35bbbda6e81c13e5a0e4166b8de2e8af.ppt
- Количество слайдов: 24
 Development of an EGFR/KRAS testing service for Non-Small Cell Lung Cancer (NSCLC) Joel Tracey 1, Caroline Clark 1, Christine Bell 1, Keith Kerr 2, Marianne Nicholson 3, Aileen Osborne 1, Zosia Miedzybrodzka 1, Kevin Kelly 1 1 Department of Medical Genetics, Polwarth Building, Aberdeen Royal Infirmary, Aberdeen 2 Department of Pathology, Aberdeen Royal Infirmary, Aberdeen 3 Clinical Oncology, Aberdeen Royal Infirmary, Aberdeen
Development of an EGFR/KRAS testing service for Non-Small Cell Lung Cancer (NSCLC) Joel Tracey 1, Caroline Clark 1, Christine Bell 1, Keith Kerr 2, Marianne Nicholson 3, Aileen Osborne 1, Zosia Miedzybrodzka 1, Kevin Kelly 1 1 Department of Medical Genetics, Polwarth Building, Aberdeen Royal Infirmary, Aberdeen 2 Department of Pathology, Aberdeen Royal Infirmary, Aberdeen 3 Clinical Oncology, Aberdeen Royal Infirmary, Aberdeen
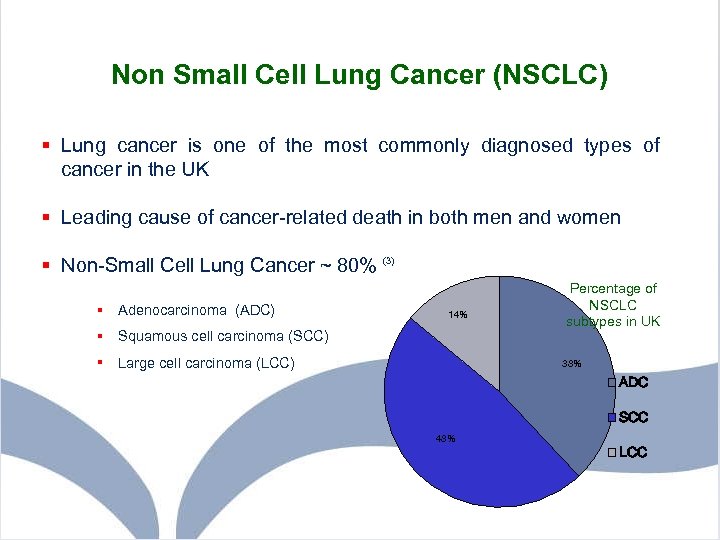 Non Small Cell Lung Cancer (NSCLC) § Lung cancer is one of the most commonly diagnosed types of cancer in the UK § Leading cause of cancer-related death in both men and women § Non-Small Cell Lung Cancer ~ 80% (3) § Adenocarcinoma (ADC) 14% § Squamous cell carcinoma (SCC) § Large cell carcinoma (LCC) Percentage of NSCLC subtypes in UK 38% ADC SCC 48% LCC
Non Small Cell Lung Cancer (NSCLC) § Lung cancer is one of the most commonly diagnosed types of cancer in the UK § Leading cause of cancer-related death in both men and women § Non-Small Cell Lung Cancer ~ 80% (3) § Adenocarcinoma (ADC) 14% § Squamous cell carcinoma (SCC) § Large cell carcinoma (LCC) Percentage of NSCLC subtypes in UK 38% ADC SCC 48% LCC
 EGFR and KRAS in NSCLC § Acquired mutations in the EGFR and KRAS genes are important in the development of NSCLC § Mutations result in inappropriately activated proteins – tumour cells become ‘addicted’ to growth signals § EGFR and KRAS mutations most common in adenocarcinoma § EGFR and KRAS mutations are mutually exclusive
EGFR and KRAS in NSCLC § Acquired mutations in the EGFR and KRAS genes are important in the development of NSCLC § Mutations result in inappropriately activated proteins – tumour cells become ‘addicted’ to growth signals § EGFR and KRAS mutations most common in adenocarcinoma § EGFR and KRAS mutations are mutually exclusive
