440aa8517fbe04a226ca900f009a5cc1.ppt
- Количество слайдов: 27
 Developing multiplex PCR assays for human identity testing - is there overlap with pathogen screening? Dr. Peter M Vallone Human Identification Project U. S. National Institute of Standards and Technology So. GAT XX Warsaw, Poland 12 -13 June 2007
Developing multiplex PCR assays for human identity testing - is there overlap with pathogen screening? Dr. Peter M Vallone Human Identification Project U. S. National Institute of Standards and Technology So. GAT XX Warsaw, Poland 12 -13 June 2007
 Human Identity Project at NIST • Genotype samples with commercial assays (nucleic acid based) • Produce Standard Reference Materials • Training and Interlaboratory Studies • Develop novel multiplex assays for genotyping
Human Identity Project at NIST • Genotype samples with commercial assays (nucleic acid based) • Produce Standard Reference Materials • Training and Interlaboratory Studies • Develop novel multiplex assays for genotyping
 Goals • Interest in further understanding of PCR • Learn more about the So. GAT mission • Building multiplex PCR assays – Upper limits of multiplexing? – Robust performance – Speed up assay development • Can this be of use to your community? • Further my knowledge of assay design outside of forensic applications
Goals • Interest in further understanding of PCR • Learn more about the So. GAT mission • Building multiplex PCR assays – Upper limits of multiplexing? – Robust performance – Speed up assay development • Can this be of use to your community? • Further my knowledge of assay design outside of forensic applications
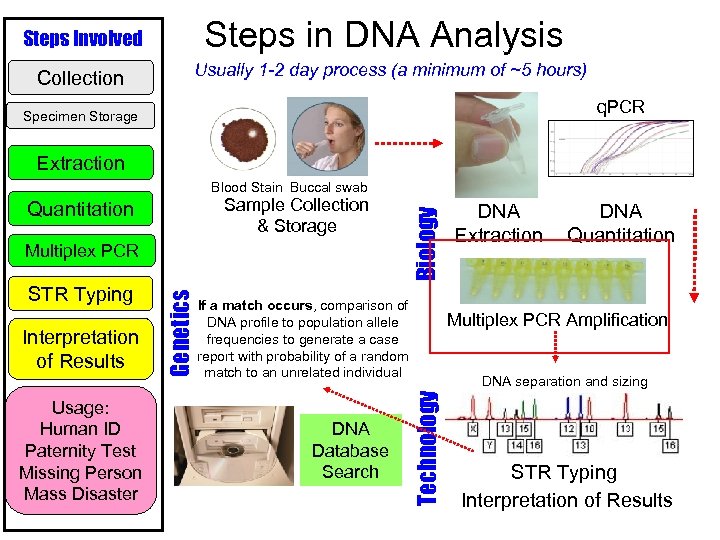 Steps in DNA Analysis Steps Involved Collection Usually 1 -2 day process (a minimum of ~5 hours) q. PCR Specimen Storage Extraction Sample Collection & Storage STR Typing Interpretation of Results Usage: Human ID Paternity Test Missing Person Mass Disaster Genetics Multiplex PCR If a match occurs, comparison of DNA profile to population allele frequencies to generate a case report with probability of a random match to an unrelated individual DNA Database Search DNA Extraction DNA Quantitation Multiplex PCR Amplification DNA separation and sizing Technology Quantitation Biology Blood Stain Buccal swab STR Typing Interpretation of Results
Steps in DNA Analysis Steps Involved Collection Usually 1 -2 day process (a minimum of ~5 hours) q. PCR Specimen Storage Extraction Sample Collection & Storage STR Typing Interpretation of Results Usage: Human ID Paternity Test Missing Person Mass Disaster Genetics Multiplex PCR If a match occurs, comparison of DNA profile to population allele frequencies to generate a case report with probability of a random match to an unrelated individual DNA Database Search DNA Extraction DNA Quantitation Multiplex PCR Amplification DNA separation and sizing Technology Quantitation Biology Blood Stain Buccal swab STR Typing Interpretation of Results
