ef8795f4bb72551e8b4855fa45bfccaf.ppt
- Количество слайдов: 32

Diagnosis of Genital Ulcer Disease (GUD) by PCR Cheng Y. Chen, Ph. D. Laboratory Reference and Research Branch Division of STD Prevention CDC March 2008

Genital Ulcer Disease (GUD) Chancroid: Haemophilus ducreyi (HD) Syphilis: Treponema pallidum (TP) Genital herpes: Herpes Simplex Virus (HSV-1, -2) Lymphogranuloma venereum (LGV): Chlamydia trachomatis (L-serovars) Granuloma inguinale (Donovanosis): Klebsiella (Calymmatobacterium) granulomatis

Background Estimated annual global incidence of GUD > 20 million cases Sexually acquired GUD is associated with increased risk for HIV acquisition and transmission GUD diagnosis based solely on clinical presentation is not always reliable (complicated by HIV infection) The most common cause of GUD can vary geographically and temporally Treatment varies based on GUD etiology Effective diagnosis and treatment of GUD could reduce the incidence of HIV infection

Laboratory Diagnosis of GUD Syphilis: serology, dark-field, DFA stain, PCR Chancroid: culture, PCR Genital herpes: serology, tissue culture, PCR LGV: culture, serology, genotyping (PCR-RFLP, sequencing) Donovanosis: Giemsa stain, PCR-restriction digest

Advantages of PCR § Detects infecting organisms directly § Sensitivity and reproducibility § Dynamic range of detection § Relatively fast § Suitable for non-invasive specimens (urine and self-collected vaginal swabs) § Can be applied to non-cultivatable organisms

Why Develop a Multiplex-PCR (M-PCR) for GUD ? § Multiple diagnoses in one assay (chancroid, § § § syphilis, genital herpes type 1, 2, and LGV) Cost effective No commercial kit available Patient management Etiology prevalence/surveillance/epidemology Development and evaluation of syndromic management algorithms
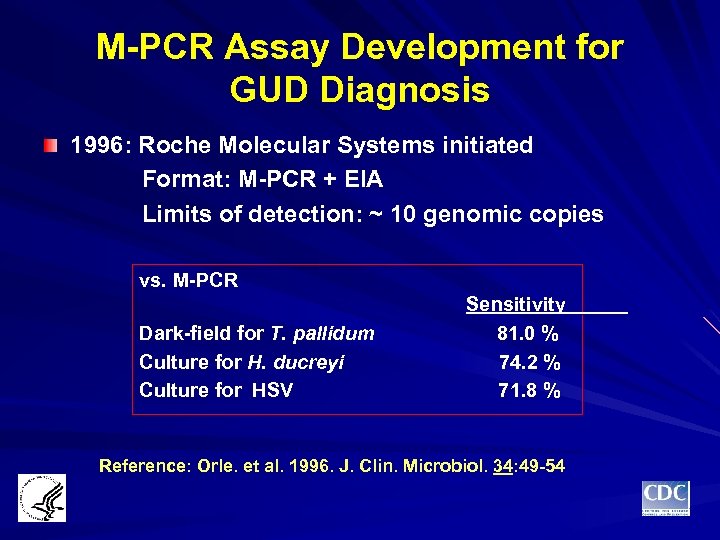
M-PCR Assay Development for GUD Diagnosis 1996: Roche Molecular Systems initiated Format: M-PCR + EIA Limits of detection: ~ 10 genomic copies vs. M-PCR Dark-field for T. pallidum Culture for H. ducreyi Culture for HSV Sensitivity 81. 0 % 74. 2 % 71. 8 % Reference: Orle. et al. 1996. J. Clin. Microbiol. 34: 49 -54