 Treatment of NSCLC § Surgery - possible in about 20% of cases (4) § Cytotoxic chemotherapy and/or radiotherapy - mostly ineffective § Survival rate poor (7% alive 5 years after diagnosis) (4) § Tyrosine Kinase Inhibitors (TKI) – new type of chemotherapeutic agent – fewer side-effects than cytotoxic chemotherapy § Target and block growth factor signals – e. g. Epidermal Growth Factor Receptor (EGFR)
Treatment of NSCLC § Surgery - possible in about 20% of cases (4) § Cytotoxic chemotherapy and/or radiotherapy - mostly ineffective § Survival rate poor (7% alive 5 years after diagnosis) (4) § Tyrosine Kinase Inhibitors (TKI) – new type of chemotherapeutic agent – fewer side-effects than cytotoxic chemotherapy § Target and block growth factor signals – e. g. Epidermal Growth Factor Receptor (EGFR)
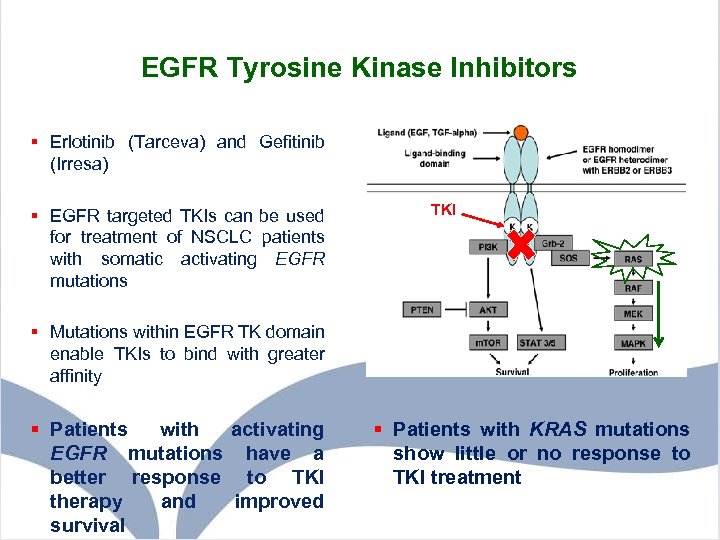 EGFR Tyrosine Kinase Inhibitors § Erlotinib (Tarceva) and Gefitinib (Irresa) § EGFR targeted TKIs can be used for treatment of NSCLC patients with somatic activating EGFR mutations TKI § Mutations within EGFR TK domain enable TKIs to bind with greater affinity § Patients with activating EGFR mutations have a better response to TKI therapy and improved survival § Patients with KRAS mutations show little or no response to TKI treatment
EGFR Tyrosine Kinase Inhibitors § Erlotinib (Tarceva) and Gefitinib (Irresa) § EGFR targeted TKIs can be used for treatment of NSCLC patients with somatic activating EGFR mutations TKI § Mutations within EGFR TK domain enable TKIs to bind with greater affinity § Patients with activating EGFR mutations have a better response to TKI therapy and improved survival § Patients with KRAS mutations show little or no response to TKI treatment
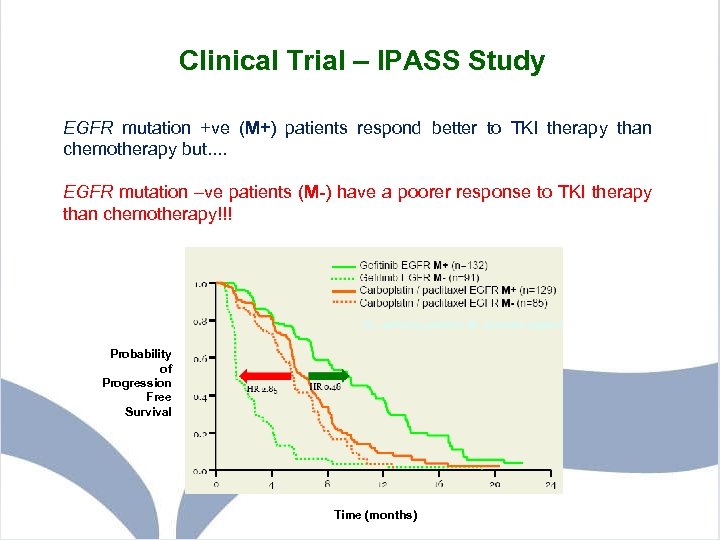 Clinical Trial – IPASS Study EGFR mutation +ve (M+) patients respond better to TKI therapy than chemotherapy but. . EGFR mutation –ve patients (M-) have a poorer response to TKI therapy than chemotherapy!!! Probability of Progression Free Survival Time (months)
Clinical Trial – IPASS Study EGFR mutation +ve (M+) patients respond better to TKI therapy than chemotherapy but. . EGFR mutation –ve patients (M-) have a poorer response to TKI therapy than chemotherapy!!! Probability of Progression Free Survival Time (months)