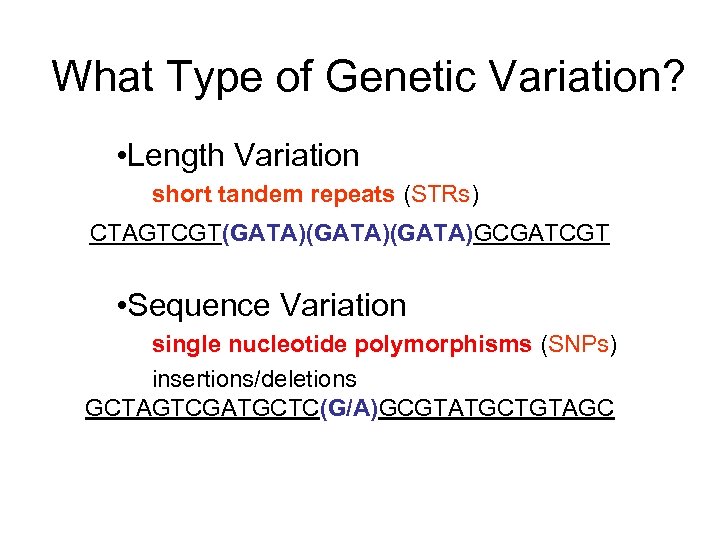 What Type of Genetic Variation? • Length Variation short tandem repeats (STRs) CTAGTCGT(GATA)(GATA)GCGATCGT • Sequence Variation single nucleotide polymorphisms (SNPs) insertions/deletions GCTAGTCGATGCTC(G/A)GCGTATGCTGTAGC
What Type of Genetic Variation? • Length Variation short tandem repeats (STRs) CTAGTCGT(GATA)(GATA)GCGATCGT • Sequence Variation single nucleotide polymorphisms (SNPs) insertions/deletions GCTAGTCGATGCTC(G/A)GCGTATGCTGTAGC
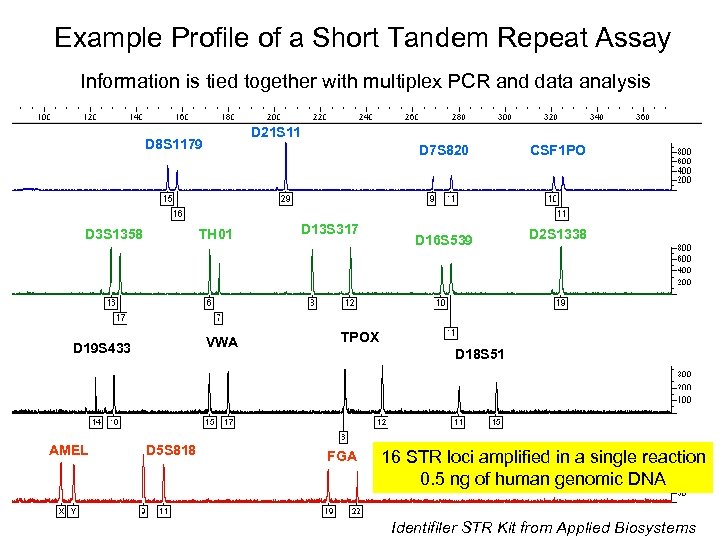 Example Profile of a Short Tandem Repeat Assay Information is tied together with multiplex PCR and data analysis D 21 S 11 D 8 S 1179 D 3 S 1358 TH 01 VWA D 19 S 433 AMEL D 7 S 820 D 5 S 818 D 13 S 317 CSF 1 PO D 16 S 539 D 2 S 1338 TPOX D 18 S 51 FGA 16 STR loci amplified in a single reaction 0. 5 ng of human genomic DNA Identifiler STR Kit from Applied Biosystems
Example Profile of a Short Tandem Repeat Assay Information is tied together with multiplex PCR and data analysis D 21 S 11 D 8 S 1179 D 3 S 1358 TH 01 VWA D 19 S 433 AMEL D 7 S 820 D 5 S 818 D 13 S 317 CSF 1 PO D 16 S 539 D 2 S 1338 TPOX D 18 S 51 FGA 16 STR loci amplified in a single reaction 0. 5 ng of human genomic DNA Identifiler STR Kit from Applied Biosystems
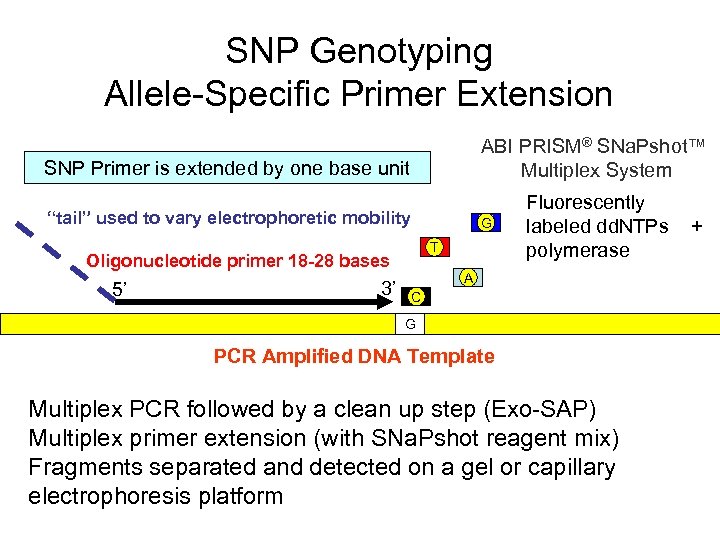 SNP Genotyping Allele-Specific Primer Extension ABI PRISM® SNa. Pshot™ Multiplex System SNP Primer is extended by one base unit “tail” used to vary electrophoretic mobility G T Oligonucleotide primer 18 -28 bases 5’ 3’ Fluorescently labeled dd. NTPs polymerase A C G PCR Amplified DNA Template Multiplex PCR followed by a clean up step (Exo-SAP) Multiplex primer extension (with SNa. Pshot reagent mix) Fragments separated and detected on a gel or capillary electrophoresis platform +
SNP Genotyping Allele-Specific Primer Extension ABI PRISM® SNa. Pshot™ Multiplex System SNP Primer is extended by one base unit “tail” used to vary electrophoretic mobility G T Oligonucleotide primer 18 -28 bases 5’ 3’ Fluorescently labeled dd. NTPs polymerase A C G PCR Amplified DNA Template Multiplex PCR followed by a clean up step (Exo-SAP) Multiplex primer extension (with SNa. Pshot reagent mix) Fragments separated and detected on a gel or capillary electrophoresis platform +
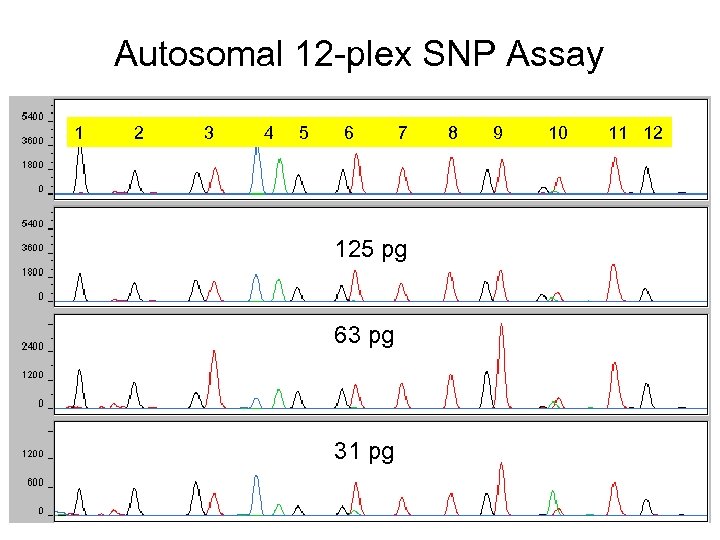 Autosomal 12 -plex SNP Assay 1 2 3 4 5 6 7 250 pg 125 pg 63 pg 31 pg 8 9 10 11 12
Autosomal 12 -plex SNP Assay 1 2 3 4 5 6 7 250 pg 125 pg 63 pg 31 pg 8 9 10 11 12
 Forensic Assays • Limited sample (0. 5 – 1 ng of genomic DNA) • Multiplex (10 or more loci/amplicons in a single reaction) • Robust amplification (results must hold up in court) • Issues with degradation, inhibition and efficient sample extraction • Standardized testing throughout the forensic community
