Genome-Assembly-Haibao.pptx
- Количество слайдов: 54

de novo Genome Assembly Haibao Tang J. Craig Venter Institute Plant Informatics Workshop July 16, 2013
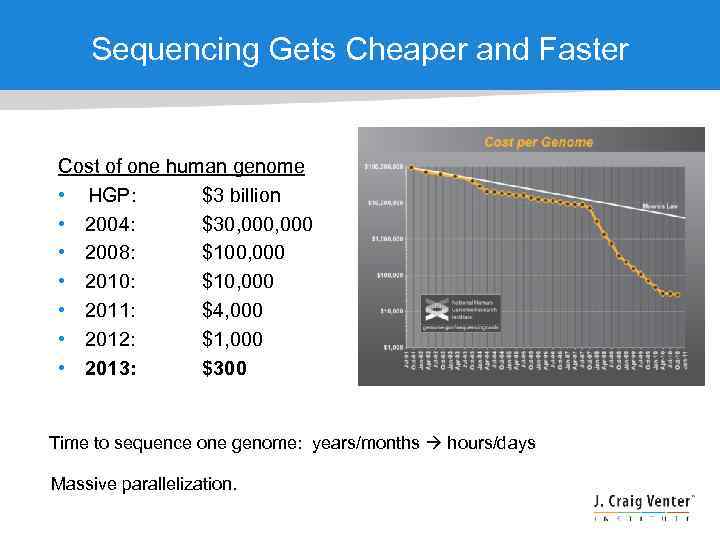
Sequencing Gets Cheaper and Faster Cost of one human genome • HGP: $3 billion • 2004: $30, 000 • 2008: $100, 000 • 2010: $10, 000 • 2011: $4, 000 • 2012: $1, 000 • 2013: $300 Time to sequence one genome: years/months hours/days Massive parallelization.
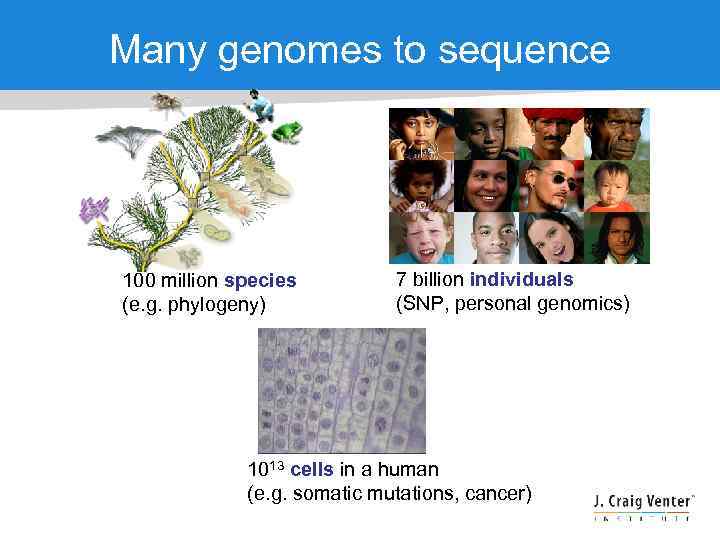
Many genomes to sequence 100 million species (e. g. phylogeny) 7 billion individuals (SNP, personal genomics) 1013 cells in a human (e. g. somatic mutations, cancer)
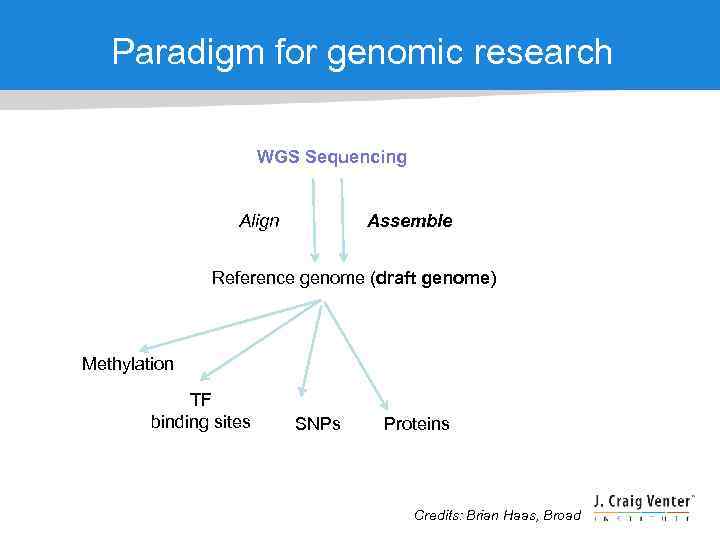
Paradigm for genomic research WGS Sequencing Align Assemble Reference genome (draft genome) Methylation TF binding sites SNPs Proteins Credits: Brian Haas, Broad
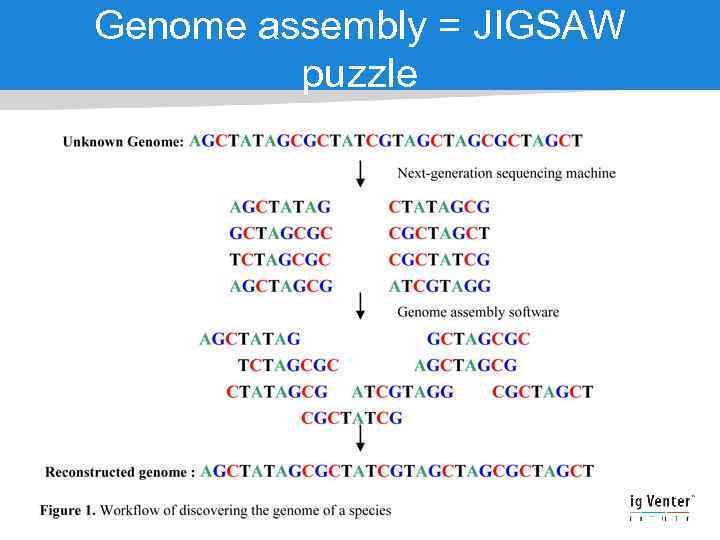
Genome assembly = JIGSAW puzzle

