4481a6709fecee7ffd443b14a5827007.ppt
- Количество слайдов: 41

Developing quantitative slide-based assays to assess target inhibition in oncology drug discovery and development Pathology Visions October 24 -27, 2010 Doug Bowman Millennium Pharmaceuticals © 2010 Millennium Pharmaceuticals Inc. , The Takeda Oncology Company

Outline ▐ ▐ ▐ Imaging @ Millennium Technology development & integration Applications in Oncology ▌ ▌ ▌ Assess in vivo potency Biomarker development Assess clinical activity
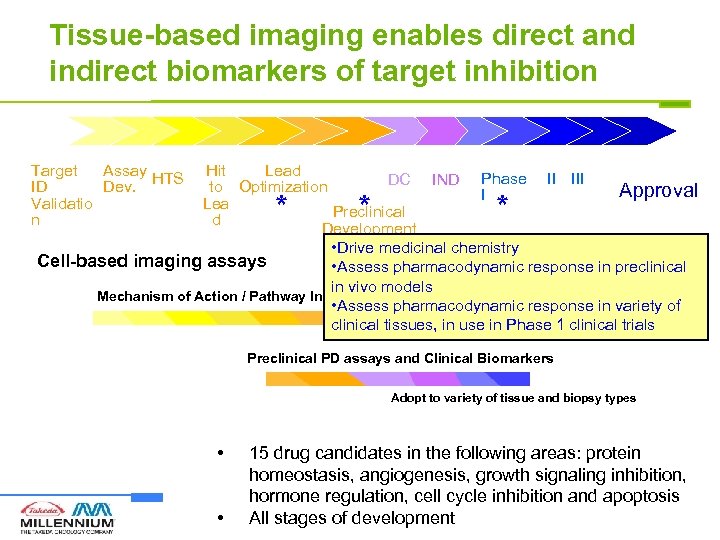
Tissue-based imaging enables direct and indirect biomarkers of target inhibition Target Assay HTS ID Dev. Validatio n Hit Lead DC IND Phase II III to Optimization Approval I Lea Preclinical d Development • Drive medicinal chemistry Cell-based imaging assays • Assess pharmacodynamic response in preclinical in vivo models Mechanism of Action / Pathway Inhibition / Terminal Outcome • Assess pharmacodynamic response in variety of clinical tissues, in use in Phase 1 clinical trials * * * Preclinical PD assays and Clinical Biomarkers Adopt to variety of tissue and biopsy types • • 15 drug candidates in the following areas: protein homeostasis, angiogenesis, growth signaling inhibition, hormone regulation, cell cycle inhibition and apoptosis All stages of development
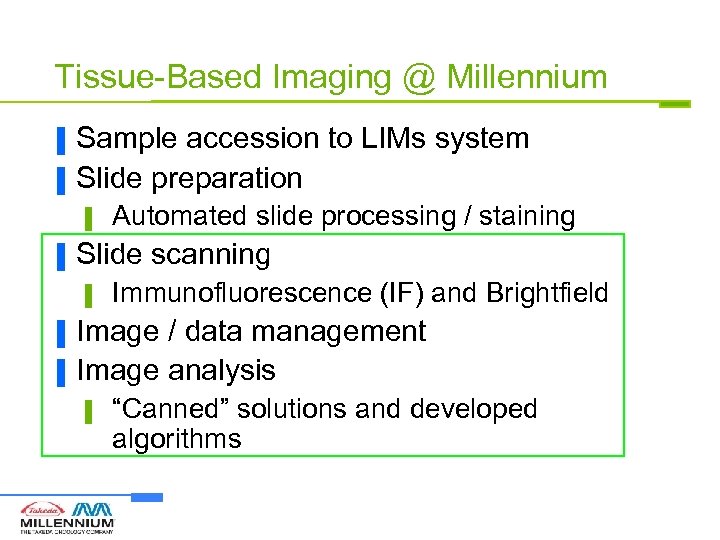
Tissue-Based Imaging @ Millennium ▐ ▐ Sample accession to LIMs system Slide preparation ▌ ▐ Slide scanning ▌ ▐ ▐ Automated slide processing / staining Immunofluorescence (IF) and Brightfield Image / data management Image analysis ▌ “Canned” solutions and developed algorithms