Forensic Assays • Limited sample (0. 5 – 1 ng of genomic DNA) • Multiplex (10 or more loci/amplicons in a single reaction) • Robust amplification (results must hold up in court) • Issues with degradation, inhibition and efficient sample extraction • Standardized testing throughout the forensic community
 In House Assay Design
In House Assay Design
 Selection of Loci • From – Literature – Collaborators – Sequencing • Characterize sequence – Map in software (Lasergene) – NCBI accession number – ~1000 bp/locus
Selection of Loci • From – Literature – Collaborators – Sequencing • Characterize sequence – Map in software (Lasergene) – NCBI accession number – ~1000 bp/locus
 Selection of Loci High quality sequence information Standardize nomenclature Strand orientation Keep track of primer design
Selection of Loci High quality sequence information Standardize nomenclature Strand orientation Keep track of primer design
 Primer Selection • Primer 3 (http: //frodo. wi. mit. edu/cgi-bin/primer 3_www. cgi) – Stand alone version on Mac OSX • Visual OMP (www. dnasoftware. com) • Standard design parameters (Tm ~60 o. C) – Amplicon length (minimize) – Restrict primers to flank target region • Take advantage of mis-priming libraries to screen primers
Primer Selection • Primer 3 (http: //frodo. wi. mit. edu/cgi-bin/primer 3_www. cgi) – Stand alone version on Mac OSX • Visual OMP (www. dnasoftware. com) • Standard design parameters (Tm ~60 o. C) – Amplicon length (minimize) – Restrict primers to flank target region • Take advantage of mis-priming libraries to screen primers
 Further Primer Screening • Auto. Dimer software – Web based version http: //yellow. nist. gov: 8444/dna. Analysis/ – Stand alone version http: //www. cstl. nist. gov/div 831/strbase//Auto. Dimer. Homepage/Auto. Dimer Program. Homepage. htm • BLAST – http: //www. ncbi. nlm. nih. gov/BLAST/ – Avoid significant homology with other chromosomal locations and/or organisms – Save search results for further review – Confirm primer binding sites
Further Primer Screening • Auto. Dimer software – Web based version http: //yellow. nist. gov: 8444/dna. Analysis/ – Stand alone version http: //www. cstl. nist. gov/div 831/strbase//Auto. Dimer. Homepage/Auto. Dimer Program. Homepage. htm • BLAST – http: //www. ncbi. nlm. nih. gov/BLAST/ – Avoid significant homology with other chromosomal locations and/or organisms – Save search results for further review – Confirm primer binding sites
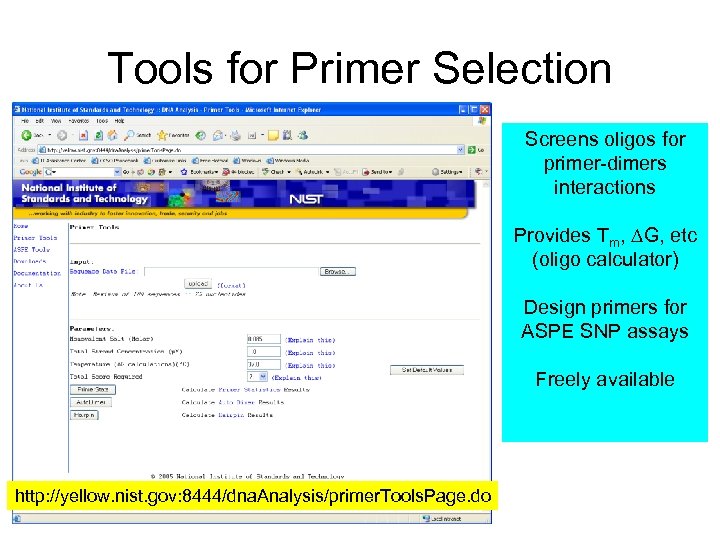 Tools for Primer Selection Screens oligos for primer-dimers interactions Provides Tm, DG, etc (oligo calculator) Design primers for ASPE SNP assays Freely available http: //yellow. nist. gov: 8444/dna. Analysis/primer. Tools. Page. do
Tools for Primer Selection Screens oligos for primer-dimers interactions Provides Tm, DG, etc (oligo calculator) Design primers for ASPE SNP assays Freely available http: //yellow. nist. gov: 8444/dna. Analysis/primer. Tools. Page. do
