Fellegi-Sunter”
Second. String distance functions • Term-based, for sets of terms S and T: – TFIDF distance – Jaccard distance – Jensen-Shannon distance • smoothing toward union of S, T reduces cost of disagreeing on common terms • unsmoothed PS, Dirichlet smoothing, Jelenik-Mercer – “Simplified Fellegi-Sunter”

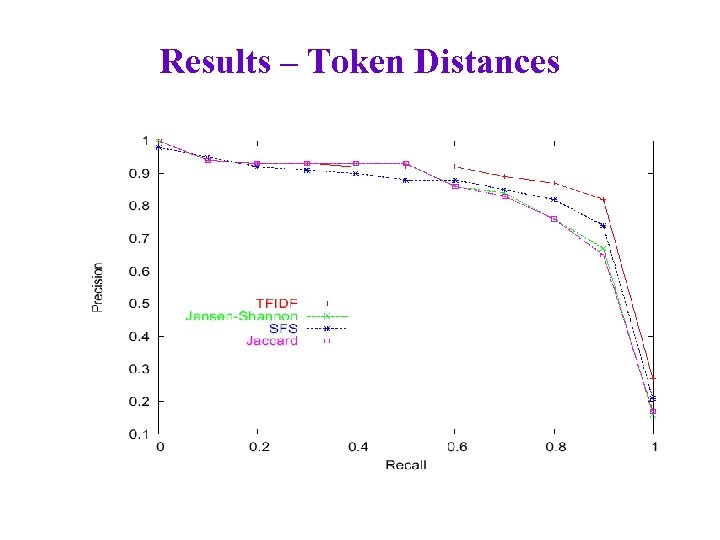 Results – Token Distances
Results – Token Distances
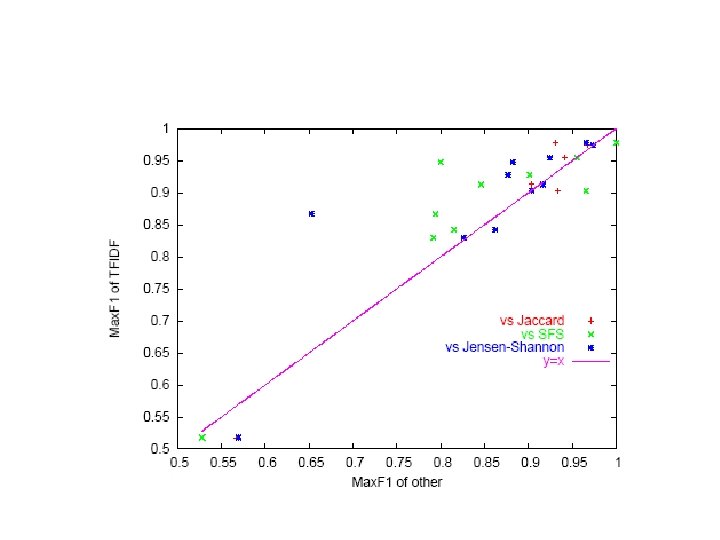
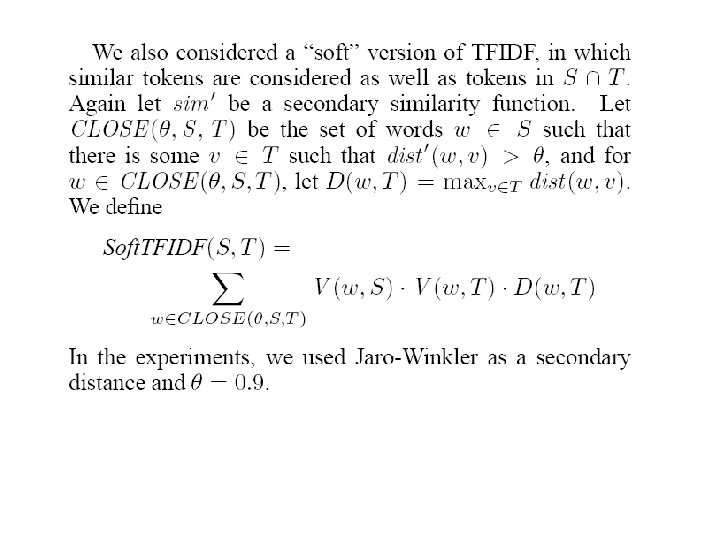
 Second. String distance functions • Hybrid term-based & edit-distance based: – Monge-Elkan’s “recursive matching scheme”, segmenting strings at