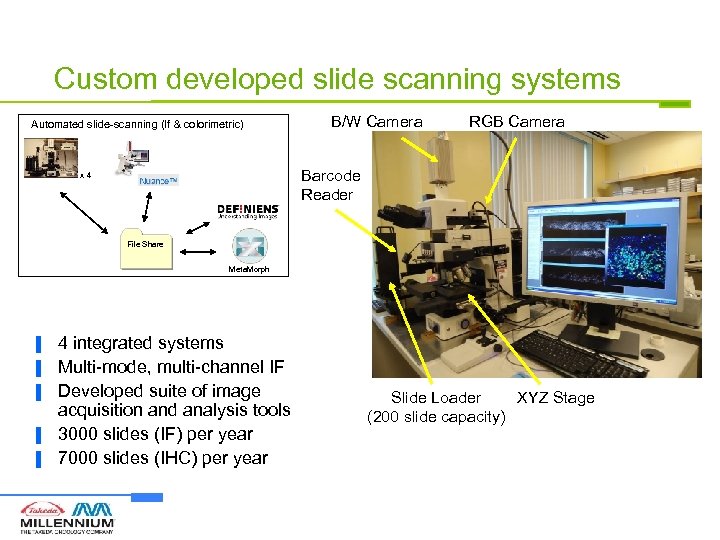
Custom developed slide scanning systems Automated slide-scanning (If & colorimetric) B/W Camera RGB Camera Barcode Reader x 4 File Share Meta. Morph ▐ ▐ ▐ 4 integrated systems Multi-mode, multi-channel IF Developed suite of image acquisition and analysis tools 3000 slides (IF) per year 7000 slides (IHC) per year Slide Loader XYZ Stage (200 slide capacity)

High resolution and efficient scanning of clinical samples 20 x objective 100 um • Multi-mode • Multi-channel IF
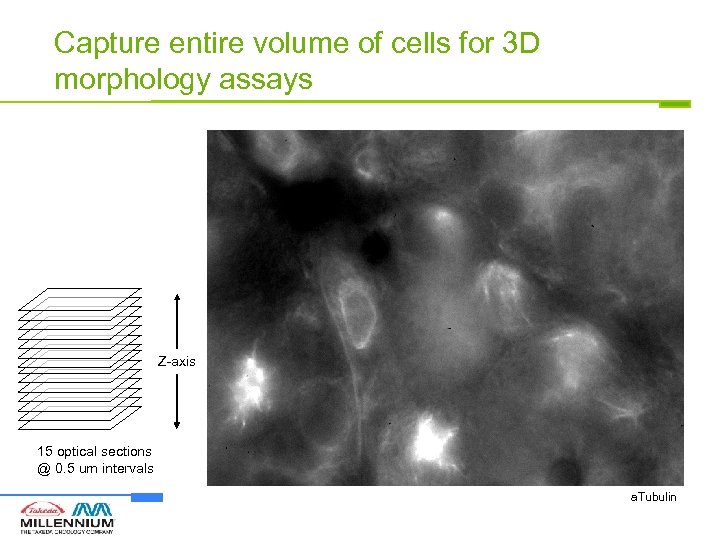
Capture entire volume of cells for 3 D morphology assays Z-axis 15 optical sections @ 0. 5 um intervals a. Tubulin
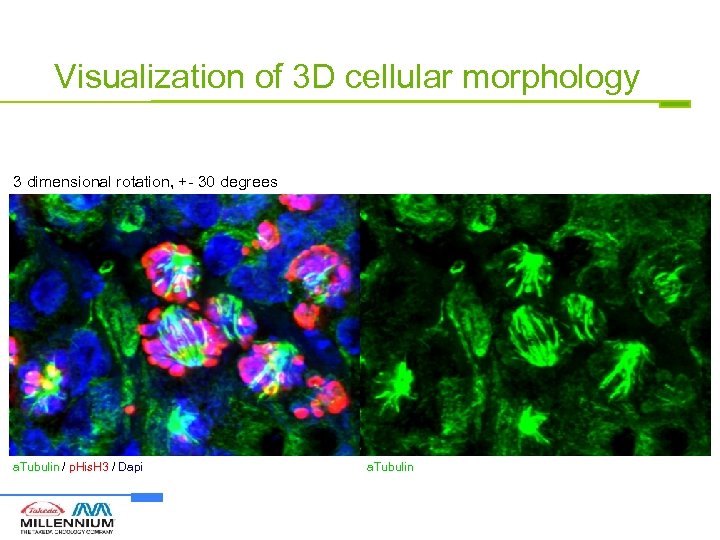
Visualization of 3 D cellular morphology 3 dimensional rotation, +- 30 degrees a. Tubulin / p. His. H 3 / Dapi a. Tubulin
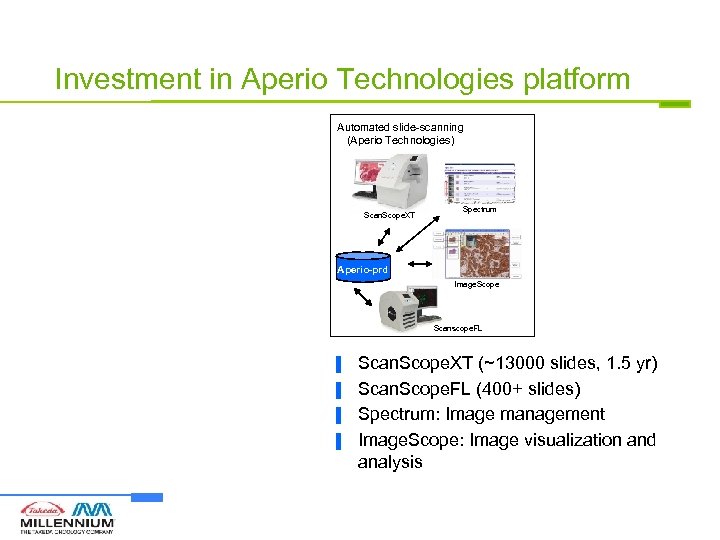
Investment in Aperio Technologies platform Automated slide-scanning (Aperio Technologies) Scan. Scope. XT Spectrum Aperio-prd Image. Scope Scanscope. FL ▐ ▐ Scan. Scope. XT (~13000 slides, 1. 5 yr) Scan. Scope. FL (400+ slides) Spectrum: Image management Image. Scope: Image visualization and analysis
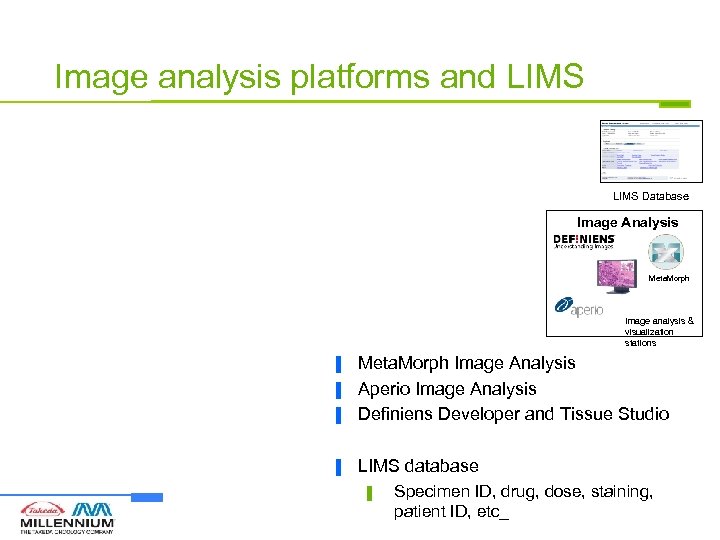
Image analysis platforms and LIMS Database Image Analysis Meta. Morph Image analysis & visualization stations ▐ Meta. Morph Image Analysis Aperio Image Analysis Definiens Developer and Tissue Studio ▐ LIMS database ▐ ▐ ▌ Specimen ID, drug, dose, staining, patient ID, etc_
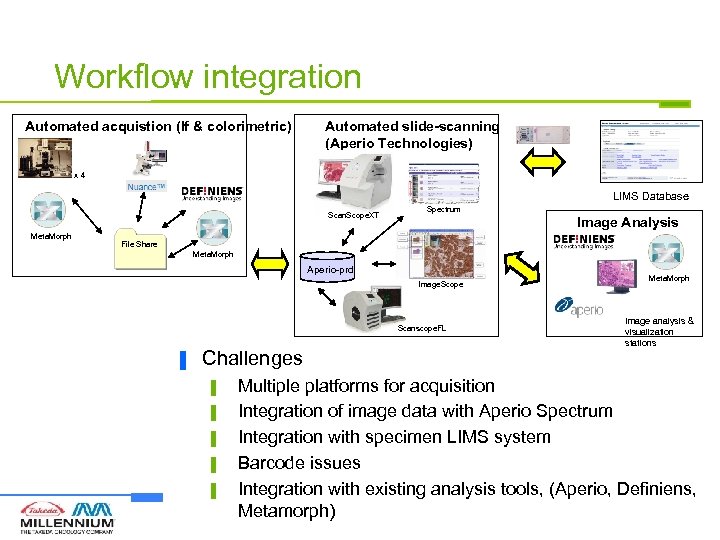