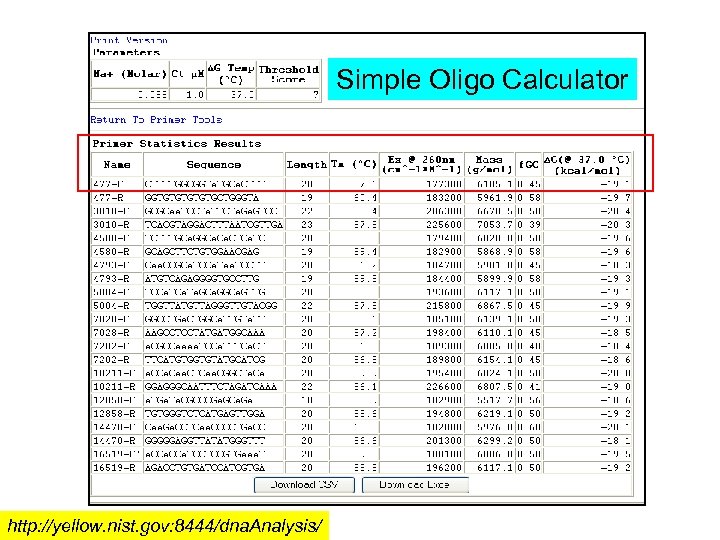 Simple Oligo Calculator http: //yellow. nist. gov: 8444/dna. Analysis/
Simple Oligo Calculator http: //yellow. nist. gov: 8444/dna. Analysis/
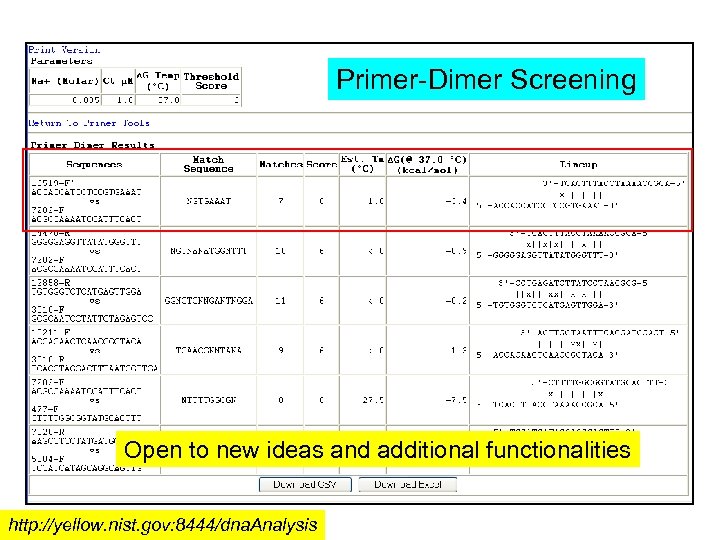 Primer-Dimer Screening Open to new ideas and additional functionalities http: //yellow. nist. gov: 8444/dna. Analysis
Primer-Dimer Screening Open to new ideas and additional functionalities http: //yellow. nist. gov: 8444/dna. Analysis
 Assay Development • UV quantitation of primers (ensure reproducibility) • Run PCR in singleplex reactions • Samples for assay development – Well characterized quality – pristine – Contain a sampling of sequence variants – Sufficient quantities for testing – Well characterized template concentration (q. PCR) – define sensitivity limits
Assay Development • UV quantitation of primers (ensure reproducibility) • Run PCR in singleplex reactions • Samples for assay development – Well characterized quality – pristine – Contain a sampling of sequence variants – Sufficient quantities for testing – Well characterized template concentration (q. PCR) – define sensitivity limits
 General PCR Conditions • Attempt to keep conditions a constant – 1 x PCR buffer – 1 Unit of polymerase (Taq. Gold) – 2 m. M Mg++ – 250 m. M d. NTPs – 0. 16 mg/m. L BSA – 0. 2 m. M of each PCR primer – 0. 5 – 1 ng of template DNA (80 -200 copies of target)
General PCR Conditions • Attempt to keep conditions a constant – 1 x PCR buffer – 1 Unit of polymerase (Taq. Gold) – 2 m. M Mg++ – 250 m. M d. NTPs – 0. 16 mg/m. L BSA – 0. 2 m. M of each PCR primer – 0. 5 – 1 ng of template DNA (80 -200 copies of target)
 Assay Development • Use singleplex data to evaluate multiplex performance • Optimization is empirical – Balance PCR primer concentration – Replace inefficient primers – Identify and replace artifact causing primers • Integrate the lessons learned back into the informatics/strategy pipeline
Assay Development • Use singleplex data to evaluate multiplex performance • Optimization is empirical – Balance PCR primer concentration – Replace inefficient primers – Identify and replace artifact causing primers • Integrate the lessons learned back into the informatics/strategy pipeline
 Literature from our Mplex Work • • • • • Butler, J. M. (2005) Constructing STR multiplex assays. Methods in Molecular Biology: Forensic DNA Typing Protocols (Carracedo, A. , ed. ), Humana Press: Totowa, New Jersey, 297: 53 -66. [preprint] Butler, J. M. , David, V. A. , O’Brien, S. J. , Menotti-Raymond, M. (2002) The Meow. Plex: a new DNA test using tetranucleotide STR markers for the domestic cat. Profiles in DNA, Promega Corporation, Volume 5, No. 2, pp. 7– 10. http: //www. promega. com/profiles/502/Profiles. In. DNA_502_07. pdf