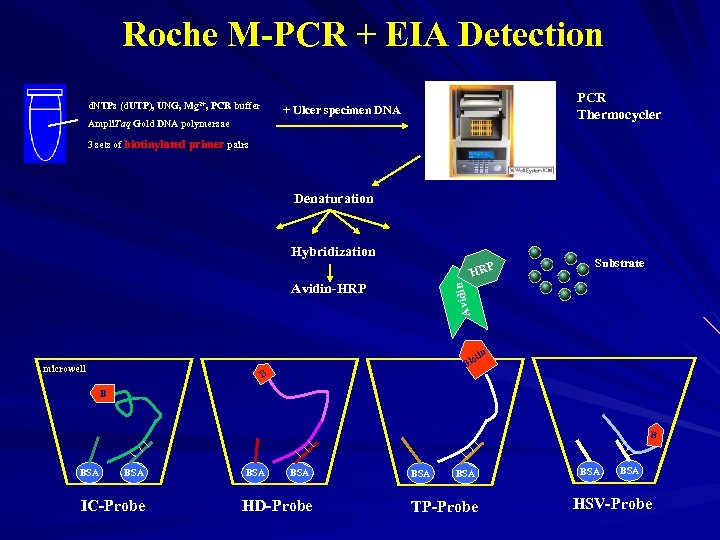
Roche M-PCR + EIA Detection d. NTPs (d. UTP), UNG, Mg 2+, PCR buffer PCR Thermocycler + Ulcer specimen DNA Ampli. Taq Gold DNA polymersae 3 sets of biotinylated primer pairs Denaturation Hybridization HRP Avidi n Avidin-HRP Substrate n oti microwell Bi B BSA BSA IC-Probe BSA HD-Probe BSA TP-Probe BSA HSV-Probe
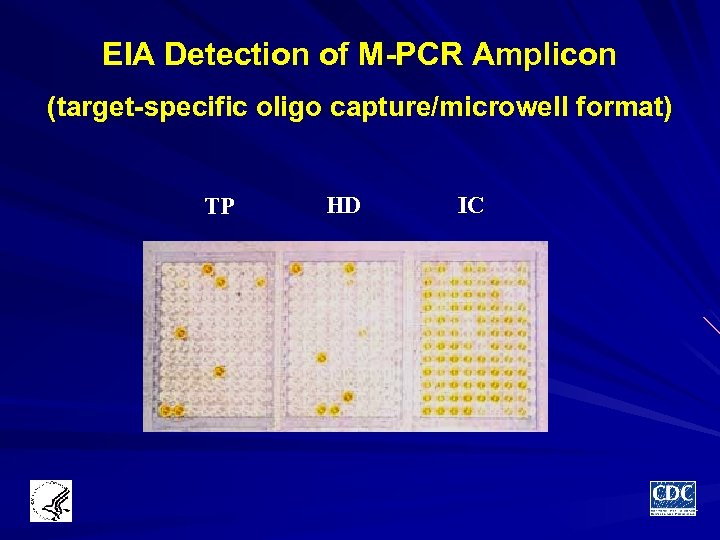
EIA Detection of M-PCR Amplicon (target-specific oligo capture/microwell format) TP HD IC
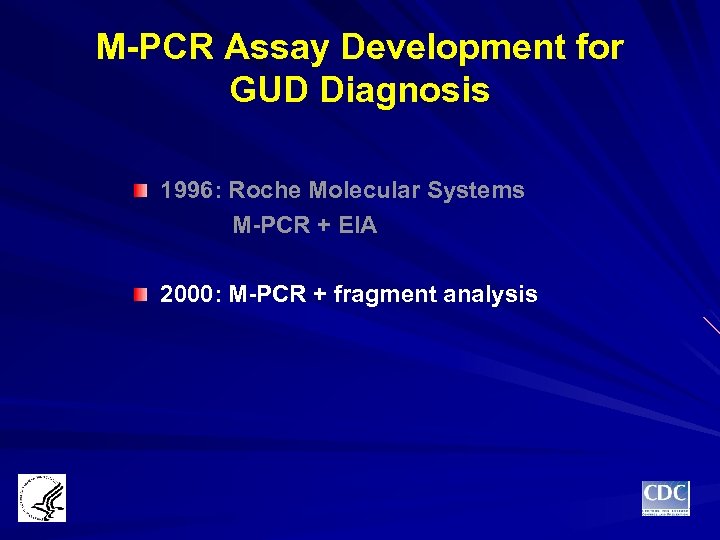
M-PCR Assay Development for GUD Diagnosis 1996: Roche Molecular Systems M-PCR + EIA 2000: M-PCR + fragment analysis
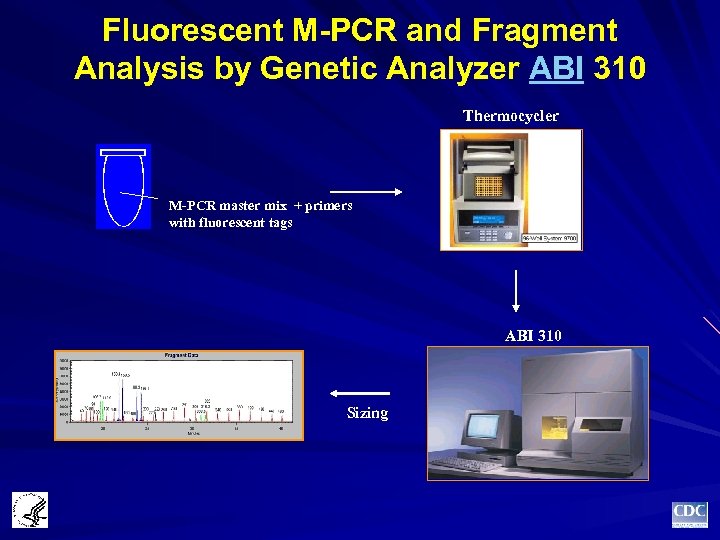
Fluorescent M-PCR and Fragment Analysis by Genetic Analyzer ABI 310 Thermocycler M-PCR master mix + primers with fluorescent tags ABI 310 Sizing
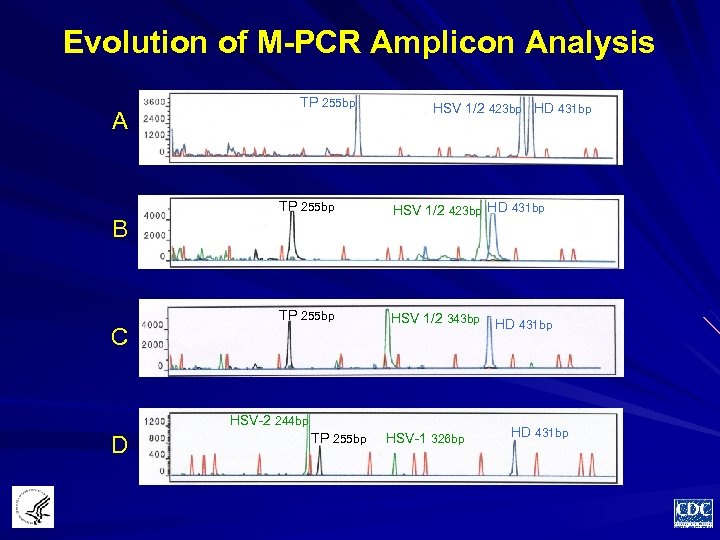