Workflow integration Automated acquistion (If & colorimetric) Automated slide-scanning (Aperio Technologies) x 4 LIMS Database Scan. Scope. XT Meta. Morph Spectrum Image Analysis File Share Meta. Morph Aperio-prd Image. Scope Scanscope. FL ▐ Challenges ▌ ▌ ▌ Meta. Morph Image analysis & visualization stations Multiple platforms for acquisition Integration of image data with Aperio Spectrum Integration with specimen LIMS system Barcode issues Integration with existing analysis tools, (Aperio, Definiens, Metamorph)
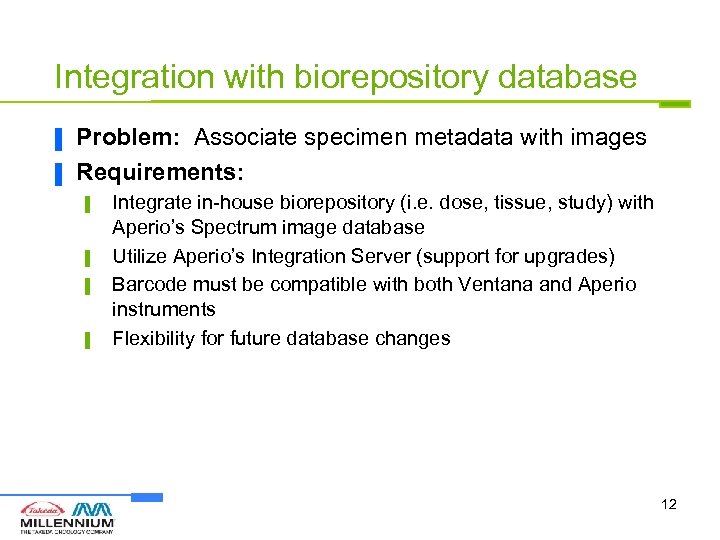
Integration with biorepository database ▐ ▐ Problem: Associate specimen metadata with images Requirements: ▌ ▌ Integrate in-house biorepository (i. e. dose, tissue, study) with Aperio’s Spectrum image database Utilize Aperio’s Integration Server (support for upgrades) Barcode must be compatible with both Ventana and Aperio instruments Flexibility for future database changes 12
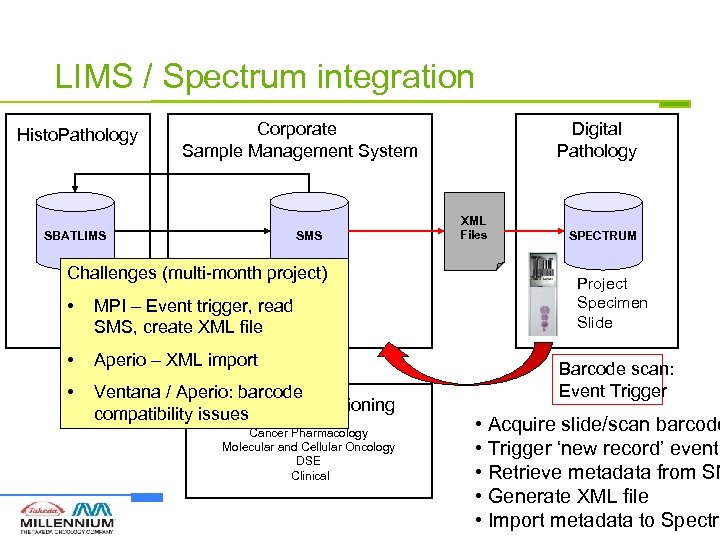
LIMS / Spectrum integration Histo. Pathology Corporate Sample Management System SBATLIMS SMS Challenges (multi-month project) • MPI – Event trigger, read SMS, create XML file • Aperio – XML import • Ventana / Aperio: barcode Sample Accessioning compatibility issues Cancer Pharmacology Molecular and Cellular Oncology DSE Clinical Digital Pathology XML Files SPECTRUM Project Specimen Slide Barcode scan: Event Trigger • Acquire slide/scan barcode • Trigger ‘new record’ event • Retrieve metadata from SM • Generate XML file • Import metadata to Spectru
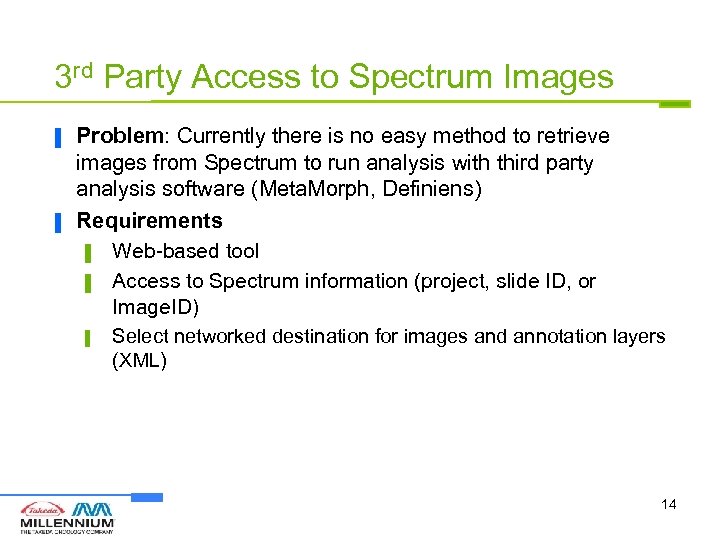
