c5e497db857af3c57fbc46ba17d6256e.ppt
- Количество слайдов: 18
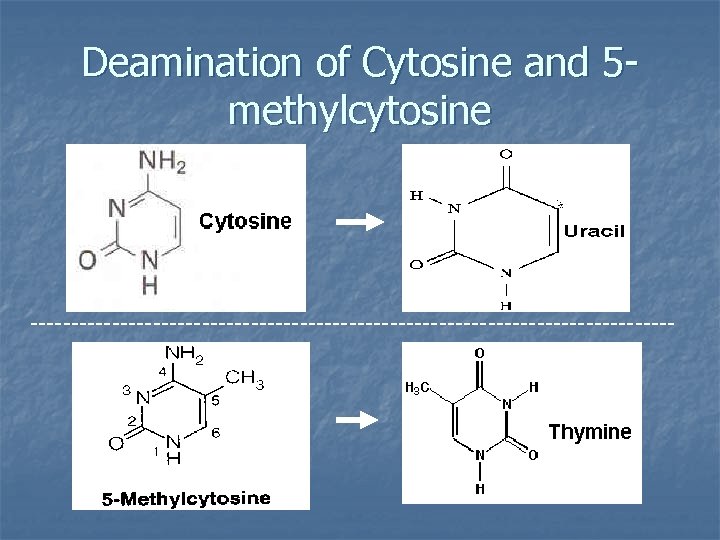 Deamination of Cytosine and 5 methylcytosine ----------------------------------------
Deamination of Cytosine and 5 methylcytosine ----------------------------------------
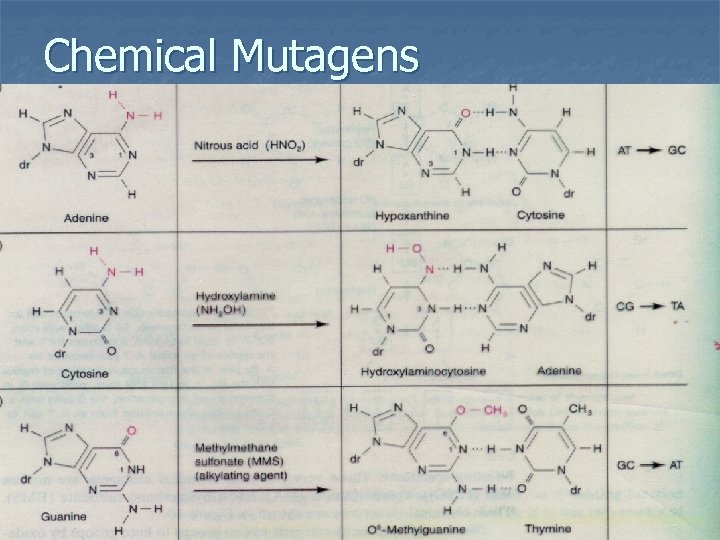 Chemical Mutagens
Chemical Mutagens
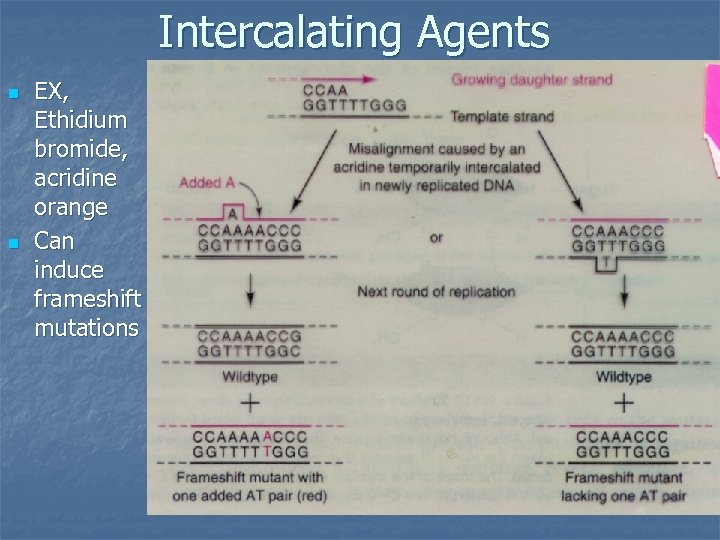 Intercalating Agents n n EX, Ethidium bromide, acridine orange Can induce frameshift mutations
Intercalating Agents n n EX, Ethidium bromide, acridine orange Can induce frameshift mutations
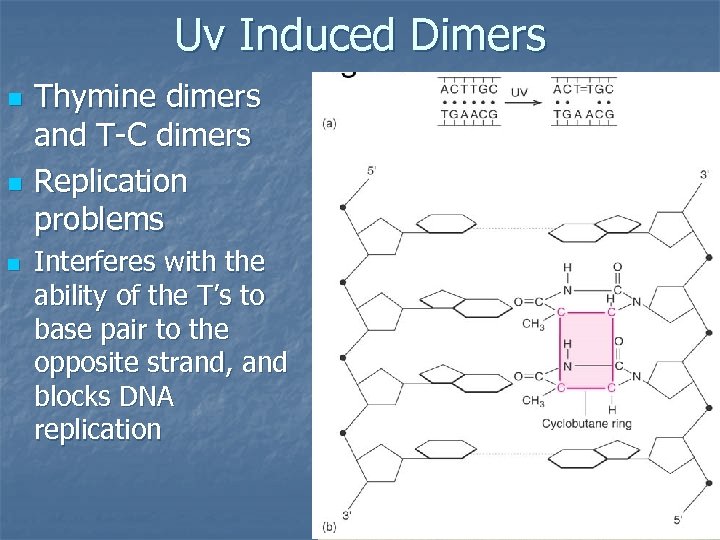 Uv Induced Dimers n n n Thymine dimers and T-C dimers Replication problems Interferes with the ability of the T’s to base pair to the opposite strand, and blocks DNA replication
Uv Induced Dimers n n n Thymine dimers and T-C dimers Replication problems Interferes with the ability of the T’s to base pair to the opposite strand, and blocks DNA replication
 Other Mutagens n Transposable elements—”jumping genes”. Major frameshift mutations n Factors in evolution n n Mutator genes—mutations increase mutation rate. Four potent mutator genes Mutant DNA pol III 3’ 5’ exonuclease activity n Mutant methylation enzymes(ex dam) n Mutant enzymes in excision repair system n Mutant enzymes in SOS system n