Evolution of M-PCR Amplicon Analysis A TP 255 bp HSV 1/2 423 bp HD 431 bp TP 255 bp HSV 1/2 343 bp B C HSV-2 244 bp D TP 255 bp HSV-1 326 bp HD 431 bp

Real-time PCR Instruments

Advantages of Real-Time PCR § Sensitivity and reproducibility § Dynamic range of detection § Permits rapid target Id § Suitable for urine and self-collected vaginal swabs § Highly specific; FQ probes (hybridization) § Eliminates post-PCR processing (closed tube) § Allows multiplexing (up to 6 target detection) § Permits quantification (viral load) § High throughput (96 -well plate) § Potential point-of-care application
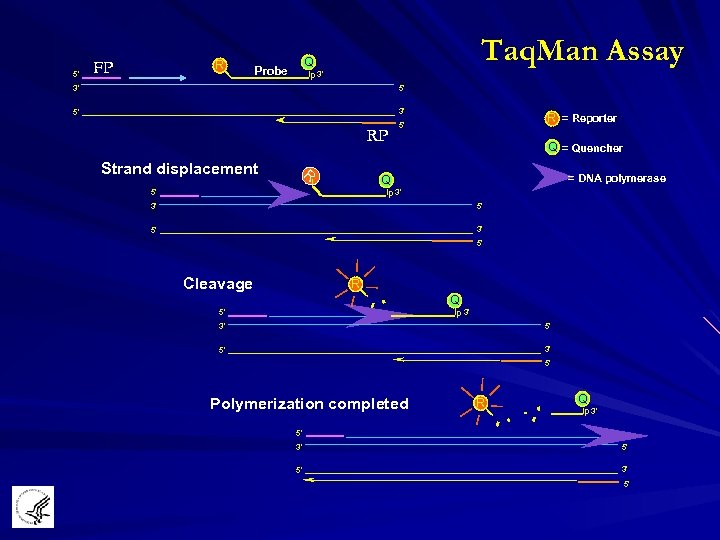
5' FP R Taq. Man Assay Q Probe p 3' 3' 5' 5' 3' RP Strand displacement R R = Reporter 5' Q = Quencher = DNA polymerase Q p 3' 5' 5' 3' 5' Cleavage R Q p 3' 5' 5' 3' 5' Polymerization completed R Q p 3' 5' 5' 3' 5'
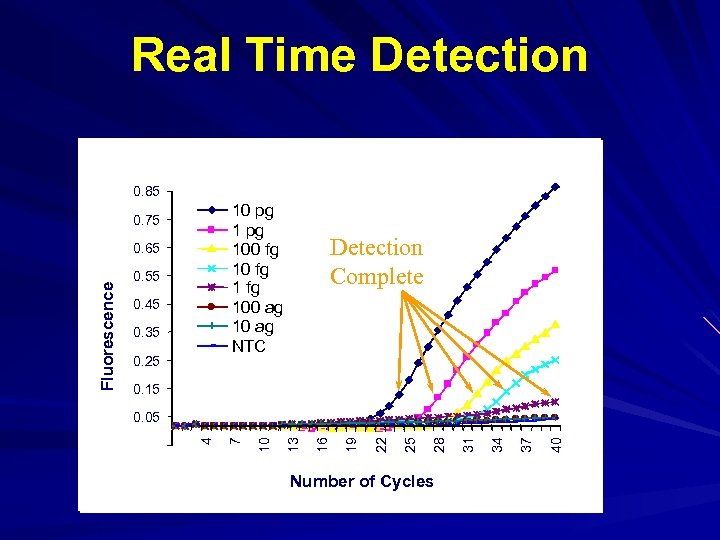
Real Time Detection 0. 85 10 pg 100 fg 100 ag 10 ag NTC 0. 75 0. 55 0. 45 0. 35 0. 25 Detection Complete 0. 15 Number of Cycles 40 37 34 31 28 25 22 19 16 13 10 -0. 05 7 0. 05 4 Fluorescence 0. 65