3 rd Party Access to Spectrum Images ▐ ▐ Problem: Currently there is no easy method to retrieve images from Spectrum to run analysis with third party analysis software (Meta. Morph, Definiens) Requirements ▌ Web-based tool ▌ Access to Spectrum information (project, slide ID, or Image. ID) ▌ Select networked destination for images and annotation layers (XML) 14
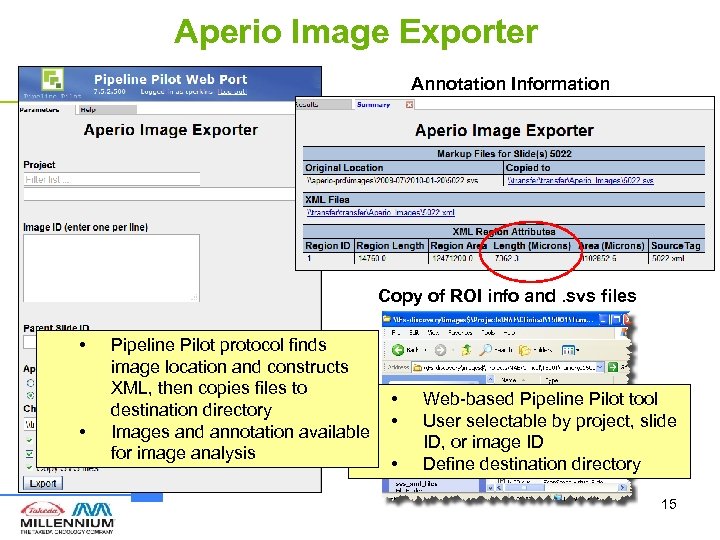
Aperio Image Exporter Annotation Information Copy of ROI info and. svs files • • Pipeline Pilot protocol finds image location and constructs XML, then copies files to destination directory Images and annotation available for image analysis • • • Web-based Pipeline Pilot tool User selectable by project, slide ID, or image ID Define destination directory 15
Application examples ▐ ▐ ▐ Direct and indirect pathway biomarkers Preclinical biomarkers Clinical biomarkers
▐ Preclinical biomarker ▌ ▌ ▌ Lead optimization efforts to measure potency of compounds Hit target, affect pathway Guide clinical: understand temporal response of biomarker for optimal sampling point and to help define clinical sampling
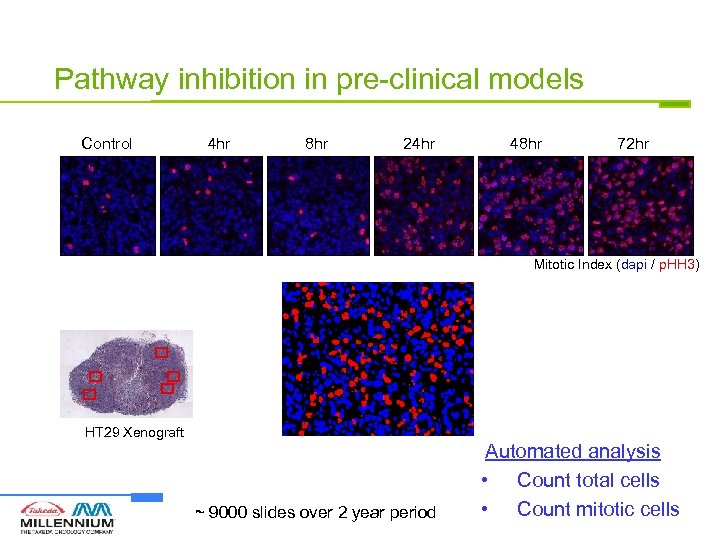
Pathway inhibition in pre-clinical models Control 4 hr 8 hr 24 hr 48 hr 72 hr Mitotic Index (dapi / p. HH 3) HT 29 Xenograft ~ 9000 slides over 2 year period Automated analysis • Count total cells • Count mitotic cells
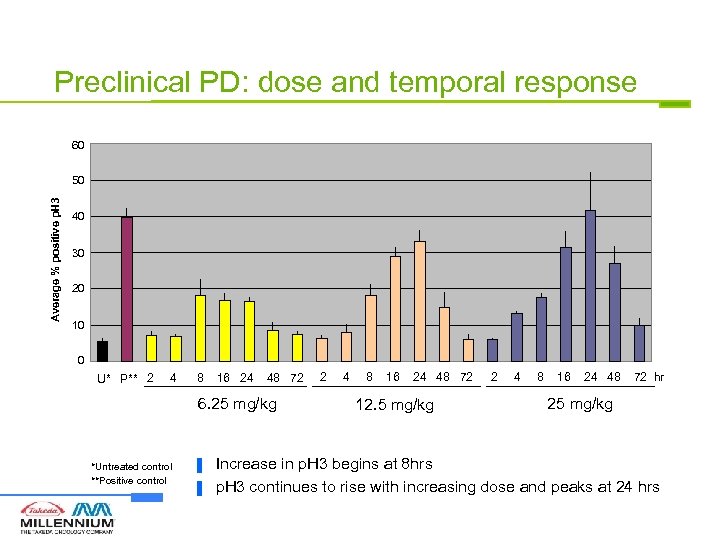
Preclinical PD: dose and temporal response 60 Average % positive p. H 3 50 40 30 20 10 0 U* P** 2 4 8 16 24 48 72 6. 25 mg/kg *Untreated control **Positive control ▐ ▐ 2 4 8 16 24 48 72 12. 5 mg/kg 2 4 8 16 24 48 72 hr 25 mg/kg Increase in p. H 3 begins at 8 hrs p. H 3 continues to rise with increasing dose and peaks at 24 hrs