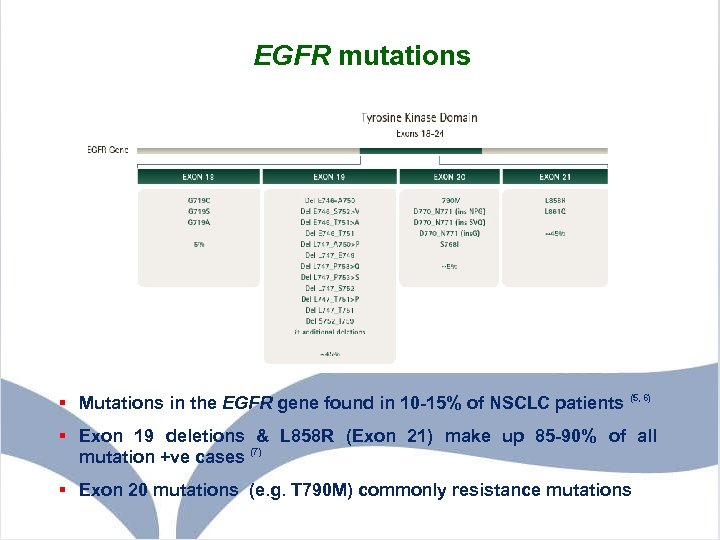 EGFR mutations § Mutations in the EGFR gene found in 10 -15% of NSCLC patients (5, 6) § Exon 19 deletions & L 858 R (Exon 21) make up 85 -90% of all mutation +ve cases (7) § Exon 20 mutations (e. g. T 790 M) commonly resistance mutations
EGFR mutations § Mutations in the EGFR gene found in 10 -15% of NSCLC patients (5, 6) § Exon 19 deletions & L 858 R (Exon 21) make up 85 -90% of all mutation +ve cases (7) § Exon 20 mutations (e. g. T 790 M) commonly resistance mutations
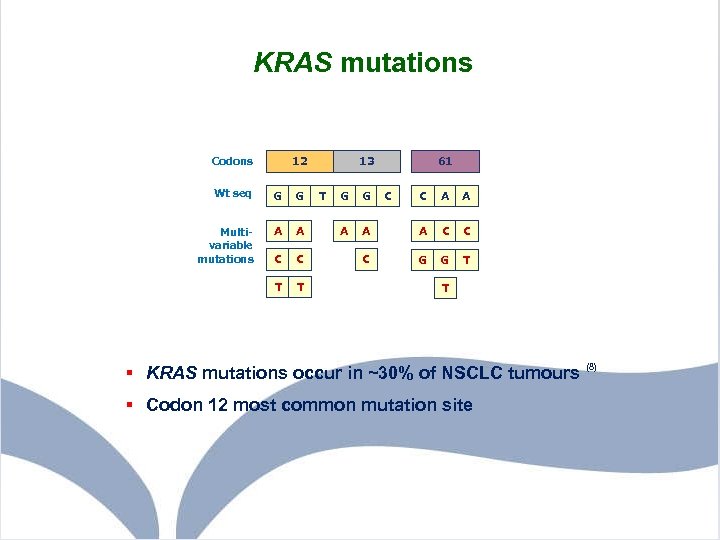 KRAS mutations 12 Codons Wt seq G G Multivariable mutations A A C C T T 13 T G G A 61 C C A A C C C G G T T § KRAS mutations occur in ~30% of NSCLC tumours (8) § Codon 12 most common mutation site
KRAS mutations 12 Codons Wt seq G G Multivariable mutations A A C C T T 13 T G G A 61 C C A A C C C G G T T § KRAS mutations occur in ~30% of NSCLC tumours (8) § Codon 12 most common mutation site
 Project Aims 1. Develop and set-up methods for EGFR and KRAS analysis 2. Determine best methods for analysis of EGFR and KRAS mutations 3. Develop and validate appropriate methodologies for testing
Project Aims 1. Develop and set-up methods for EGFR and KRAS analysis 2. Determine best methods for analysis of EGFR and KRAS mutations 3. Develop and validate appropriate methodologies for testing
 Samples § 48 Adenocarcinoma patient samples (ARI Pathology Dept) § 4 EGFR +ve control samples (Holland) § All were FFPE lung tumour samples (cores, slides & rolls) § 14 DNA samples for KRAS analysis (Transgenomic Inc) § Mutations in codon 12, 13 and 61 § Varied mutation levels (3% to 33%)
Samples § 48 Adenocarcinoma patient samples (ARI Pathology Dept) § 4 EGFR +ve control samples (Holland) § All were FFPE lung tumour samples (cores, slides & rolls) § 14 DNA samples for KRAS analysis (Transgenomic Inc) § Mutations in codon 12, 13 and 61 § Varied mutation levels (3% to 33%)
 Challenges with NSCLC testing using FFPE samples • Frequently low sample quantity = Low DNA yield • Variable tumour content within sample (<5% to 100%) • Poor DNA quality - Degradation (<300 bp), PCR inhibitors • Genetic heterogeneity – inter- and intra-tumour variation • Pathology departments involvement at this stage important to maximise % tumour – macrodissection