Butler, J. M. , Schoske, R. , Vallone, P. M. Highly multiplexed assays for measuring polymorphisms on the Y-chromosome. (2003) Progress in Forensic Genetics 9 (Brinkmann, B. and Carracedo, A. , eds. ), Elsevier Science: Amsterdam, The Netherlands, International Congress Series 1239, pp. 301 -305. Butler, J. M. , Shen, Y. , Mc. Cord, B. R. (2003) The development of reduced size STR amplicons as tools for analysis of degraded DNA. J. Forensic Sci 48(5) 1054 -1064. Butler, J. M. , Schoske, R. , Vallone, P. M. , Kline, M. C. , Redd, A. J. , Hammer, M. F. (2002) A novel multiplex for simultaneous amplification of 20 Y chromosome STR markers. Forensic Sci. Int. 129: 10 -24. Butler, J. M. , C. M. Ruitberg, Vallone, P. M. (2001) Capillary electrophoresis as a tool for optimization of multiplex PCR reactions, Fresenius J. Anal. Chem. 369: 200 -205. Coble, M. D. and Butler, J. M. (2005) Characterization of new mini. STR loci to aid analysis of degraded DNA. J. Forensic Sci. 50: 43 -53. Devaney, J. M. Pettit, E. L. , Kaler, S. G. , Vallone, P. M. , Butler, J. M. , Marino, M. A. (2001) Genotyping of two mutations in the HFE gene usingle-base extension and high-performance liquid chromatography. Anal. Chem. 73: 620 -624. Just, R. S. , Irwin, J. A. , O'Callaghan, J. E. , Saunier, J. L. , Coble, M. D. , Vallone, P. M. , Butler, J. M. , Barritt, S. M. , Parsons, T. J. (2004) Toward increased utility of mt. DNA in forensic identifications. Forensic Sci. Int. 146 S: S 147 -S 149. Menotti-Raymond, M. A. , David, V. A. , Wachter, L. A. , Butler, J. M. , O’Brien, S. J. (2005) An STR forensic typing system for genetic individualization of domestic cat (Felis catus) samples. J. Forensic Sci. 50(5): 1061 -1070. Schoske, R. , Butler, J. M. , Vallone, P. M. , Kline, M. C. , Prinz, M. , Redd, A. J. , Hammer, M. F. (2001) Development of Y STR megaplex assays. Proceedings of the Twelve International Symposium on Human Identification 2001, Promega Corporation. http: //www. promega. com/geneticidproc/ussymp 12 proc/contents/butler. PDF Schoske, R. , Vallone, P. M. , Ruitberg, C. M. , Butler, J. M. (2003) Multiplex PCR design strategy used for the simultaneous amplification of 10 Y chromosome short tandem repeat (STR) loci. Anal. Bioanal. Chem. , 375: 333 -343. Schoske, R. (2003) The design, optimization and testing of Y chromosome short tandem repeat megaplexes. Ph. D. dissertation, American University, 270 pp. Schoske, R. , Vallone, P. M. , Kline, M. C. , Redman, J. W. , Butler, J. M. (2004) High-throughput Y‑STR typing of U. S. populations with 27 regions of the Y chromosome using two multiplex PCR assays. Forensic Sci. Int. 139: 107 -121. Vallone, P. M. , Just, R. S. , Coble, M. D. , Butler, J. M. , Parsons, T. J. (2004) A multiplex allele-specific primer extension assay forensically informative SNPs distributed throughout the mitochondrial genome. Int. J. Legal Med. , 118: 