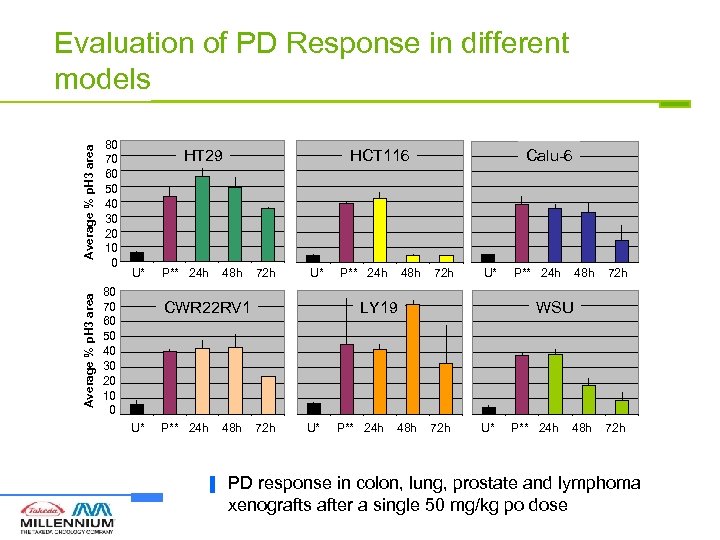
Average % p. H 3 area Evaluation of PD Response in different models 80 70 60 50 40 30 20 10 0 HT 29 U* 80 70 60 50 40 30 20 10 0 P** 24 h HCT 116 48 h 72 h U* CWR 22 RV 1 U* P** 24 h ▐ 48 h P** 24 h 48 h Calu-6 72 h U* LY 19 72 h U* P** 24 h 48 h 72 h WSU 72 h U* P** 24 h PD response in colon, lung, prostate and lymphoma xenografts after a single 50 mg/kg po dose

▐ MLN 8237: Aurora A Kinase inhibitor ▌ ▌ Pharmacodynamic evaluation in Phase 1 clinical studies in advanced solid tumors Includes image-based PD biomarker strategy to assess activity
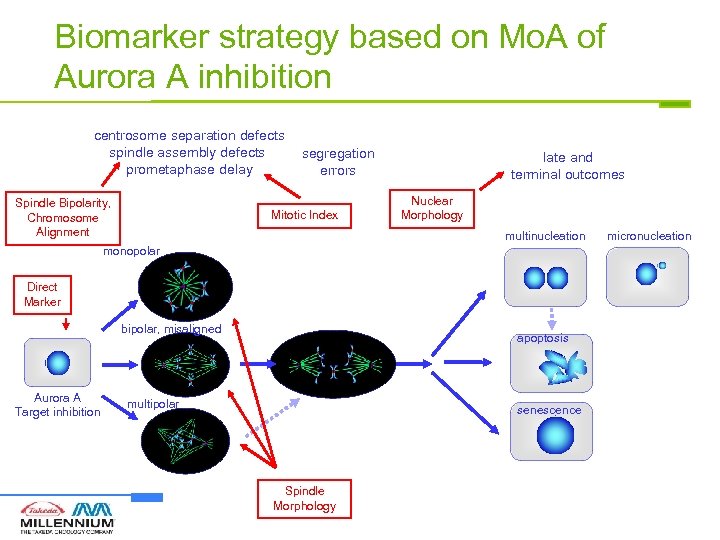
Biomarker strategy based on Mo. A of Aurora A inhibition centrosome separation defects spindle assembly defects prometaphase delay Spindle Bipolarity, Chromosome Alignment segregation errors Mitotic Index late and terminal outcomes Nuclear Morphology multinucleation monopolar Direct Marker bipolar, misaligned Aurora A Target inhibition apoptosis multipolar senescence Spindle Morphology micronucleation
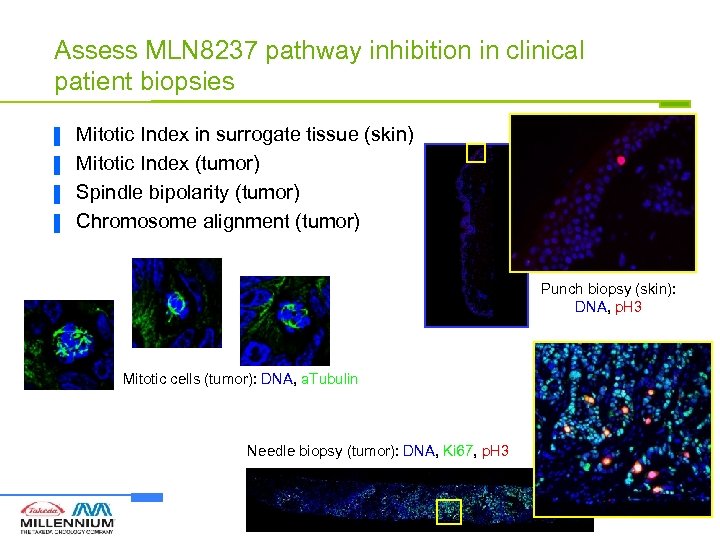
Assess MLN 8237 pathway inhibition in clinical patient biopsies ▐ ▐ Mitotic Index in surrogate tissue (skin) Mitotic Index (tumor) Spindle bipolarity (tumor) Chromosome alignment (tumor) Punch biopsy (skin): DNA, p. H 3 Mitotic cells (tumor): DNA, a. Tubulin Needle biopsy (tumor): DNA, Ki 67, p. H 3
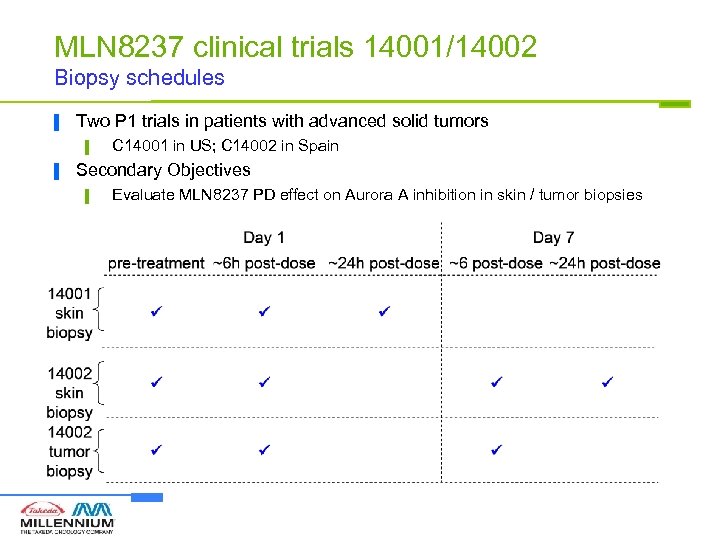