How difficult is a JIGSAW puzzle Yes, but you must deal with • Millions of pieces • Lots of malformed pieces • Often missing pieces • Pieces mixed from another puzzle • Lots of similar blue sky pieces… • If de novo you… don’t even know the final picture Credits: Jason Miller, JCVI

A gigantic JIGSAW puzzle
Outline • Assembly preparation – reads, libraries, etc. • Assembly – OLC assemblers vs. de Bruijn (K-mer) graph assemblers • Assembly QC - Identify data or assembly issues • Assembly curation - Further scaffolding, build chromosomes

1. Reads

Reads sampled from genomes
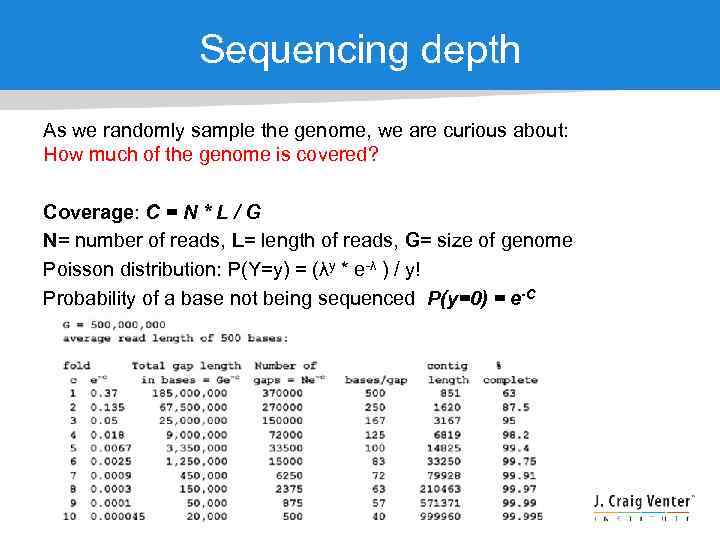
Sequencing depth As we randomly sample the genome, we are curious about: How much of the genome is covered? Coverage: C = N * L / G N= number of reads, L= length of reads, G= size of genome Poisson distribution: P(Y=y) = (λy * e-λ ) / y! Probability of a base not being sequenced P(y=0) = e-C
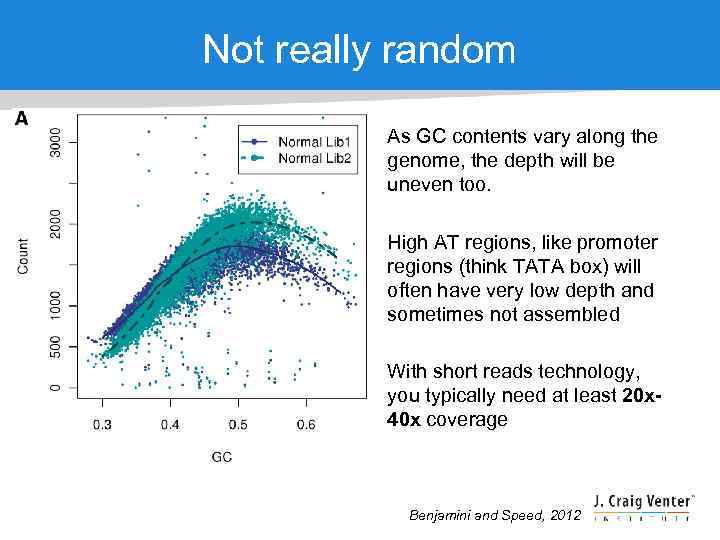
Not really random As GC contents vary along the genome, the depth will be uneven too. High AT regions, like promoter regions (think TATA box) will often have very low depth and sometimes not assembled With short reads technology, you typically need at least 20 x 40 x coverage Benjamini and Speed, 2012