Challenges with NSCLC testing using FFPE samples • Frequently low sample quantity = Low DNA yield • Variable tumour content within sample (<5% to 100%) • Poor DNA quality - Degradation (<300 bp), PCR inhibitors • Genetic heterogeneity – inter- and intra-tumour variation • Pathology departments involvement at this stage important to maximise % tumour – macrodissection
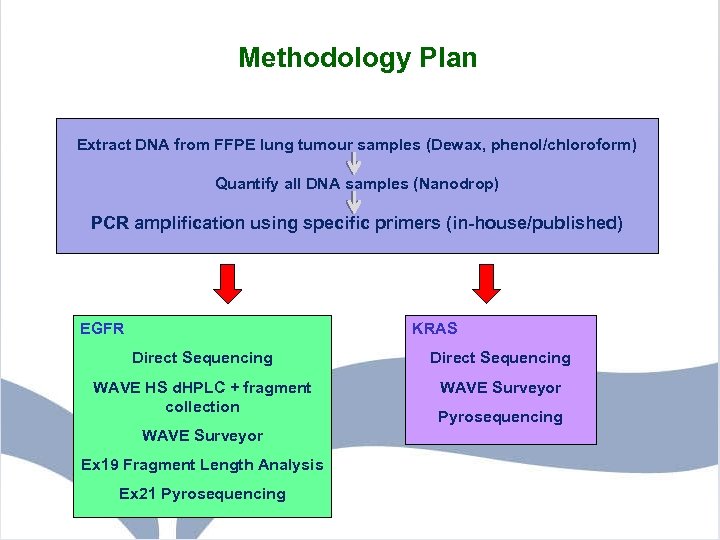 Methodology Plan Extract DNA from FFPE lung tumour samples (Dewax, phenol/chloroform) Quantify all DNA samples (Nanodrop) PCR amplification using specific primers (in-house/published) Quantify all DNA samples PCR amplification using specific primers EGFR KRAS Direct Sequencing WAVE HS d. HPLC + fragment collection WAVE Surveyor Ex 19 Fragment Length Analysis Ex 21 Pyrosequencing
Methodology Plan Extract DNA from FFPE lung tumour samples (Dewax, phenol/chloroform) Quantify all DNA samples (Nanodrop) PCR amplification using specific primers (in-house/published) Quantify all DNA samples PCR amplification using specific primers EGFR KRAS Direct Sequencing WAVE HS d. HPLC + fragment collection WAVE Surveyor Ex 19 Fragment Length Analysis Ex 21 Pyrosequencing
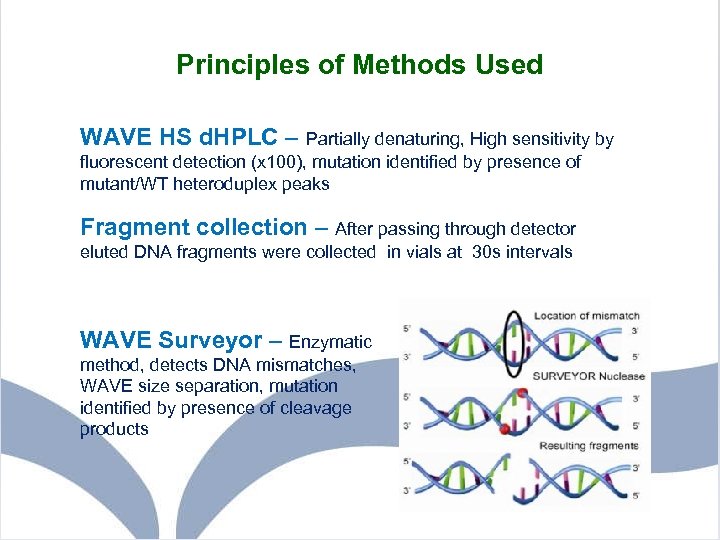 Principles of Methods Used WAVE HS d. HPLC – Partially denaturing, High sensitivity by fluorescent detection (x 100), mutation identified by presence of mutant/WT heteroduplex peaks Fragment collection – After passing through detector eluted DNA fragments were collected in vials at 30 s intervals WAVE Surveyor – Enzymatic method, detects DNA mismatches, WAVE size separation, mutation identified by presence of cleavage products
Principles of Methods Used WAVE HS d. HPLC – Partially denaturing, High sensitivity by fluorescent detection (x 100), mutation identified by presence of mutant/WT heteroduplex peaks Fragment collection – After passing through detector eluted DNA fragments were collected in vials at 30 s intervals WAVE Surveyor – Enzymatic method, detects DNA mismatches, WAVE size separation, mutation identified by presence of cleavage products
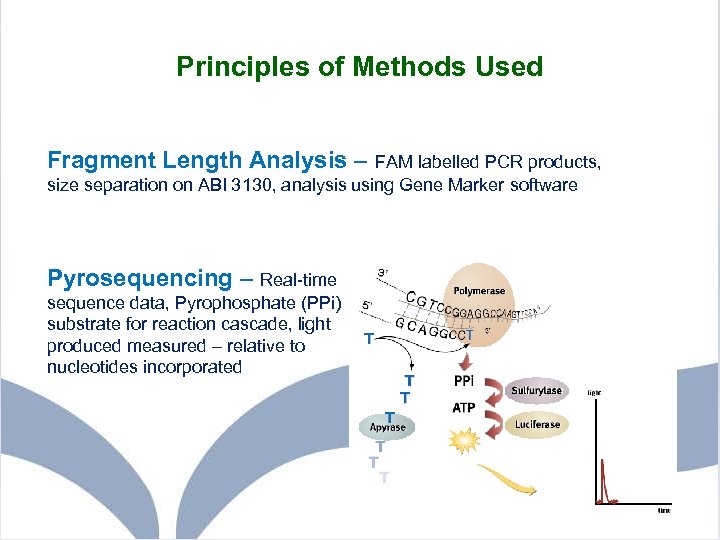 Principles of Methods Used Fragment Length Analysis – FAM labelled PCR products, size separation on ABI 3130, analysis using Gene Marker software Pyrosequencing – Real-time sequence data, Pyrophosphate (PPi) substrate for reaction cascade, light produced measured – relative to nucleotides incorporated