MLN 8237 clinical trials 14001/14002 Biopsy schedules ▐ Two P 1 trials in patients with advanced solid tumors ▌ ▐ C 14001 in US; C 14002 in Spain Secondary Objectives ▌ Evaluate MLN 8237 PD effect on Aurora A inhibition in skin / tumor biopsies
PT 703, a case study to highlight pharmacodynamic assays used ▐ ▐ 33 year old woman with neural sheath sarcoma 150 mg QD dose group (Spain) Completed 4 cycles of treatment Usable tissue and high dose make this a good case study
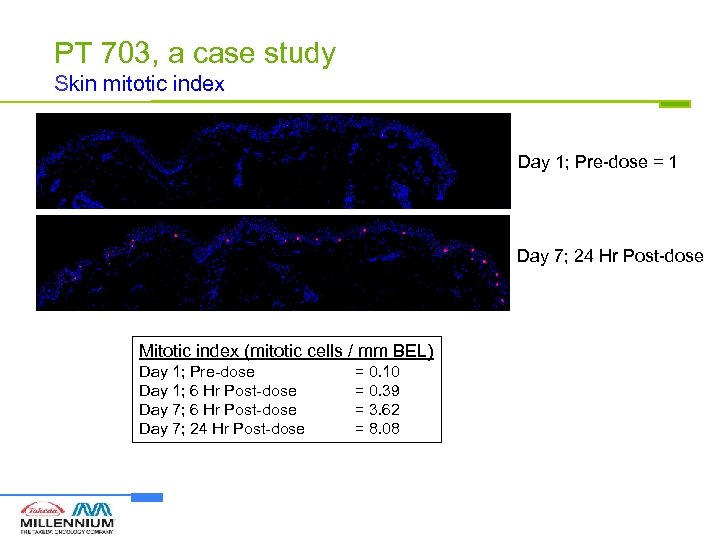
PT 703, a case study Skin mitotic index Day 1; Pre-dose = 1 Day 7; 24 Hr Post-dose Mitotic index (mitotic cells / mm BEL) Day 1; Pre-dose Day 1; 6 Hr Post-dose Day 7; 24 Hr Post-dose = 0. 10 = 0. 39 = 3. 62 = 8. 08
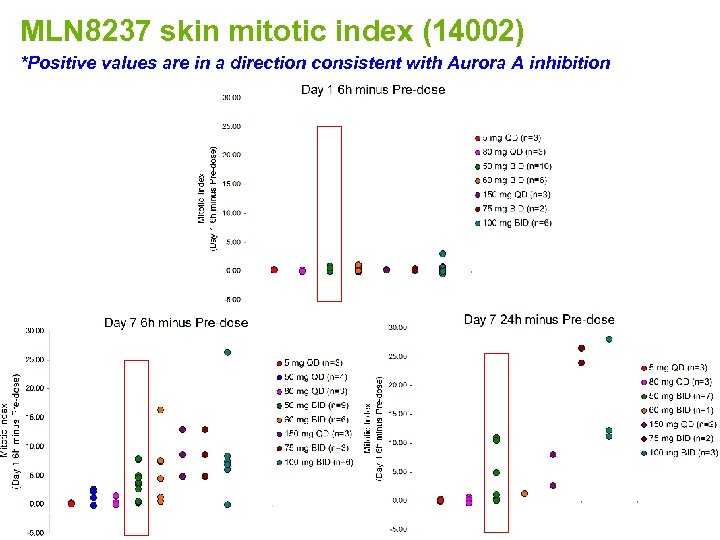
MLN 8237 skin mitotic index (14002) *Positive values are in a direction consistent with Aurora A inhibition
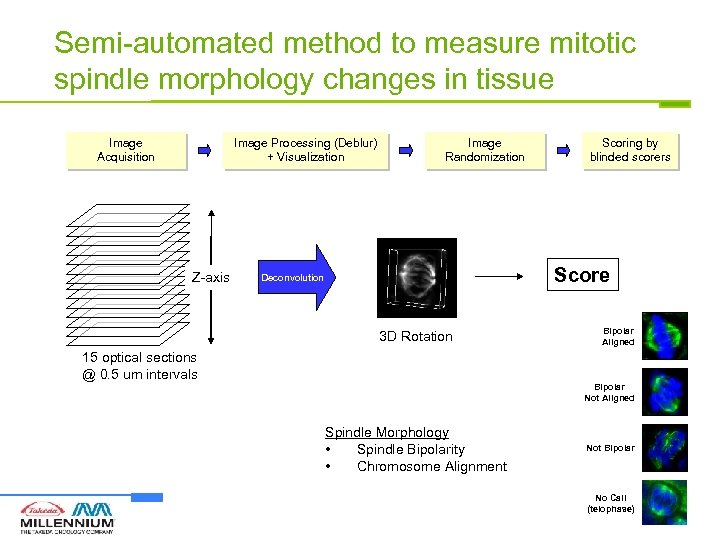
Semi-automated method to measure mitotic spindle morphology changes in tissue Image Acquisition Image Processing (Deblur) + Visualization Z-axis Image Randomization Scoring by blinded scorers Score Deconvolution 3 D Rotation 15 optical sections @ 0. 5 um intervals Bipolar Aligned Bipolar Not Aligned Spindle Morphology • Spindle Bipolarity • Chromosome Alignment Not Bipolar No Call (telophase)
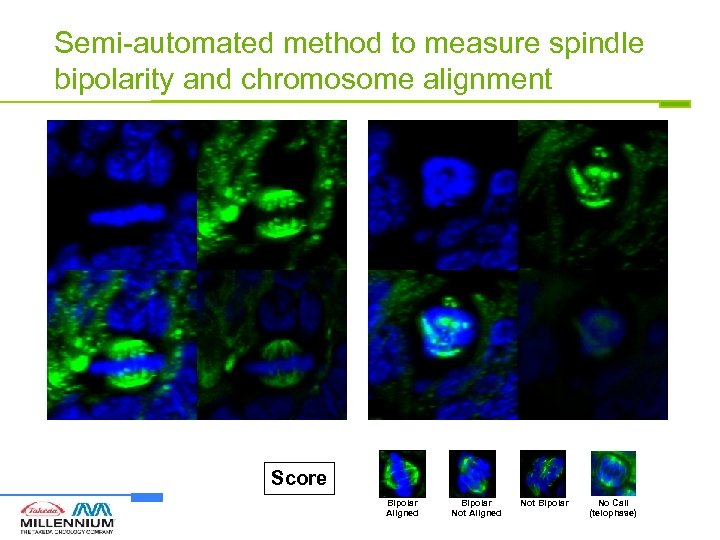
Semi-automated method to measure spindle bipolarity and chromosome alignment Score Bipolar Aligned Bipolar Not Aligned Not Bipolar No Call (telophase)
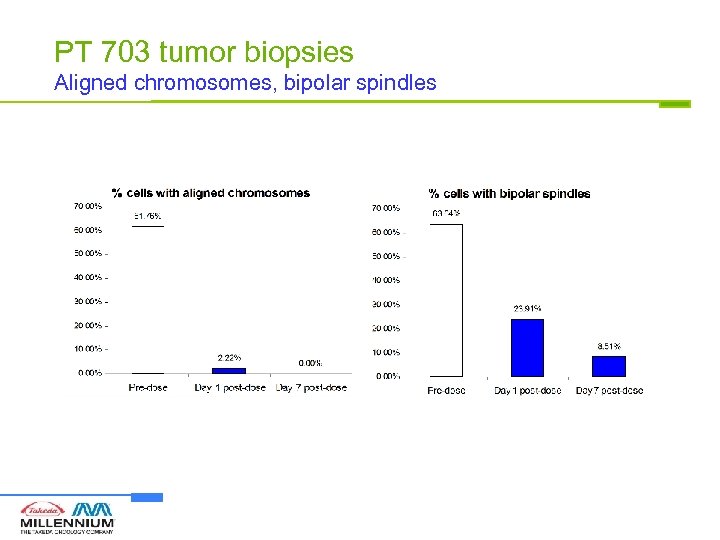
PT 703 tumor biopsies Aligned chromosomes, bipolar spindles
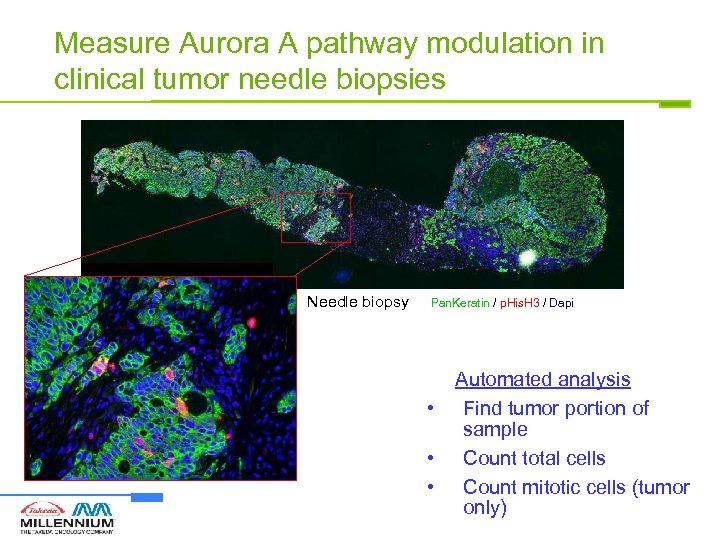