147 -157. Vallone, P. M. and Butler, J. M. (2004) Multiplexed assays for evaluation of Y-SNP markers in U. S. populations. Progress in Forensic Genetics 10, Elsevier Science: Amsterdam, The Netherlands, International Congress Series 1261, 85 -87. Vallone, P. M. and Butler, J. M. (2004) Y-SNP typing of U. S. African American and Caucasian samples using allele-specific hybridization and primer extension. J. Forensic Sci. 49(4): 723‑ 732. Vallone, P. M. , Decker, A. E. , Butler, J. M. (2005) Allele frequencies for 70 autosomal SNP loci with U. S. Caucasian, African American, and Hispanic samples. Forensic Sci. Int. 149: 279 -286. Vallone, P. M. , Decker, A. E. , Coble, M. D. , Butler, J. M. (2006) Evaluation of an autosomal SNP 12 -plex assay. Progress in Forensic Genetics 11, Elsevier Science: Amsterdam, The Netherlands, International Congress Series 1288, 61 -63. Vallone, P. M. , Fahr, K. , Kostrzewa, M. (2005) Genotyping SNPs using a UV photocleavable oligonucleotide in MALDI-TOF MS. Methods in Molecular Biology: Forensic DNA Typing Protocols (Carracedo, A. , ed. ), Humana Press: Totowa, New Jersey, 297: 169 -178. Multiplex Assays Developed at NIST Various Y-SNPs (6 -plexes) Y-STR 10 -plex & 20 -plex Mitochondrial SNP 11 -plex Autosomal SNP 12 -plexes Coming soon: Autosomal STR 26 -plex http: //www. cstl. nist. gov/biotech/strbase/NISTpub. htm
Literature from our Mplex Work • • • • • Butler, J. M. (2005) Constructing STR multiplex assays. Methods in Molecular Biology: Forensic DNA Typing Protocols (Carracedo, A. , ed. ), Humana Press: Totowa, New Jersey, 297: 53 -66. [preprint] Butler, J. M. , David, V. A. , O’Brien, S. J. , Menotti-Raymond, M. (2002) The Meow. Plex: a new DNA test using tetranucleotide STR markers for the domestic cat. Profiles in DNA, Promega Corporation, Volume 5, No. 2, pp. 7– 10. http: //www. promega. com/profiles/502/Profiles. In. DNA_502_07. pdf Butler, J. M. , Schoske, R. , Vallone, P. M. Highly multiplexed assays for measuring polymorphisms on the Y-chromosome. (2003) Progress in Forensic Genetics 9 (Brinkmann, B. and Carracedo, A. , eds. ), Elsevier Science: Amsterdam, The Netherlands, International Congress Series 1239, pp. 301 -305. Butler, J. M. , Shen, Y. , Mc. Cord, B. R. (2003) The development of reduced size STR amplicons as tools for analysis of degraded DNA. J. Forensic Sci 48(5) 1054 -1064. Butler, J. M. , Schoske, R. , Vallone, P. M. , Kline, M. C. , Redd, A. J. , Hammer, M. F. (2002) A novel multiplex for simultaneous amplification of 20 Y chromosome STR markers. Forensic Sci. Int. 129: 10 -24. Butler, J. M. , C. M. Ruitberg, Vallone, P. M. (2001) Capillary electrophoresis as a tool for optimization of multiplex PCR reactions, Fresenius J. Anal. Chem. 369: 200 -205. Coble, M. D. and Butler, J. M. (2005) Characterization of new mini. STR loci to aid analysis of degraded DNA. J. Forensic