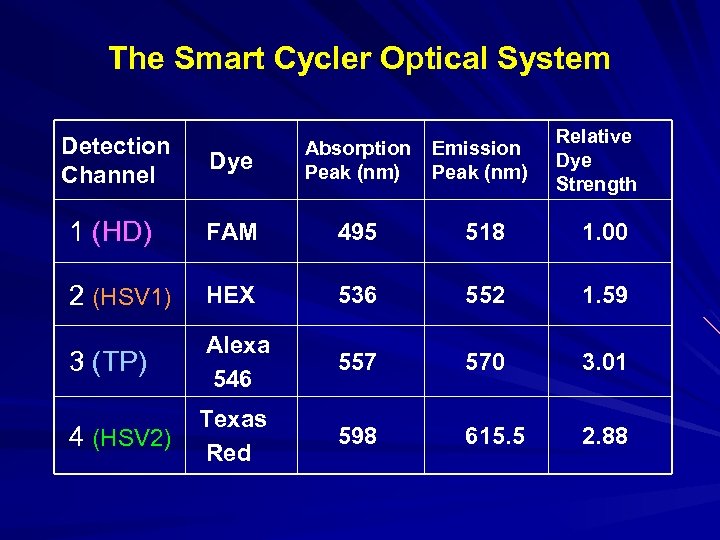
The Smart Cycler Optical System Detection Channel Relative Dye Strength Dye Absorption Peak (nm) Emission Peak (nm) 1 (HD) FAM 495 518 1. 00 2 (HSV 1) HEX 536 552 1. 59 3 (TP) Alexa 546 557 570 3. 01 4 (HSV 2) Texas Red 598 615. 5 2. 88
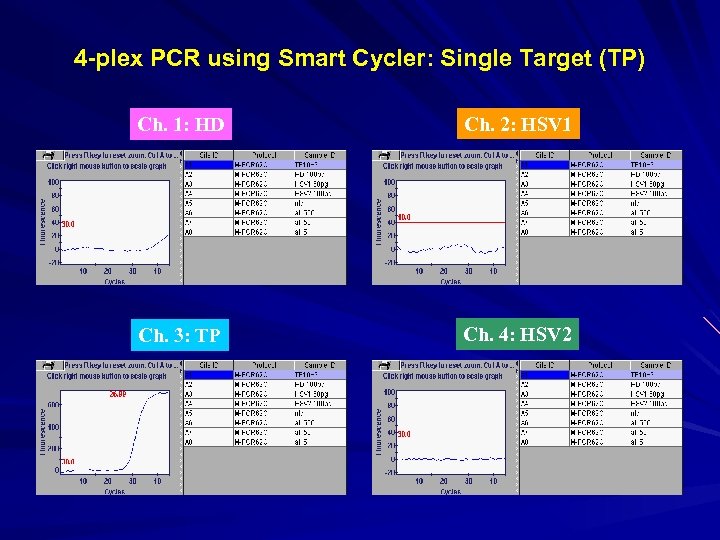
4 -plex PCR using Smart Cycler: Single Target (TP) Ch. 1: HD Ch. 2: HSV 1 Ch. 3: TP Ch. 4: HSV 2
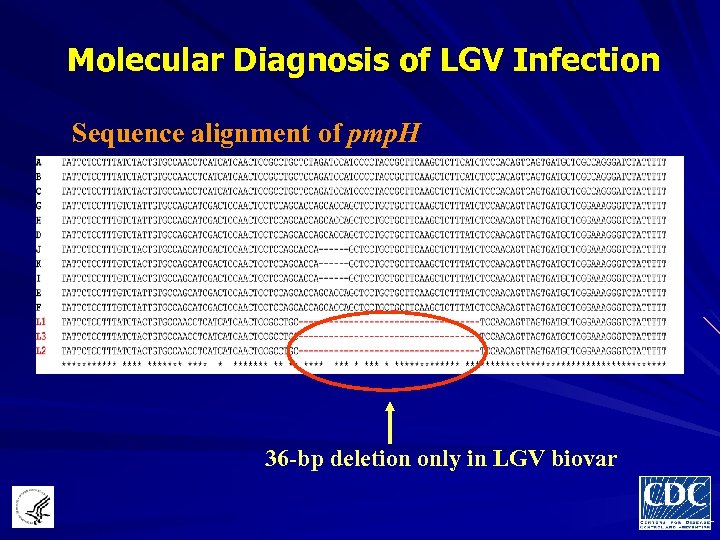
Molecular Diagnosis of LGV Infection Sequence alignment of pmp. H 36 -bp deletion only in LGV biovar
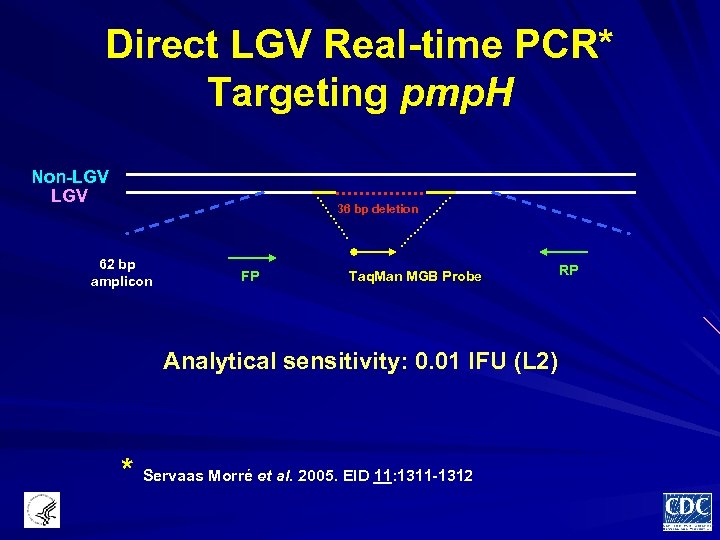
Direct LGV Real-time PCR* Targeting pmp. H Non-LGV 36 bp deletion 62 bp amplicon FP Taq. Man MGB Probe Analytical sensitivity: 0. 01 IFU (L 2) * Servaas Morré et al. 2005. EID 11: 1311 -1312 RP
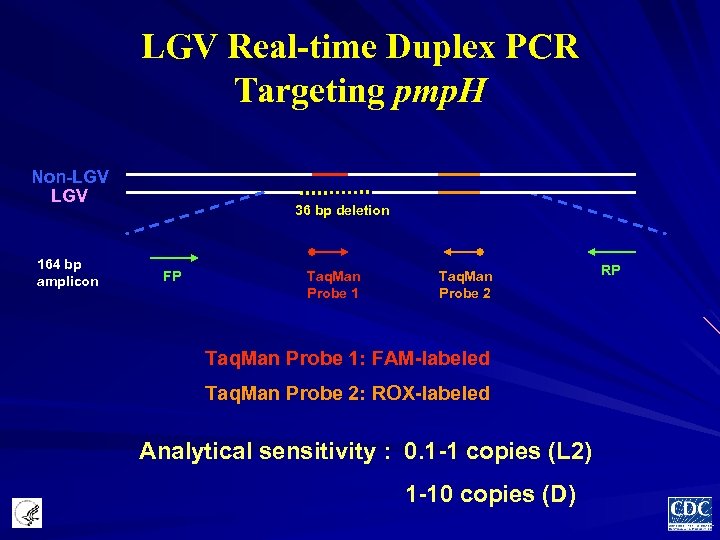