Other Mutagens n Transposable elements—”jumping genes”. Major frameshift mutations n Factors in evolution n n Mutator genes—mutations increase mutation rate. Four potent mutator genes Mutant DNA pol III 3’ 5’ exonuclease activity n Mutant methylation enzymes(ex dam) n Mutant enzymes in excision repair system n Mutant enzymes in SOS system n
 Reversions n n Mutations in an mutant can restore the wild type function (reversion, back mutation, or reverse mutation) Spontaneous or induced If mutation occurs at the site of the original=True reversion Wild type restored by mutation at another site= second site mutation Second site in same gene= intragenic suppression n Second site in another gene=intergenic suppression n
Reversions n n Mutations in an mutant can restore the wild type function (reversion, back mutation, or reverse mutation) Spontaneous or induced If mutation occurs at the site of the original=True reversion Wild type restored by mutation at another site= second site mutation Second site in same gene= intragenic suppression n Second site in another gene=intergenic suppression n
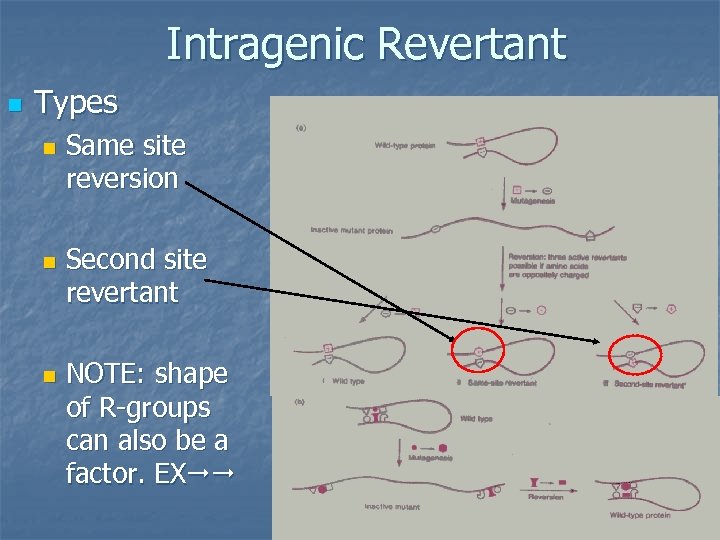 Intragenic Revertant n Types n n n Same site reversion Second site revertant NOTE: shape of R-groups can also be a factor. EX
Intragenic Revertant n Types n n n Same site reversion Second site revertant NOTE: shape of R-groups can also be a factor. EX
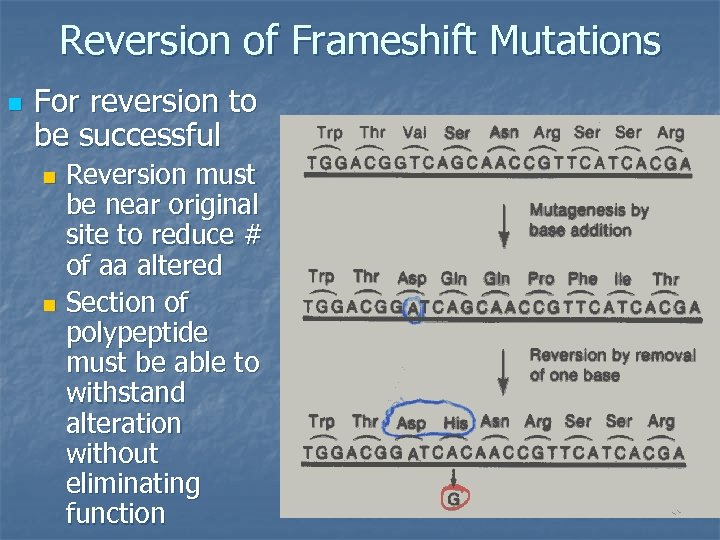 Reversion of Frameshift Mutations n For reversion to be successful Reversion must be near original site to reduce # of aa altered n Section of polypeptide must be able to withstand alteration without eliminating function n