token boundaries (rather than separators like commas) – Soft. TFIDF • Like TFIDF but consider not just tokens in both S and T, but tokens in S “close to” something in T (“close to” relative to some distance metric) • Downweight close tokens slightly
Second. String distance functions • Hybrid term-based & edit-distance based: – Monge-Elkan’s “recursive matching scheme”, segmenting strings at token boundaries (rather than separators like commas) – Soft. TFIDF • Like TFIDF but consider not just tokens in both S and T, but tokens in S “close to” something in T (“close to” relative to some distance metric) • Downweight close tokens slightly
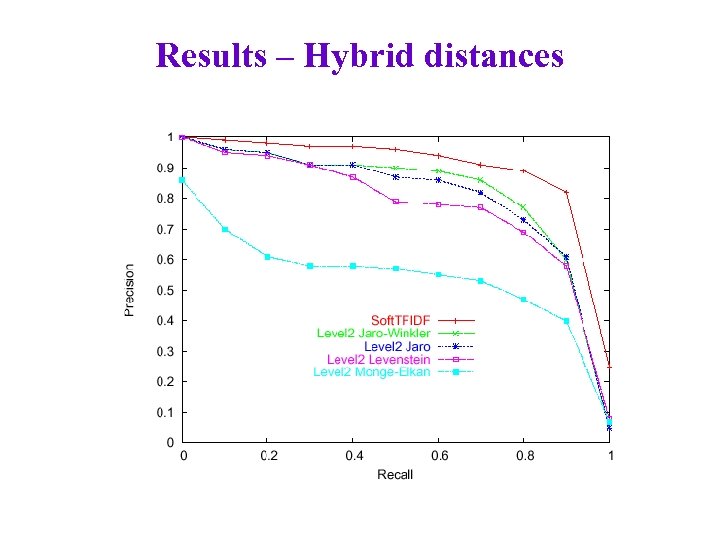 Results – Hybrid distances
Results – Hybrid distances
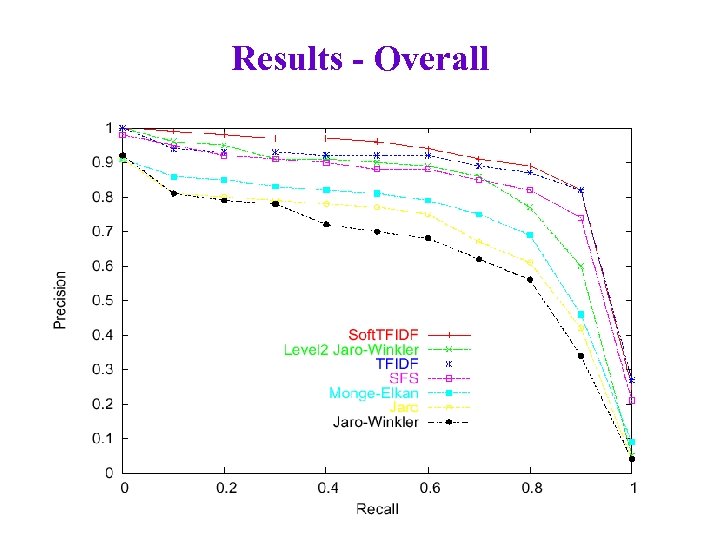 Results - Overall
Results - Overall
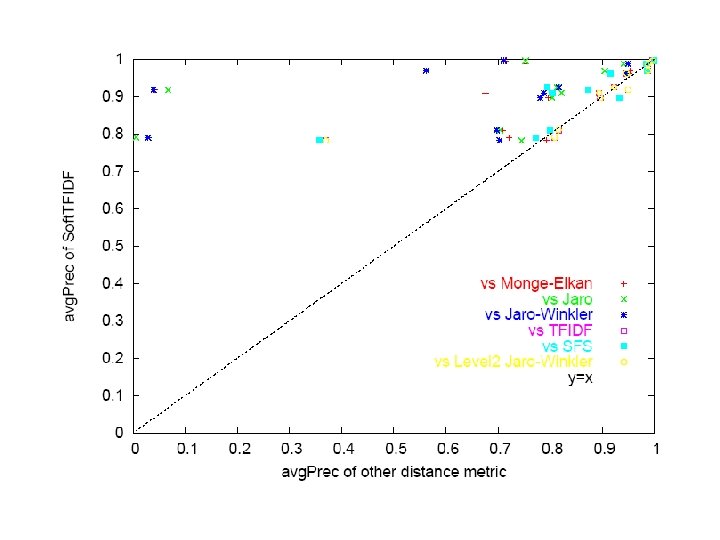
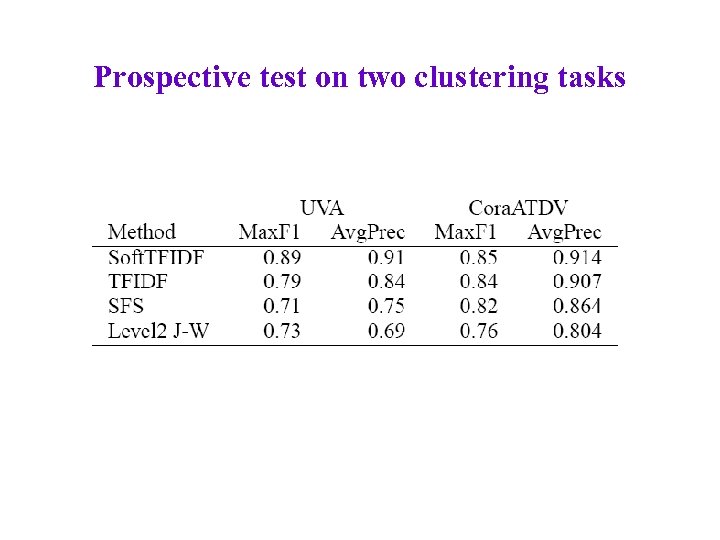 Prospective test on two clustering tasks
Prospective test on two clustering tasks
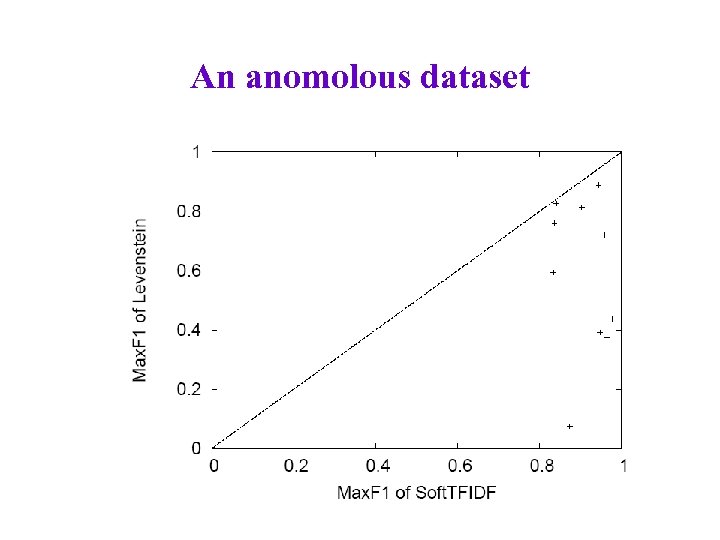 An anomolous dataset
An anomolous dataset
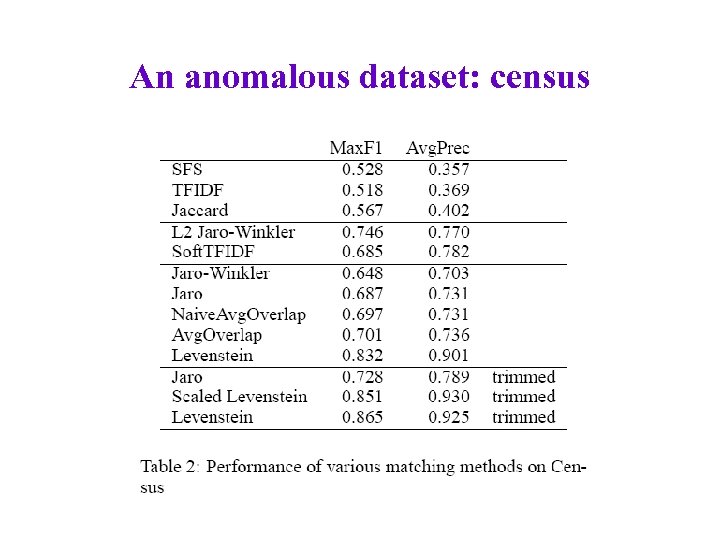 An anomalous dataset: census
An anomalous dataset: census
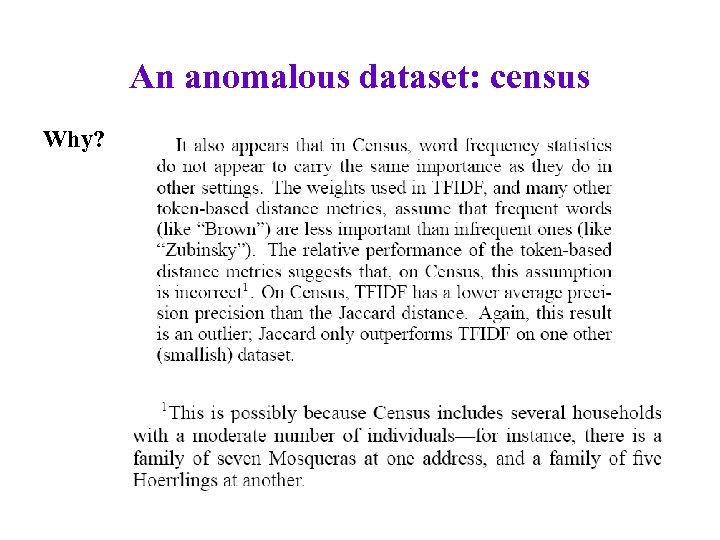 An anomalous dataset: census Why?
An anomalous dataset: census Why?