Sci. 50: 43 -53. Devaney, J. M. Pettit, E. L. , Kaler, S. G. , Vallone, P. M. , Butler, J. M. , Marino, M. A. (2001) Genotyping of two mutations in the HFE gene usingle-base extension and high-performance liquid chromatography. Anal. Chem. 73: 620 -624. Just, R. S. , Irwin, J. A. , O'Callaghan, J. E. , Saunier, J. L. , Coble, M. D. , Vallone, P. M. , Butler, J. M. , Barritt, S. M. , Parsons, T. J. (2004) Toward increased utility of mt. DNA in forensic identifications. Forensic Sci. Int. 146 S: S 147 -S 149. Menotti-Raymond, M. A. , David, V. A. , Wachter, L. A. , Butler, J. M. , O’Brien, S. J. (2005) An STR forensic typing system for genetic individualization of domestic cat (Felis catus) samples. J. Forensic Sci. 50(5): 1061 -1070. Schoske, R. , Butler, J. M. , Vallone, P. M. , Kline, M. C. , Prinz, M. , Redd, A. J. , Hammer, M. F. (2001) Development of Y STR megaplex assays. Proceedings of the Twelve International Symposium on Human Identification 2001, Promega Corporation. http: //www. promega. com/geneticidproc/ussymp 12 proc/contents/butler. PDF Schoske, R. , Vallone, P. M. , Ruitberg, C. M. , Butler, J. M. (2003) Multiplex PCR design strategy used for the simultaneous amplification of 10 Y chromosome short tandem repeat (STR) loci. Anal. Bioanal. Chem. , 375: 333 -343. Schoske, R. (2003) The design, optimization and testing of Y chromosome short tandem repeat megaplexes. Ph. D. dissertation, American University, 270 pp. Schoske, R. , Vallone, P. M. , Kline, M. C. , Redman, J. W. , Butler, J. M. (2004) High-throughput Y‑STR typing of U. S. populations with 27 regions of the Y chromosome using two multiplex PCR assays. Forensic Sci. Int. 139: 107 -121. Vallone, P. M. , Just, R. S. , Coble, M. D. , Butler, J. M. , Parsons, T. J. (2004) A multiplex allele-specific primer extension assay forensically informative SNPs distributed throughout the mitochondrial genome. Int. J. Legal Med. , 118: 147 -157. Vallone, P. M. and Butler, J. M. (2004) Multiplexed assays for evaluation of Y-SNP markers in U. S. populations. Progress in Forensic Genetics 10, Elsevier Science: Amsterdam, The Netherlands, International Congress Series 1261, 85 -87. Vallone, P. M. and Butler, J. M. (2004) Y-SNP typing of U. S. African American and Caucasian samples using allele-specific hybridization and primer extension. J. Forensic Sci. 49(4): 723‑ 732. Vallone, P. M. , Decker, A. E. , Butler, J. M. (2005) Allele frequencies for 70 autosomal SNP loci with U. S. Caucasian, African American, and Hispanic samples. Forensic Sci. Int. 149: 279 -286. Vallone, P. M. , Decker, A. E. , Coble, M. D. , Butler, J. M. (2006) Evaluation of an autosomal SNP 12 -plex assay. Progress in Forensic Genetics 11, Elsevier Science: Amsterdam, The Netherlands, International Congress Series 1288, 61 -63. Vallone, P. M. , Fahr, K. , Kostrzewa, M. (2005) Genotyping SNPs using a UV photocleavable oligonucleotide in MALDI-TOF MS. Methods in Molecular Biology: Forensic DNA Typing Protocols (Carracedo, A. , ed. ), Humana Press: Totowa, New Jersey, 297: 169 -178. Multiplex Assays Developed at NIST Various Y-SNPs (6 -plexes) Y-STR 10 -plex & 20 -plex Mitochondrial SNP 11 -plex Autosomal SNP 12 -plexes Coming soon: Autosomal STR 26 -plex http: //www. cstl. nist. gov/biotech/strbase/NISTpub. htm