Reversion of Frameshift Mutations n For reversion to be successful Reversion must be near original site to reduce # of aa altered n Section of polypeptide must be able to withstand alteration without eliminating function n
 Intergenic Suppression n Refers to a chnge in another gene which suppresses or eliminates the mutant phenotype. EX Multisubunit proteins—Mutation in one subunit may be masked by mutation in another subunit (ex. restoring hydrophobic patches) n Suppression via suppressor t. RNAs n
Intergenic Suppression n Refers to a chnge in another gene which suppresses or eliminates the mutant phenotype. EX Multisubunit proteins—Mutation in one subunit may be masked by mutation in another subunit (ex. restoring hydrophobic patches) n Suppression via suppressor t. RNAs n
 Suppressor t. RNA n Nonsense mutation- aa codon “Stop” n n n Some bacteria can “read through” these mutations (though protein function may be altered). HOW? Mutant t. RNA that has an anticodon that recognizes “Stop” as a reading codon. n n Ex. AAG UAG aa encoded depends on which t. RNA is mutated Not every suppressor restores normal function
Suppressor t. RNA n Nonsense mutation- aa codon “Stop” n n n Some bacteria can “read through” these mutations (though protein function may be altered). HOW? Mutant t. RNA that has an anticodon that recognizes “Stop” as a reading codon. n n Ex. AAG UAG aa encoded depends on which t. RNA is mutated Not every suppressor restores normal function