LGV Real-time Duplex PCR Targeting pmp. H Non-LGV 164 bp amplicon 36 bp deletion FP Taq. Man Probe 1 Taq. Man Probe 2 Taq. Man Probe 1: FAM-labeled Taq. Man Probe 2: ROX-labeled Analytical sensitivity : 0. 1 -1 copies (L 2) 1 -10 copies (D) RP
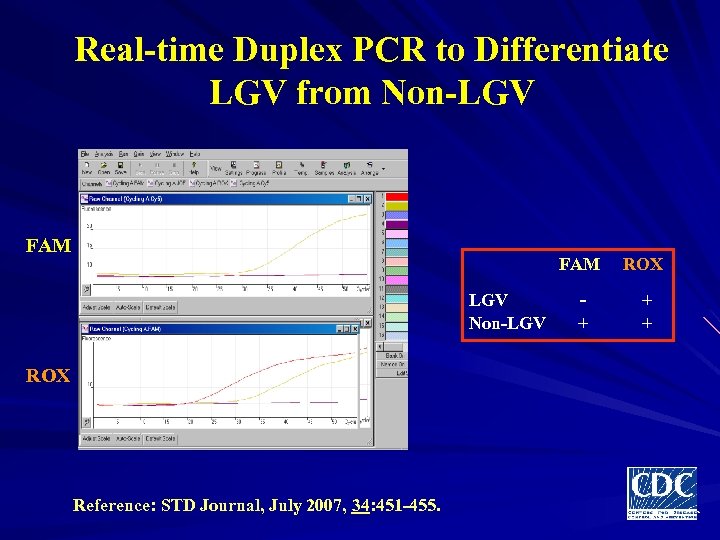
Real-time Duplex PCR to Differentiate LGV from Non-LGV FAM LGV Non-LGV ROX Reference: STD Journal, July 2007, 34: 451 -455. ROX - + + +
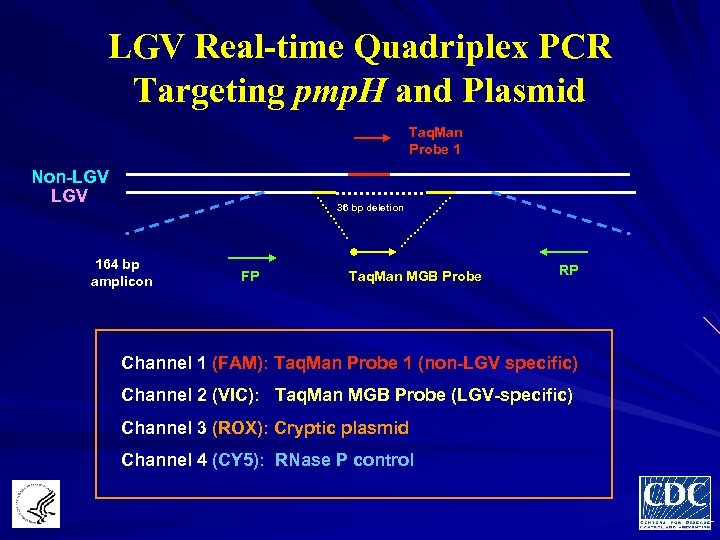
LGV Real-time Quadriplex PCR Targeting pmp. H and Plasmid Taq. Man Probe 1 Non-LGV 36 bp deletion 164 bp amplicon FP Taq. Man MGB Probe RP Channel 1 (FAM): Taq. Man Probe 1 (non-LGV specific) Channel 2 (VIC): Taq. Man MGB Probe (LGV-specific) Channel 3 (ROX): Cryptic plasmid Channel 4 (CY 5): RNase P control
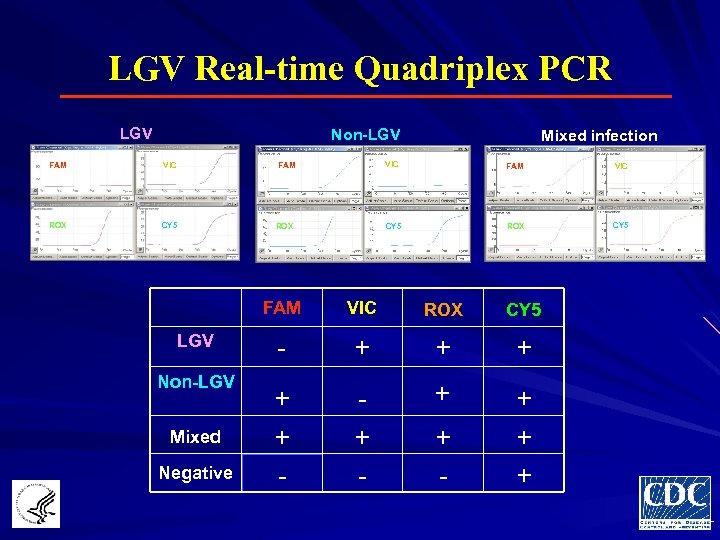
LGV Real-time Quadriplex PCR LGV Non-LGV Mixed infection FAM VIC ROX CY 5 FAM LGV Non-LGV Mixed Negative VIC ROX CY 5 - + + + -
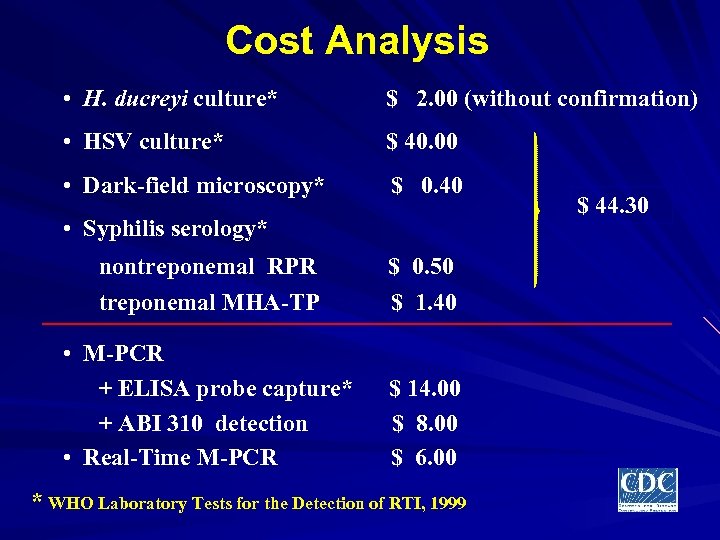