Measure Aurora A pathway modulation in clinical tumor needle biopsies Needle biopsy Pan. Keratin / p. His. H 3 / Dapi • • • Automated analysis Find tumor portion of sample Count total cells Count mitotic cells (tumor only)
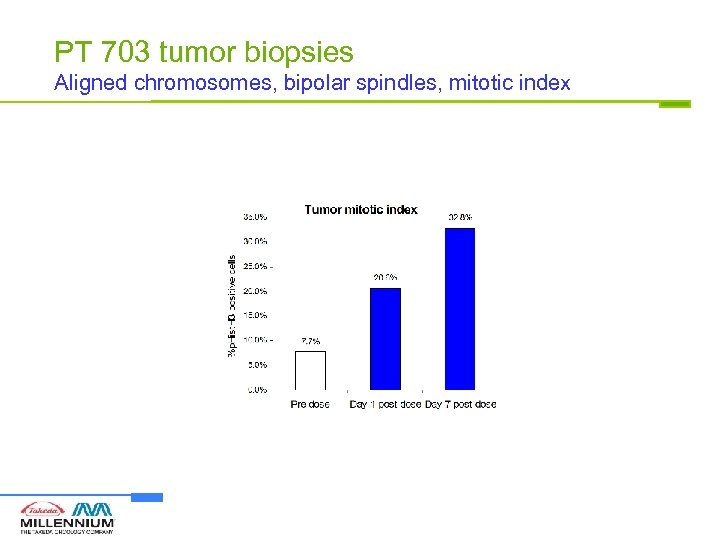
PT 703 tumor biopsies Aligned chromosomes, bipolar spindles, mitotic index
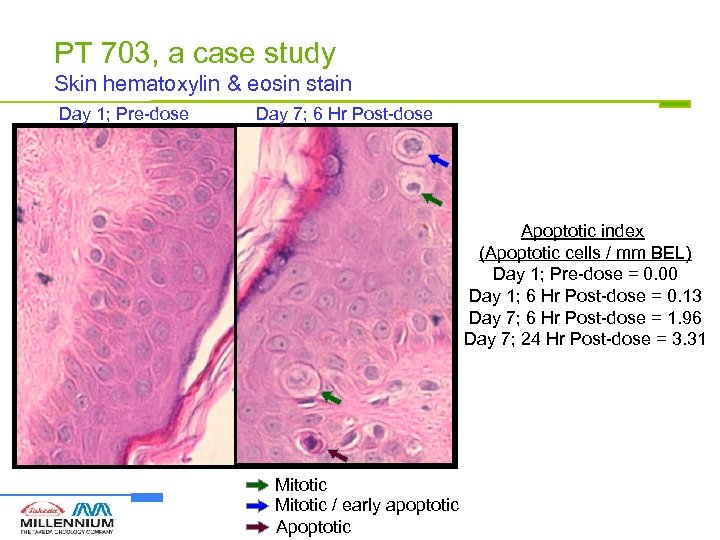
PT 703, a case study Skin hematoxylin & eosin stain Day 1; Pre-dose Day 7; 6 Hr Post-dose Apoptotic index (Apoptotic cells / mm BEL) Day 1; Pre-dose = 0. 00 Day 1; 6 Hr Post-dose = 0. 13 Day 7; 6 Hr Post-dose = 1. 96 Day 7; 24 Hr Post-dose = 3. 31 Mitotic / early apoptotic Apoptotic
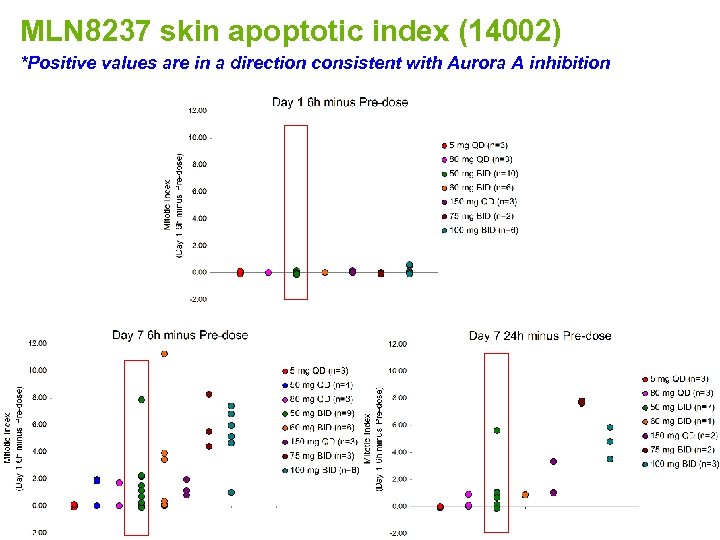
MLN 8237 skin apoptotic index (14002) *Positive values are in a direction consistent with Aurora A inhibition
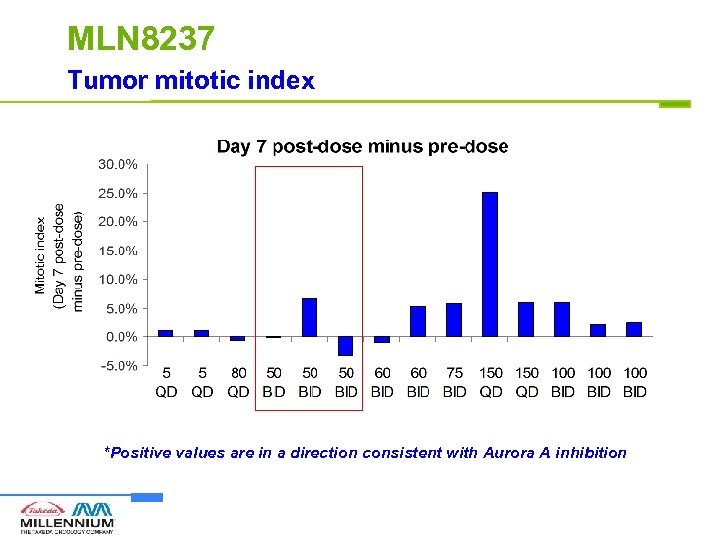
MLN 8237 Tumor mitotic index *Positive values are in a direction consistent with Aurora A inhibition
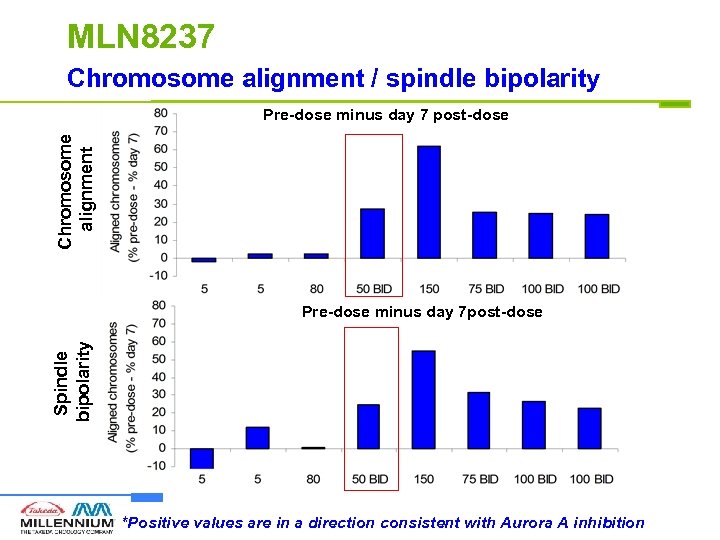
MLN 8237 Chromosome alignment / spindle bipolarity Chromosome alignment Pre-dose minus day 7 post-dose Spindle bipolarity Pre-dose minus day 7 post-dose *Positive values are in a direction consistent with Aurora A inhibition
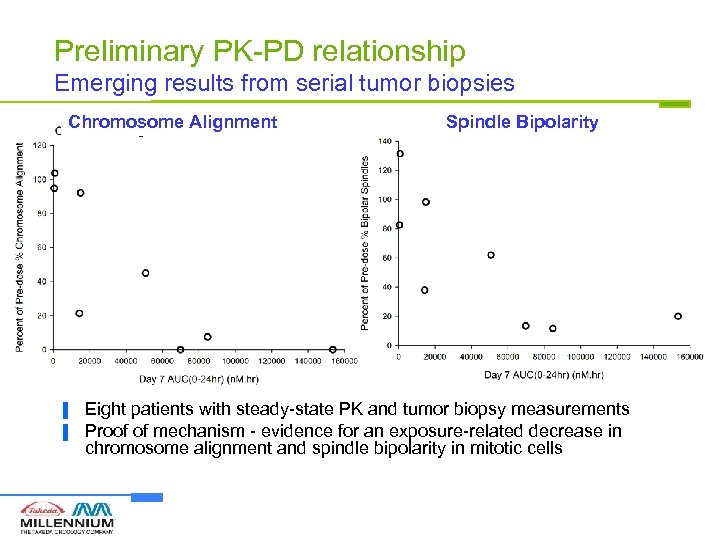
Preliminary PK-PD relationship Emerging results from serial tumor biopsies Chromosome Alignment ▐ ▐ Spindle Bipolarity Eight patients with steady-state PK and tumor biopsy measurements Proof of mechanism - evidence for an exposure-related decrease in chromosome alignment and spindle bipolarity in mitotic cells