 Suppressor Mutants (cont’d) n n n EX. UUG (Leu) UAG (Stop) (AUC anticodon) A mutation in a t. RNA resulting in “AUC” allows that t. RNA to recognize “Stop”. Can get suppression or partial suppression NOTE: must be 2 copies of t. RNA mutated. Why? n In any cell containing mutator, must also be a wild type Suppressors allow survival, even if suboptimal
Suppressor Mutants (cont’d) n n n EX. UUG (Leu) UAG (Stop) (AUC anticodon) A mutation in a t. RNA resulting in “AUC” allows that t. RNA to recognize “Stop”. Can get suppression or partial suppression NOTE: must be 2 copies of t. RNA mutated. Why? n In any cell containing mutator, must also be a wild type Suppressors allow survival, even if suboptimal
 Termination of Translation in Suppressor Strains n Problem: Must be a means of terminating translation. n HOW? n Release factors still present, will compete for the “Stop “ site n Many genes are double-terminated n EX. UAG-UAA
Termination of Translation in Suppressor Strains n Problem: Must be a means of terminating translation. n HOW? n Release factors still present, will compete for the “Stop “ site n Many genes are double-terminated n EX. UAG-UAA
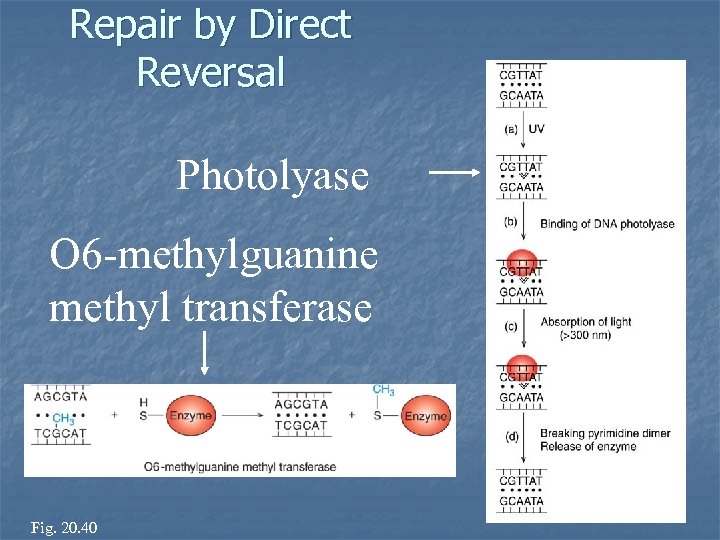 Repair by Direct Reversal Photolyase O 6 -methylguanine methyl transferase Fig. 20. 39 Fig. 20. 40
Repair by Direct Reversal Photolyase O 6 -methylguanine methyl transferase Fig. 20. 39 Fig. 20. 40