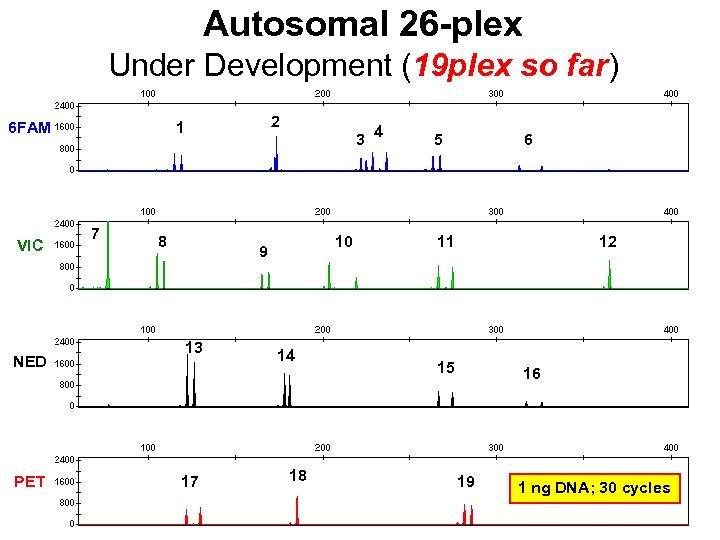 Autosomal 26 -plex Under Development (19 plex so far) VIC NED PET 2 1 6 FAM 7 3 8 10 9 13 17 14 18 4 5 6 11 12 15 16 19 1 ng DNA; 30 cycles
Autosomal 26 -plex Under Development (19 plex so far) VIC NED PET 2 1 6 FAM 7 3 8 10 9 13 17 14 18 4 5 6 11 12 15 16 19 1 ng DNA; 30 cycles
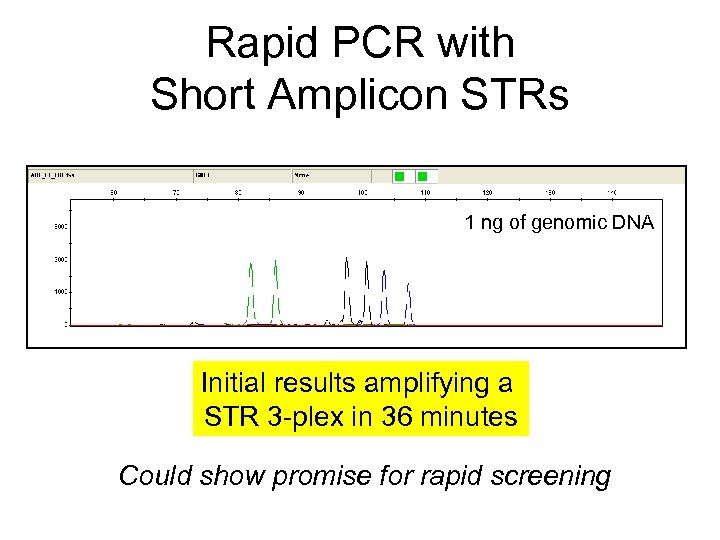 Rapid PCR with Short Amplicon STRs 1 ng of genomic DNA Initial results amplifying a STR 3 -plex in 36 minutes Could show promise for rapid screening
Rapid PCR with Short Amplicon STRs 1 ng of genomic DNA Initial results amplifying a STR 3 -plex in 36 minutes Could show promise for rapid screening
 Areas of Concern/Differences • Speed ~1 day – – Extraction 1 hour Quantitation 1 hour PCR ~2. 5 hours Capillary/Gel separation 1 hour • Faster mutation rate – unique variants (or closely spaced sites) • We use q. PCR for quantitation not detection, but multiplex assay design strategy may apply
Areas of Concern/Differences • Speed ~1 day – – Extraction 1 hour Quantitation 1 hour PCR ~2. 5 hours Capillary/Gel separation 1 hour • Faster mutation rate – unique variants (or closely spaced sites) • We use q. PCR for quantitation not detection, but multiplex assay design strategy may apply
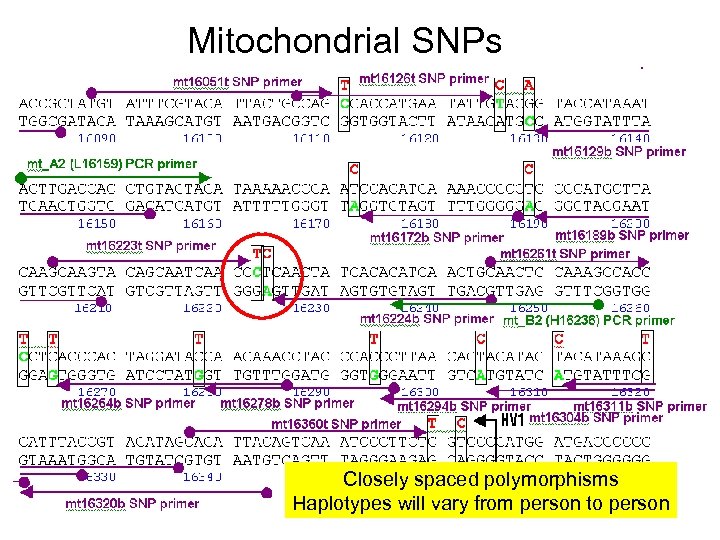 Mitochondrial SNPs Closely spaced polymorphisms Haplotypes will vary from person to person
Mitochondrial SNPs Closely spaced polymorphisms Haplotypes will vary from person to person
 Summary • Evolving strategy for developing multiplex PCR assays • Robust assay development assists in interlaboratory testing • What aspects can be applied to pathogen screening?
Summary • Evolving strategy for developing multiplex PCR assays • Robust assay development assists in interlaboratory testing • What aspects can be applied to pathogen screening?
 Acknowledgments Funding from interagency agreement 2003 -IJ-R-029 between the National Institute of Justice and the NIST Office of Law Enforcement Standards NIST Human Identity Project Team – Leading the Way in Forensic DNA… John Butler Margaret Kline Jan Redman petev@nist. gov Amy Decker Becky Hill Dave Duewer
Acknowledgments Funding from interagency agreement 2003 -IJ-R-029 between the National Institute of Justice and the NIST Office of Law Enforcement Standards NIST Human Identity Project Team – Leading the Way in Forensic DNA… John Butler Margaret Kline Jan Redman petev@nist. gov Amy Decker Becky Hill Dave Duewer