 Other results with Second. String • Distance functions over structured data records (first name, last name, street, house number) • Learning to combine distance functions • Unsupervised/semi-supervised training for distance functions over structured data
Other results with Second. String • Distance functions over structured data records (first name, last name, street, house number) • Learning to combine distance functions • Unsupervised/semi-supervised training for distance functions over structured data
 Combining Information Extraction and Similarity Computations 2) Krauthammer et al 1) Bunescu et al
Combining Information Extraction and Similarity Computations 2) Krauthammer et al 1) Bunescu et al
 Experiments • Hand-tagged 50 abstracts for gene/protein entities (pre-selected to be about human genes) • Collected dictionary of 40, 000+ protein names from on-line sources – not complete – example matching is not sufficient • Approach: use hand-coded heuristics to propose likely generalizations of existing dictionary entries. – not hand-coded or off-the-shelf similarity metrics
Experiments • Hand-tagged 50 abstracts for gene/protein entities (pre-selected to be about human genes) • Collected dictionary of 40, 000+ protein names from on-line sources – not complete – example matching is not sufficient • Approach: use hand-coded heuristics to propose likely generalizations of existing dictionary entries. – not hand-coded or off-the-shelf similarity metrics
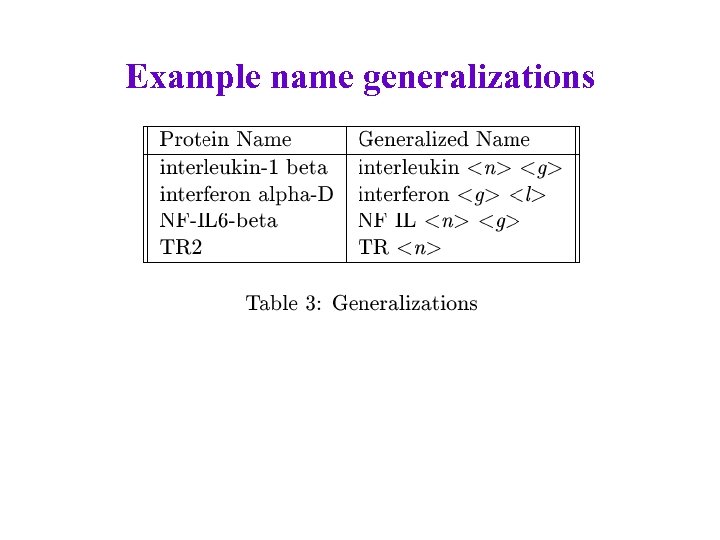 Example name generalizations
Example name generalizations
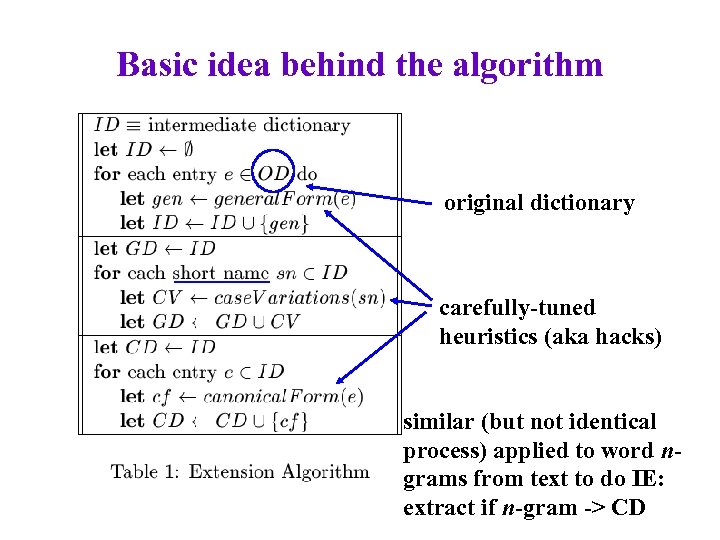 Basic idea behind the algorithm original dictionary carefully-tuned heuristics (aka hacks) similar (but not identical process) applied to word ngrams from text to do IE: extract if n-gram -> CD
Basic idea behind the algorithm original dictionary carefully-tuned heuristics (aka hacks) similar (but not identical process) applied to word ngrams from text to do IE: extract if n-gram -> CD