Principles of Methods Used Fragment Length Analysis – FAM labelled PCR products, size separation on ABI 3130, analysis using Gene Marker software Pyrosequencing – Real-time sequence data, Pyrophosphate (PPi) substrate for reaction cascade, light produced measured – relative to nucleotides incorporated
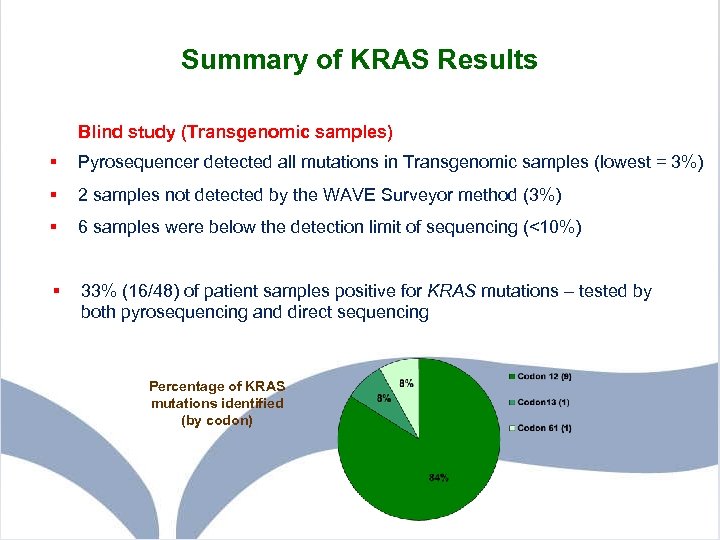 Summary of KRAS Results Blind study (Transgenomic samples) § Pyrosequencer detected all mutations in Transgenomic samples (lowest = 3%) § 2 samples not detected by the WAVE Surveyor method (3%) § 6 samples were below the detection limit of sequencing (<10%) § 33% (16/48) of patient samples positive for KRAS mutations – tested by both pyrosequencing and direct sequencing Percentage of KRAS mutations identified (by codon)
Summary of KRAS Results Blind study (Transgenomic samples) § Pyrosequencer detected all mutations in Transgenomic samples (lowest = 3%) § 2 samples not detected by the WAVE Surveyor method (3%) § 6 samples were below the detection limit of sequencing (<10%) § 33% (16/48) of patient samples positive for KRAS mutations – tested by both pyrosequencing and direct sequencing Percentage of KRAS mutations identified (by codon)
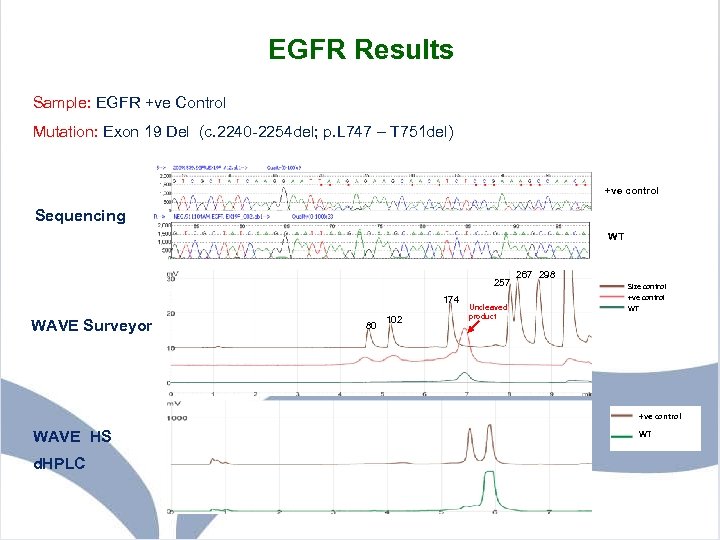 EGFR Results Sample: EGFR +ve Control Mutation: Exon 19 Del (c. 2240 -2254 del; p. L 747 – T 751 del) +ve control Sequencing WT 257 174 WAVE Surveyor 80 102 Uncleaved product 267 298 Size control +ve control WT +ve control WAVE HS d. HPLC WT
EGFR Results Sample: EGFR +ve Control Mutation: Exon 19 Del (c. 2240 -2254 del; p. L 747 – T 751 del) +ve control Sequencing WT 257 174 WAVE Surveyor 80 102 Uncleaved product 267 298 Size control +ve control WT +ve control WAVE HS d. HPLC WT
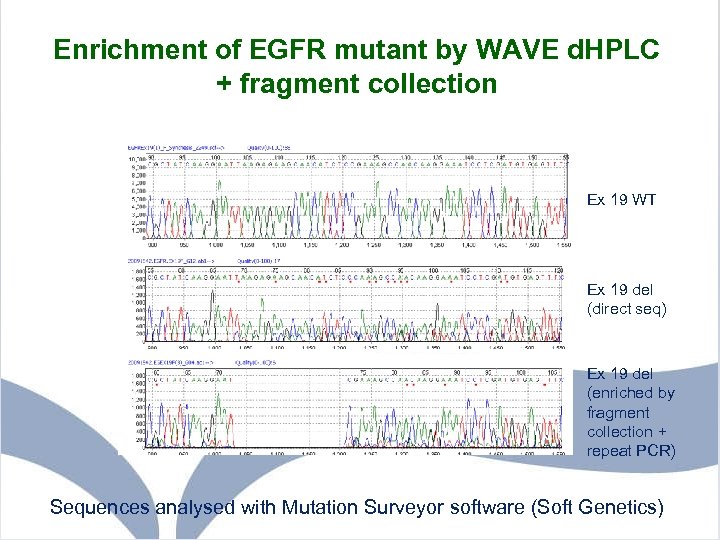 Enrichment of EGFR mutant by WAVE d. HPLC + fragment collection Ex 19 WT Ex 19 del (direct seq) Ex 19 del (enriched by fragment collection + repeat PCR) Sequences analysed with Mutation Surveyor software (Soft Genetics)
Enrichment of EGFR mutant by WAVE d. HPLC + fragment collection Ex 19 WT Ex 19 del (direct seq) Ex 19 del (enriched by fragment collection + repeat PCR) Sequences analysed with Mutation Surveyor software (Soft Genetics)
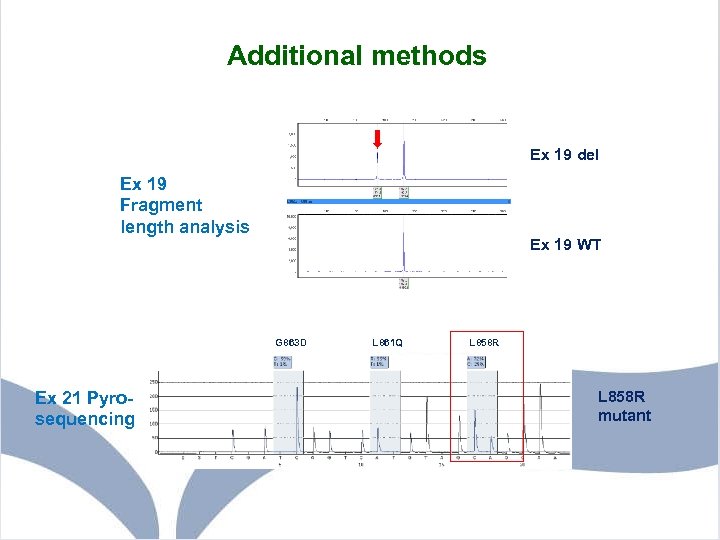 Additional methods Ex 19 del Ex 19 Fragment length analysis Ex 19 WT G 863 D Ex 21 Pyrosequencing L 861 Q L 858 R mutant