Cost Analysis • H. ducreyi culture* $ 2. 00 (without confirmation) • HSV culture* $ 40. 00 • Dark-field microscopy* $ 0. 40 • Syphilis serology* nontreponemal RPR treponemal MHA-TP $ 0. 50 $ 1. 40 • M-PCR + ELISA probe capture* + ABI 310 detection • Real-Time M-PCR $ 14. 00 $ 8. 00 $ 6. 00 * WHO Laboratory Tests for the Detection of RTI, 1999 $ 44. 30
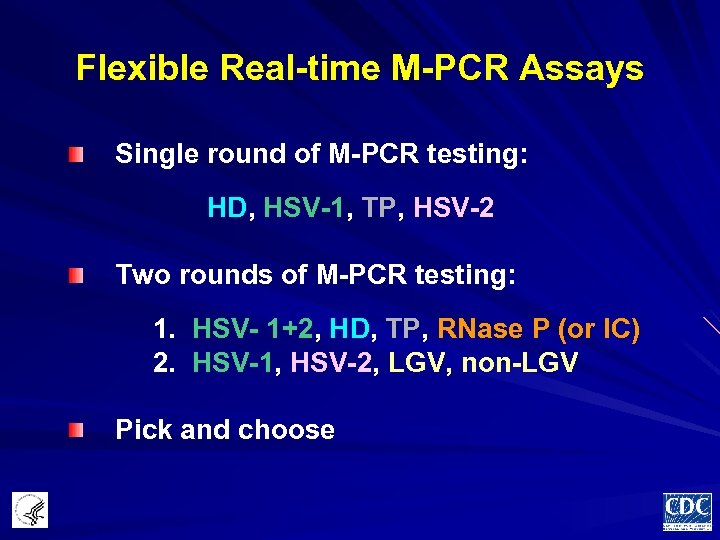
Flexible Real-time M-PCR Assays Single round of M-PCR testing: HD, HSV-1, TP, HSV-2 Two rounds of M-PCR testing: 1. HSV- 1+2, HD, TP, RNase P (or IC) 2. HSV-1, HSV-2, LGV, non-LGV Pick and choose

Nanotechnology Lab-on-a-chip using microfluidics: • filtration and concentration • cell lysis • real time PCR/M-PCR • fluorescence detection • battery powered hand-held device • result in <30 min • nanolitre-scale bio-reagents • accommodate multiple sensors • low manufacturing costs • inexpensive reagents
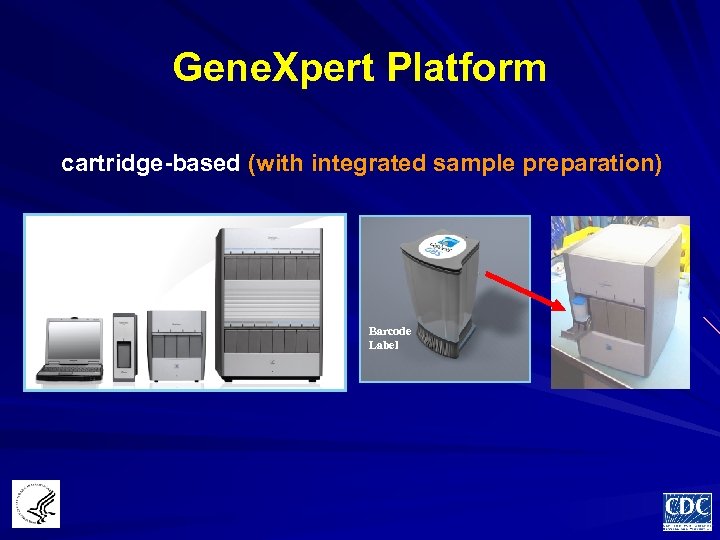
Gene. Xpert Platform cartridge-based (with integrated sample preparation) Barcode Label
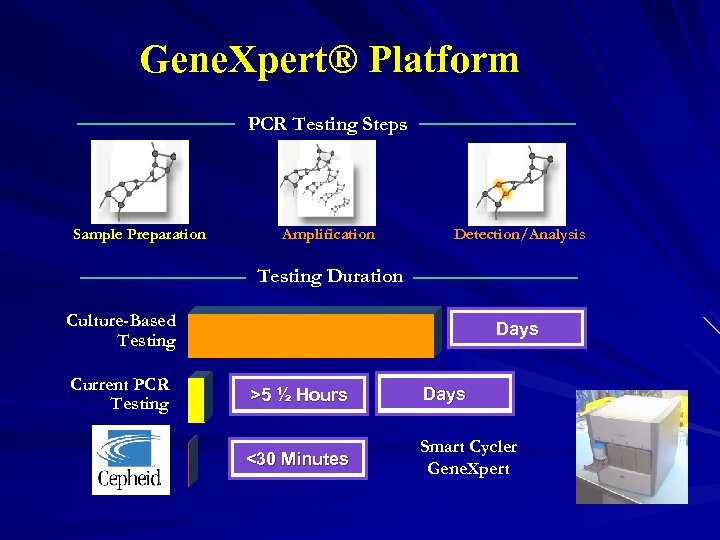
Gene. Xpert® Platform PCR Testing Steps Sample Preparation Amplification Detection/Analysis Testing Duration Culture-Based Testing Current PCR Testing Days >5 ½ Hours Days <30 Minutes Smart Cycler Gene. Xpert

Disclaimer The in-house developed multiplex-PCR assays for GUD diagnosis are not FDA-cleared and should be used for research and epidemiologic study only and not for direct patient management The findings and conclusions in this presentation are those of the author(s) and have not been formally disseminated by the Centers for Disease Control and Prevention and should not be construed to represent any agency determination or policy.

Acknowledgment CDC Kai-Hua Chi Wei Lai Ye Htun Stephen Morse Allan Pillay Hsi Liu Ron Ballard HPA, UK Sarah Alexander Iona Martin Cathy Ison Roche Judy Weiss

Contact Cheng Y. Chen, Ph. D. Laboratory Reference & Research Branch Division of STD Prevention Centers for Disease Control and Prevention 1600 Clifton Road, MS: G-39 Atlanta, GA 30333 Tel: (404) 639 -3154 E-mail: cychen@cdc. gov
ef8795f4bb72551e8b4855fa45bfccaf.ppt