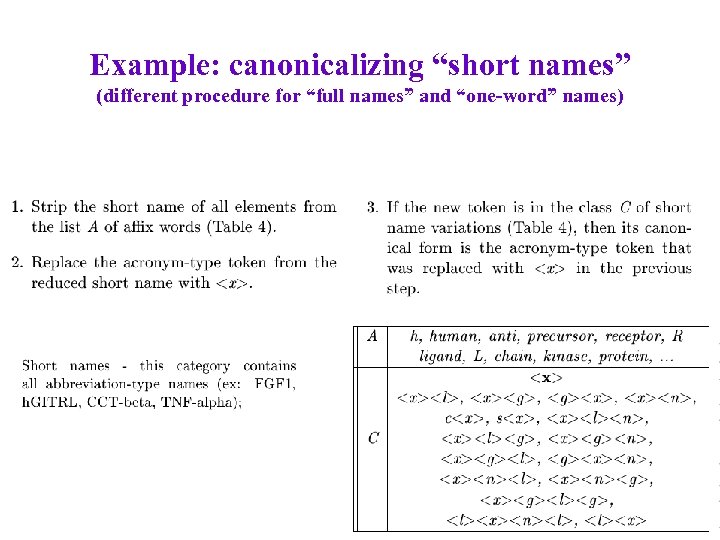 Example: canonicalizing “short names” (different procedure for “full names” and “one-word” names)
Example: canonicalizing “short names” (different procedure for “full names” and “one-word” names)
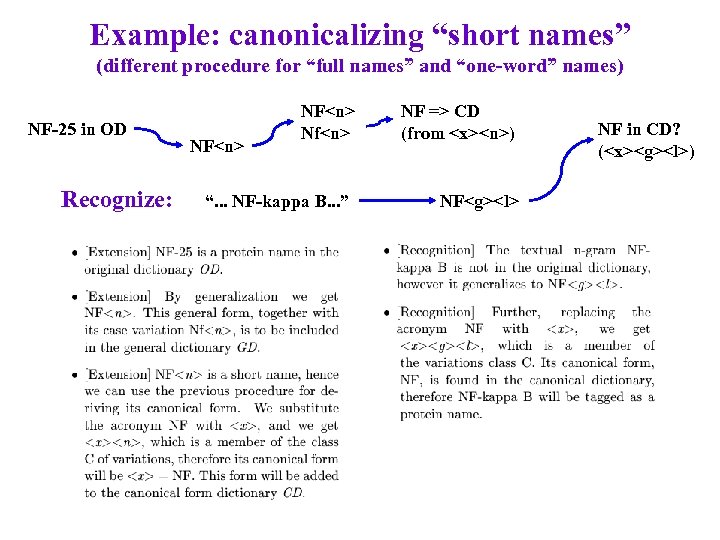 Example: canonicalizing “short names” (different procedure for “full names” and “one-word” names) NF-25 in OD Recognize: NF
Example: canonicalizing “short names” (different procedure for “full names” and “one-word” names) NF-25 in OD Recognize: NF
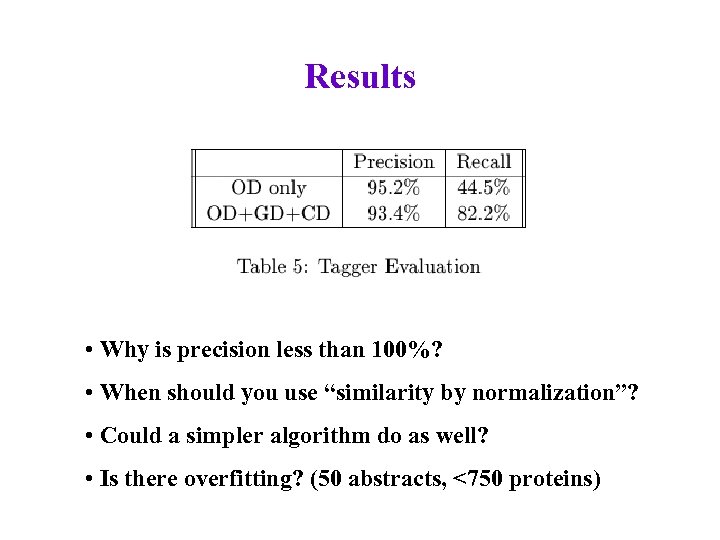 Results • Why is precision less than 100%? • When should you use “similarity by normalization”? • Could a simpler algorithm do as well? • Is there overfitting? (50 abstracts, <750 proteins)
Results • Why is precision less than 100%? • When should you use “similarity by normalization”? • Could a simpler algorithm do as well? • Is there overfitting? (50 abstracts, <750 proteins)
 . . .
. . .
 Combining Information Extraction and Similarity Computations 2) Krauthammer et al 1) Bunescu et al
Combining Information Extraction and Similarity Computations 2) Krauthammer et al 1) Bunescu et al
 Background • Common task in proteomics/genomics: – look for (soft) matches to a query sequence in a large “database” of sequences. – want to find subsequences (genes) that are highly similar (and hence probably related) – want to ignore “accidental” matches – possible technique is Smith-Waterman (local alignment) • want char-char “reward” for alignment to reflect confidence that the alignment is not due to chance
Background • Common task in proteomics/genomics: – look for (soft) matches to a query sequence in a large “database” of sequences. – want to find subsequences (genes) that are highly similar (and hence probably related) – want to ignore “accidental” matches – possible technique is Smith-Waterman (local alignment) • want char-char “reward” for alignment to reflect confidence that the alignment is not due to chance