Additional methods Ex 19 del Ex 19 Fragment length analysis Ex 19 WT G 863 D Ex 21 Pyrosequencing L 861 Q L 858 R mutant
 Summary of EGFR Results § 12. 5% (6/48) of patient samples positive for EGFR mutation (Ex 19 - 3 Deletions; Ex 20 - 1 Insertion; Ex 21 - 2 L 858 R) § All methods detected Ex 19 del mutations in 4 EGFR +ve control samples § WAVE Surveyor confirmed all mutations found by direct sequence analysis § One Ex 19 del mutant too low to report by direct sequence analysis but clear by WAVE Surveyor and Fragment length analysis § Pyrosequencer – successfully detected Ex 21 mutants § Confident no false positive results All EGFR +ve samples were KRAS –ve Mutations confirmed by multiple methods
Summary of EGFR Results § 12. 5% (6/48) of patient samples positive for EGFR mutation (Ex 19 - 3 Deletions; Ex 20 - 1 Insertion; Ex 21 - 2 L 858 R) § All methods detected Ex 19 del mutations in 4 EGFR +ve control samples § WAVE Surveyor confirmed all mutations found by direct sequence analysis § One Ex 19 del mutant too low to report by direct sequence analysis but clear by WAVE Surveyor and Fragment length analysis § Pyrosequencer – successfully detected Ex 21 mutants § Confident no false positive results All EGFR +ve samples were KRAS –ve Mutations confirmed by multiple methods
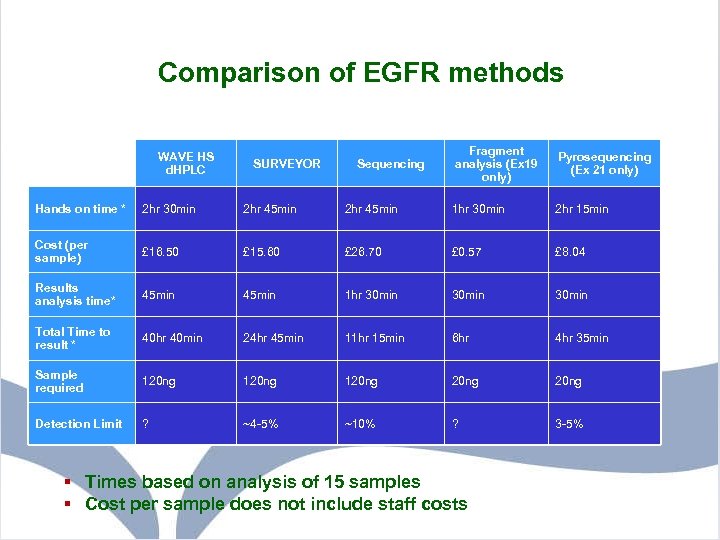 Comparison of EGFR methods WAVE HS d. HPLC SURVEYOR Sequencing Fragment analysis (Ex 19 only) Pyrosequencing (Ex 21 only) Hands on time * 2 hr 30 min 2 hr 45 min 1 hr 30 min 2 hr 15 min Cost (per sample) £ 16. 50 £ 15. 60 £ 26. 70 £ 0. 57 £ 8. 04 Results analysis time* 45 min 1 hr 30 min Total Time to result * 40 hr 40 min 24 hr 45 min 11 hr 15 min 6 hr 4 hr 35 min Sample required 120 ng 20 ng Detection Limit ? ~4 -5% ~10% ? 3 -5% § Times based on analysis of 15 samples § Cost per sample does not include staff costs
Comparison of EGFR methods WAVE HS d. HPLC SURVEYOR Sequencing Fragment analysis (Ex 19 only) Pyrosequencing (Ex 21 only) Hands on time * 2 hr 30 min 2 hr 45 min 1 hr 30 min 2 hr 15 min Cost (per sample) £ 16. 50 £ 15. 60 £ 26. 70 £ 0. 57 £ 8. 04 Results analysis time* 45 min 1 hr 30 min Total Time to result * 40 hr 40 min 24 hr 45 min 11 hr 15 min 6 hr 4 hr 35 min Sample required 120 ng 20 ng Detection Limit ? ~4 -5% ~10% ? 3 -5% § Times based on analysis of 15 samples § Cost per sample does not include staff costs