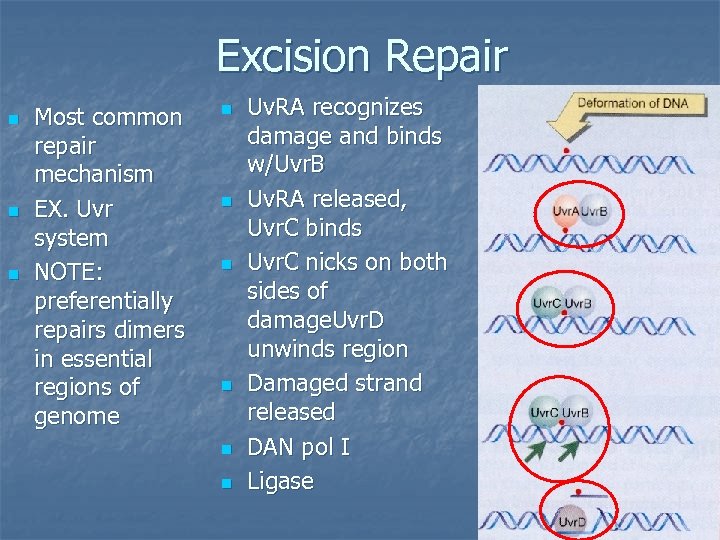 Excision Repair n n n Most common repair mechanism EX. Uvr system NOTE: preferentially repairs dimers in essential regions of genome n n n Uv. RA recognizes damage and binds w/Uvr. B Uv. RA released, Uvr. C binds Uvr. C nicks on both sides of damage. Uvr. D unwinds region Damaged strand released DAN pol I Ligase
Excision Repair n n n Most common repair mechanism EX. Uvr system NOTE: preferentially repairs dimers in essential regions of genome n n n Uv. RA recognizes damage and binds w/Uvr. B Uv. RA released, Uvr. C binds Uvr. C nicks on both sides of damage. Uvr. D unwinds region Damaged strand released DAN pol I Ligase
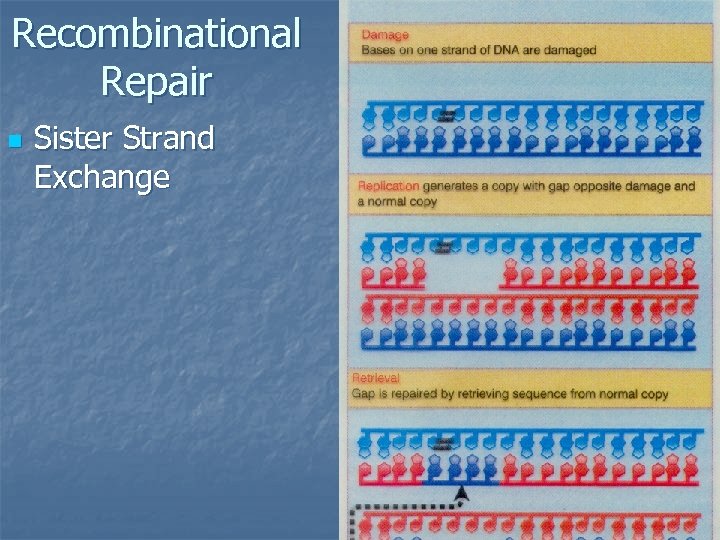 Recombinational Repair n Sister Strand Exchange
Recombinational Repair n Sister Strand Exchange
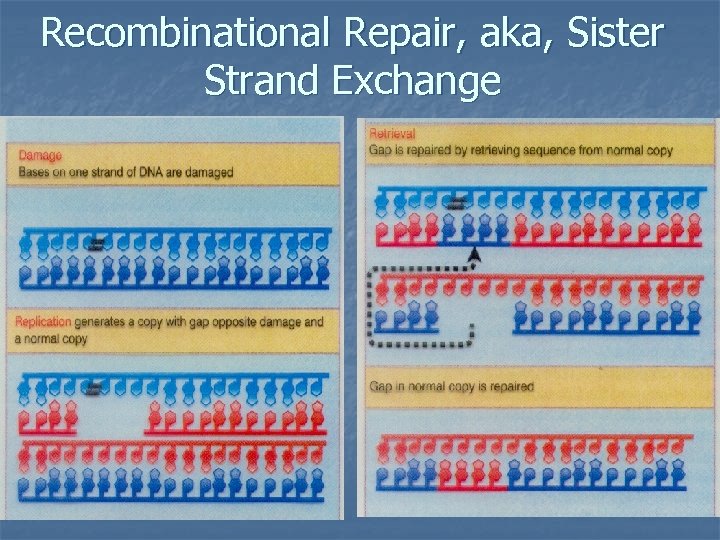 Recombinational Repair, aka, Sister Strand Exchange
Recombinational Repair, aka, Sister Strand Exchange
 SOS Response n n Is an inducible system of last resort Also called error prone replication because it inactivates the proofreading function of DNA pol III. Turned on only when DNA damage is extreme Main players: rec. A and lex. A and a battery of inducible enzymes
SOS Response n n Is an inducible system of last resort Also called error prone replication because it inactivates the proofreading function of DNA pol III. Turned on only when DNA damage is extreme Main players: rec. A and lex. A and a battery of inducible enzymes
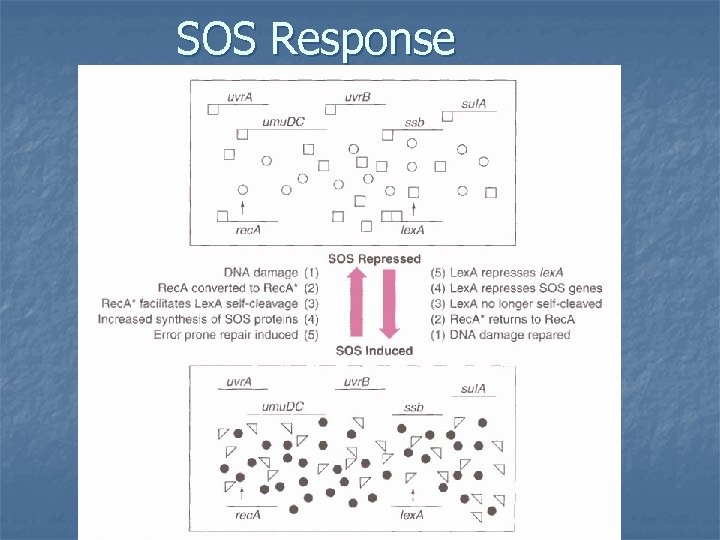 SOS Response
SOS Response