 Background • Common task in proteomics/genomics: – look for (soft) matches to a query sequence in a large “database” of sequences. – want to find subsequences (genes) that are highly similar (and hence probably related) – want to ignore “accidental” matches – possible technique is Smith-Waterman (local alignment) • want char-char “reward” for alignment to reflect confidence that the alignment is not due to chance
Background • Common task in proteomics/genomics: – look for (soft) matches to a query sequence in a large “database” of sequences. – want to find subsequences (genes) that are highly similar (and hence probably related) – want to ignore “accidental” matches – possible technique is Smith-Waterman (local alignment) • want char-char “reward” for alignment to reflect confidence that the alignment is not due to chance
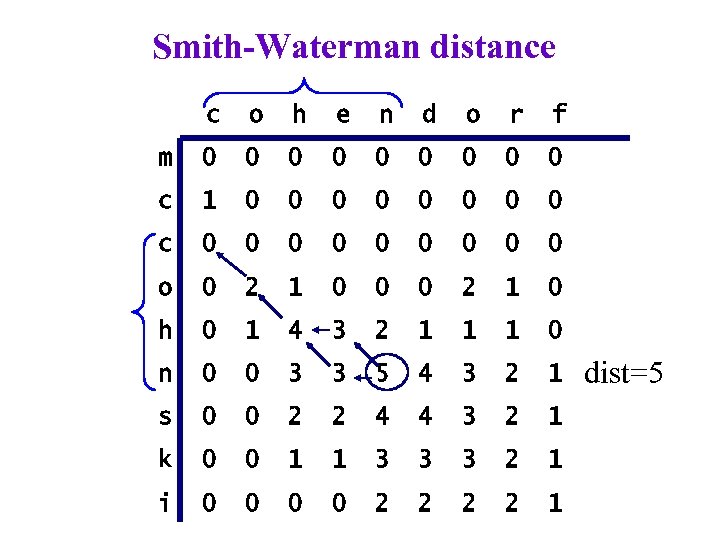 Smith-Waterman distance c o h e n d o r f m 0 0 0 0 0 c 1 0 0 0 0 c 0 0 0 0 0 o 0 2 1 0 0 0 2 1 0 h 0 1 4 3 2 1 1 1 0 n 0 0 3 3 5 4 3 2 1 s 0 0 2 2 4 4 3 2 1 k 0 0 1 1 3 3 3 2 1 i 0 0 2 2 1 dist=5
Smith-Waterman distance c o h e n d o r f m 0 0 0 0 0 c 1 0 0 0 0 c 0 0 0 0 0 o 0 2 1 0 0 0 2 1 0 h 0 1 4 3 2 1 1 1 0 n 0 0 3 3 5 4 3 2 1 s 0 0 2 2 4 4 3 2 1 k 0 0 1 1 3 3 3 2 1 i 0 0 2 2 1 dist=5
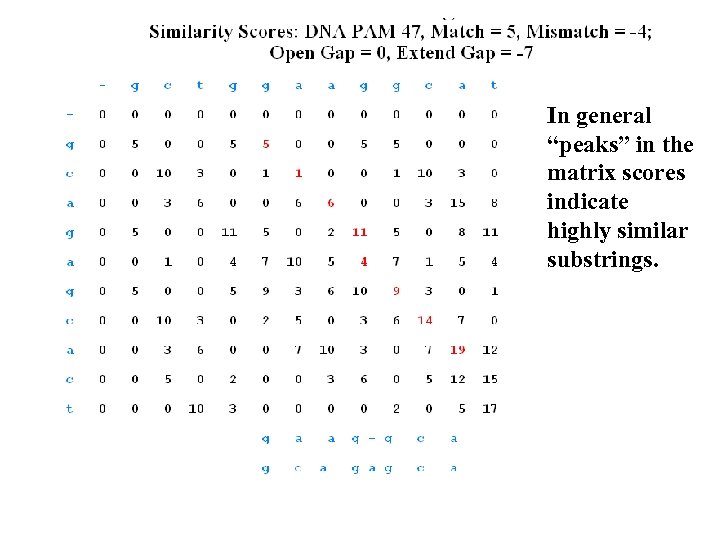 In general “peaks” in the matrix scores indicate highly similar substrings.
In general “peaks” in the matrix scores indicate highly similar substrings.
 Background • Common task in proteomics/genomics: – look for (soft) matches to a query sequence in a large “database” of sequences. – possible technique is Smith-Waterman (local alignment) • want char-char “reward” for alignment to reflect confidence that the alignment is not due to chance • based on substitutability theory for amino acids – doesn’t scale well • BLAST and FASTA: fast approximate S-W
Background • Common task in proteomics/genomics: – look for (soft) matches to a query sequence in a large “database” of sequences. – possible technique is Smith-Waterman (local alignment) • want char-char “reward” for alignment to reflect confidence that the alignment is not due to chance • based on substitutability theory for amino acids – doesn’t scale well • BLAST and FASTA: fast approximate S-W