 Conclusions § Pick-up rate of EGFR mutations consistent with published data § Direct sequencing pick-up rate higher than expected. This likely to be due to enrichment of samples for tumour tissue by macrodissection § WAVE Surveyor, fragment analysis and pyrosequencing methods may be useful as a higher sensitivity screen in conjunction with direct sequencing § Fragment collection is a viable method for enrichment of low level mutations
Conclusions § Pick-up rate of EGFR mutations consistent with published data § Direct sequencing pick-up rate higher than expected. This likely to be due to enrichment of samples for tumour tissue by macrodissection § WAVE Surveyor, fragment analysis and pyrosequencing methods may be useful as a higher sensitivity screen in conjunction with direct sequencing § Fragment collection is a viable method for enrichment of low level mutations
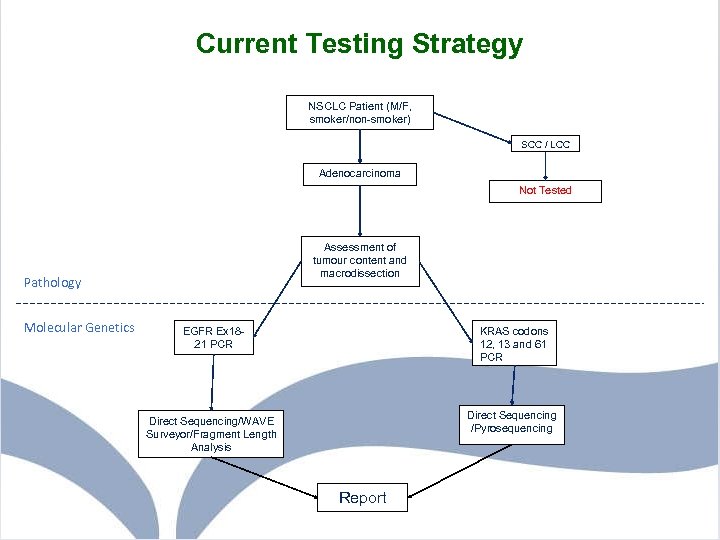 Current Testing Strategy NSCLC Patient (M/F, smoker/non-smoker) SCC / LCC Adenocarcinoma Not Tested Assessment of tumour content and macrodissection Pathology Molecular Genetics EGFR Ex 1821 PCR KRAS codons 12, 13 and 61 PCR Direct Sequencing /Pyrosequencing Direct Sequencing/WAVE Surveyor/Fragment Length Analysis Report
Current Testing Strategy NSCLC Patient (M/F, smoker/non-smoker) SCC / LCC Adenocarcinoma Not Tested Assessment of tumour content and macrodissection Pathology Molecular Genetics EGFR Ex 1821 PCR KRAS codons 12, 13 and 61 PCR Direct Sequencing /Pyrosequencing Direct Sequencing/WAVE Surveyor/Fragment Length Analysis Report
 Acknowledgements § Aberdeen Lab Caroline Clark Christine Bell Aileen Osborne Louise Carnegie Heather Greig Kevin Kelly § Transgenomic Gerald Martin § Clinical/Pathology Keith Kerr Marianne Nicholson Zosia Miedzybrodzka § Astra Zeneca For providing funding
Acknowledgements § Aberdeen Lab Caroline Clark Christine Bell Aileen Osborne Louise Carnegie Heather Greig Kevin Kelly § Transgenomic Gerald Martin § Clinical/Pathology Keith Kerr Marianne Nicholson Zosia Miedzybrodzka § Astra Zeneca For providing funding