How has the PK/PD data guided future decisions? ▐ Demonstrated proof of mechanism – MLN 8237 inhibits Aurora A in patients ▌ Clinical responses likely related to Aurora A inhibition ▌ Use of p. Hist. H 3 as marker of mitotic accumulation confirmed selectivity for Aurora A relative to Aurora B in patients ▌ Allows for rational drug development based on Aurora A mechanism • Combination selection, response marker identification ▐ ▐ Demonstrated that RP 2 D (50 mg BIDx 7 d) results in biologically active exposures ▌ Same assays applied to MLN 8054 demonstrated that biologically active exposures achieved at doses greater than the MTD (defined by somnolence) PD data informing future decisions ▌ Guide dose and schedule decisions for combination studies
Summary ▐ ▐ Developed and integrated imaging technologies for use in multiple drug discovery and development programs Leveraged tissue-based assays and technologies ▌ ▌ ▌ Drive medicinal chemistry Assess pharmacodynamic response in preclinical in vivo models Assess pharmacodynamic response in variety of clinical tissues, in use in Phase 1 clinical trials
Acknowledgements ▐ Slide-based Assay Team ▌ ▌ ▌ Krissy Burke Alice Mc. Donald Vaishali Shinde Yu Yang Brad Stringer ▐ Molecular and Cellular Oncology ▌ ▌ ▐ Takeda Development Research ▌ ▐ Research Systems / IT ▌ ▌ David Statham Chris Perkins ▐ Jeff Ecsedy Natalie Roy D’Amore Arijit Chakravarty MLN 8237 Project Team * POSTER (P 25): Details integration work and highlights example using Definiens Tissue Studio and Developer

We Aspire to Cure Cancer™ © 2010 Millennium Pharmaceuticals, Inc.
4481a6709fecee7ffd443b14a5827007.ppt