 BLAST/FASTA ideas • Find all char n-grams (“words”) in the query string. • FASTA: – Use inverted indices to find out where these words appear in the DB sequence – Use S-W only near DB sections that contain some of these words
BLAST/FASTA ideas • Find all char n-grams (“words”) in the query string. • FASTA: – Use inverted indices to find out where these words appear in the DB sequence – Use S-W only near DB sections that contain some of these words
 BLAST/FASTA ideas • Find all char n-grams (“words”) in the query string. • BLAST: – Generate variations of these words by looking for changes that would lead to strong similarities – Discard “low IDF” words (where accidental matches are likely) – Use expanded set of n-grams to focus search
BLAST/FASTA ideas • Find all char n-grams (“words”) in the query string. • BLAST: – Generate variations of these words by looking for changes that would lead to strong similarities – Discard “low IDF” words (where accidental matches are likely) – Use expanded set of n-grams to focus search
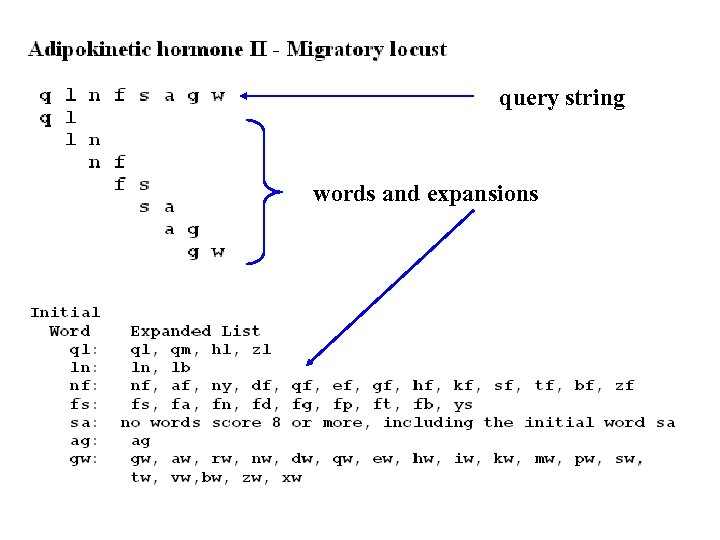 query string words and expansions
query string words and expansions
 BLAST/FASTA ideas • Find all char n-grams (“words”) in the query string. • BLAST: – Generate variations of these words by looking for changes that would lead to strong similarities – Discard “low IDF” words (where accidental matches are likely) – Use expanded set of n-grams to focus search • The BLAST program: – – Widely used, Fast implementation, Supports asking multiple queries against a database at once. . . Can one use it find soft matches of protein names (from a dictionary) in text?
BLAST/FASTA ideas • Find all char n-grams (“words”) in the query string. • BLAST: – Generate variations of these words by looking for changes that would lead to strong similarities – Discard “low IDF” words (where accidental matches are likely) – Use expanded set of n-grams to focus search • The BLAST program: – – Widely used, Fast implementation, Supports asking multiple queries against a database at once. . . Can one use it find soft matches of protein names (from a dictionary) in text?