 References 1 Ferlay J. et al. Estimates of the cancer incidence and mortality in Europe in 2006. Annals of Oncology (2007) 18: 581 -5923 2 Harkness E. F. et al. Changing trends in incidence of lung cancer by histologic type in Scotland. Int. J. Cancer (2002) 102: 179 -183 3 D’Addario G. & Felip E. Non-small-cell lung cancer: ESMO Clinical Recommendations for diagnosis, treatment and follow-up. Annals of Oncology (2008) 19 (Sup 2): ii 39 - ii 40 4 Scottish Executive Health Department. Cancer scenarios: an aid to planning cancer services in Scotland in the next decade. . 2001 The Scottish Executive: Edinburgh. 5 Janne P. A. et al. A rapid and sensitive enzymatic method for epidermal growth factor receptor mutation screening. Clin Cancer Res (2006); 12 (3): 751 - 758 6 Sequist L. V. et al. Epidermal Growth Factor Receptor mutation testing in the care of lung cancer patients. Clin Cancer Res (2006); 12 (Sup 14): 4403 s – 4408 s 7 Sequist L. V. & Lynch T. J. EGFR Tyrosine Kinase Inhibitors in lung cancer: an evolving story. Ann Rev Med (2008) 59: 429 -42 8 Do, H. et al. High resolution melting analysis for rapid and sensitive EGFR and KRAS detection in formalin fixed paraffin embedded biopsies. BMC Cancer (2008) 8: 142
References 1 Ferlay J. et al. Estimates of the cancer incidence and mortality in Europe in 2006. Annals of Oncology (2007) 18: 581 -5923 2 Harkness E. F. et al. Changing trends in incidence of lung cancer by histologic type in Scotland. Int. J. Cancer (2002) 102: 179 -183 3 D’Addario G. & Felip E. Non-small-cell lung cancer: ESMO Clinical Recommendations for diagnosis, treatment and follow-up. Annals of Oncology (2008) 19 (Sup 2): ii 39 - ii 40 4 Scottish Executive Health Department. Cancer scenarios: an aid to planning cancer services in Scotland in the next decade. . 2001 The Scottish Executive: Edinburgh. 5 Janne P. A. et al. A rapid and sensitive enzymatic method for epidermal growth factor receptor mutation screening. Clin Cancer Res (2006); 12 (3): 751 - 758 6 Sequist L. V. et al. Epidermal Growth Factor Receptor mutation testing in the care of lung cancer patients. Clin Cancer Res (2006); 12 (Sup 14): 4403 s – 4408 s 7 Sequist L. V. & Lynch T. J. EGFR Tyrosine Kinase Inhibitors in lung cancer: an evolving story. Ann Rev Med (2008) 59: 429 -42 8 Do, H. et al. High resolution melting analysis for rapid and sensitive EGFR and KRAS detection in formalin fixed paraffin embedded biopsies. BMC Cancer (2008) 8: 142