 Basic idea: • Biomedical paper • Protein name dictionary • Extracted protein name (dict. entry->text) • IE system: dictionaries+BLAST (optimized for this problem) • Protein database • Query strings • Proposed alignment (query->database) • Query algorithm: BLAST
Basic idea: • Biomedical paper • Protein name dictionary • Extracted protein name (dict. entry->text) • IE system: dictionaries+BLAST (optimized for this problem) • Protein database • Query strings • Proposed alignment (query->database) • Query algorithm: BLAST
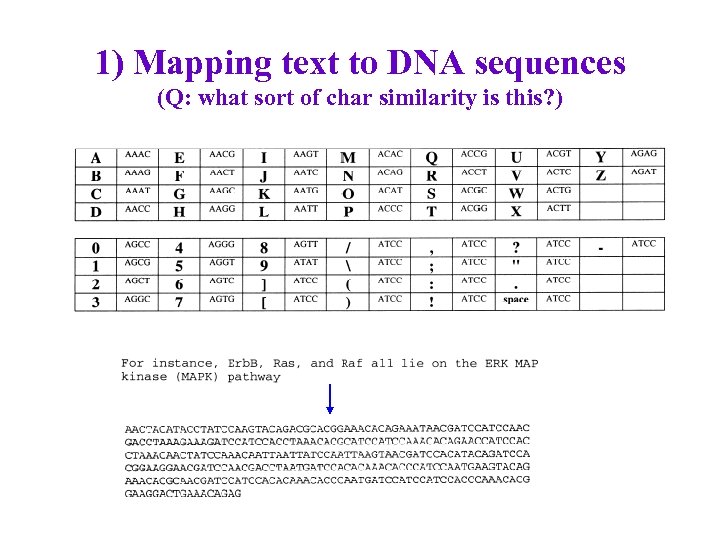 1) Mapping text to DNA sequences (Q: what sort of char similarity is this? )
1) Mapping text to DNA sequences (Q: what sort of char similarity is this? )
 2) Optimizing blast • Split protein-name database into several parts (for short, medium-length, long protein names) • Require space chars before and after “short” protein names. • Manually search (grid search? ) for better settings for certain key parameters for each protein-name subdatabase – With what data? • Evaluate on one review article, 1162 protein names – inter-annotator agreement not great (70 -85%)
2) Optimizing blast • Split protein-name database into several parts (for short, medium-length, long protein names) • Require space chars before and after “short” protein names. • Manually search (grid search? ) for better settings for certain key parameters for each protein-name subdatabase – With what data? • Evaluate on one review article, 1162 protein names – inter-annotator agreement not great (70 -85%)
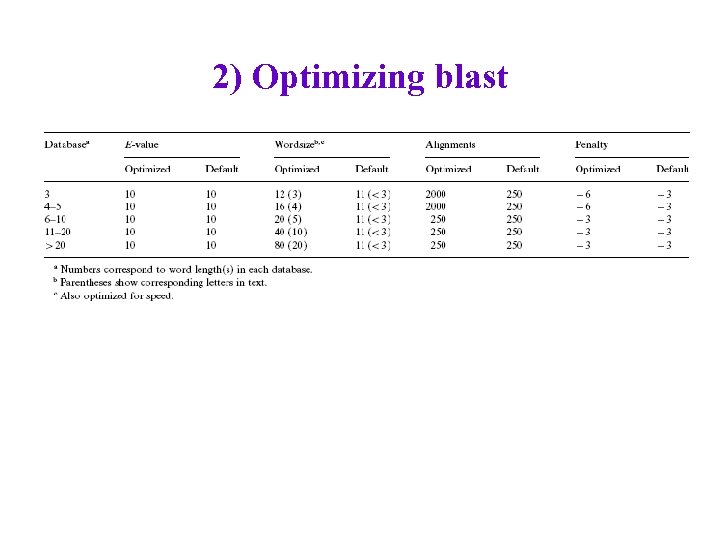 2) Optimizing blast
2) Optimizing blast
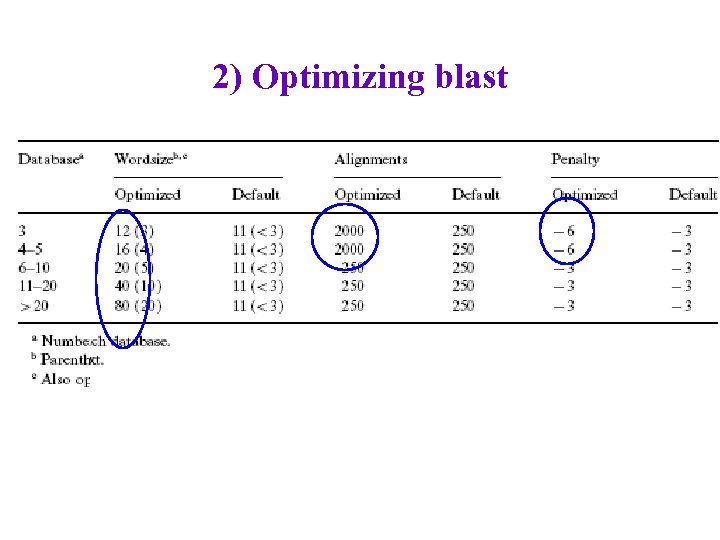 2) Optimizing blast
2) Optimizing blast
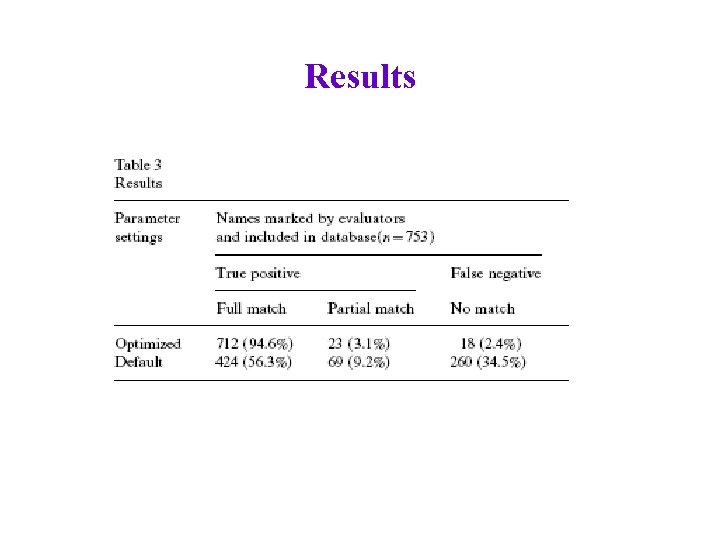 Results
Results
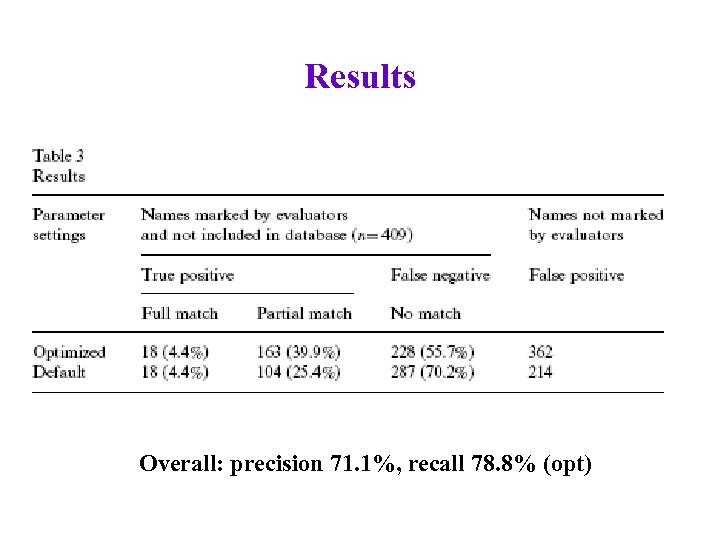 Results Overall: precision 71. 1%, recall 78. 8% (opt)
Results Overall: precision 71. 1%, recall 78. 8% (opt)