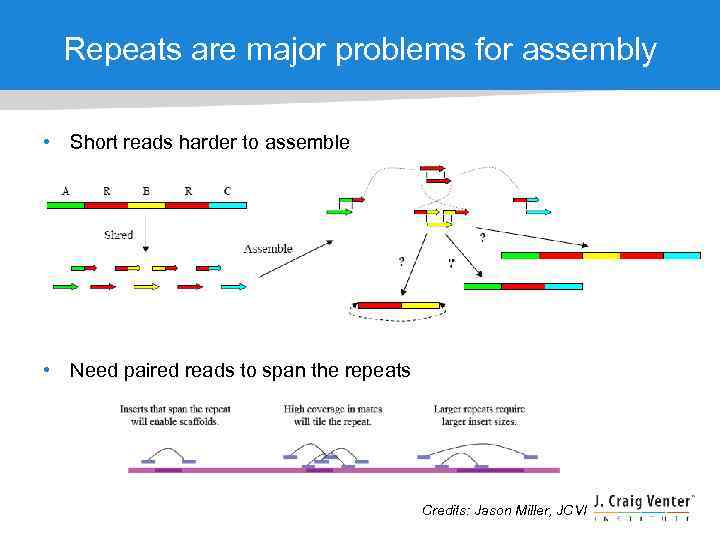
Repeats are major problems for assembly • Short reads harder to assemble • Need paired reads to span the repeats Credits: Jason Miller, JCVI
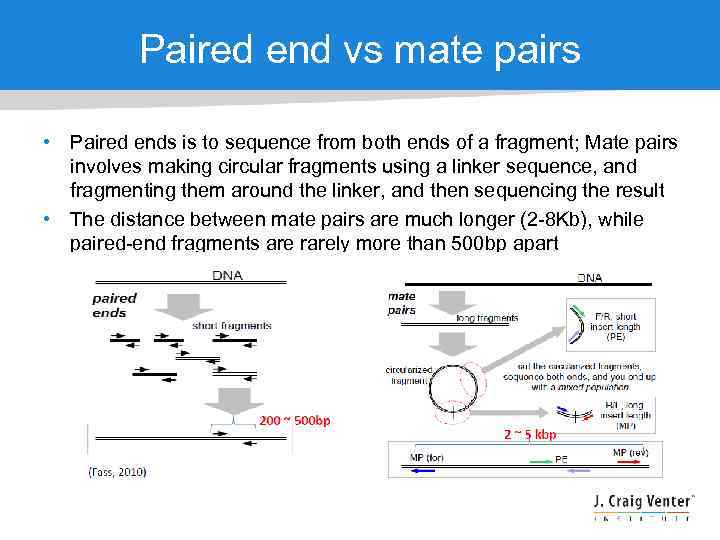
Paired end vs mate pairs • Paired ends is to sequence from both ends of a fragment; Mate pairs involves making circular fragments using a linker sequence, and fragmenting them around the linker, and then sequencing the result • The distance between mate pairs are much longer (2 -8 Kb), while paired-end fragments are rarely more than 500 bp apart

Quick checklist • • • How are my pairs oriented? (inward, outward, same strand) How is the data formatted? (sff, fastq, fasta) Are the paired reads in the same file (interleaved? ) What is the insert size and its size distribution What is the adapter sequence used when making the library? • Make sure to understand where the results come from. Consult your sequencing operator for details on the library preparation. • When in doubt you can always map them in single end mode, then visualize the results and see how the pairs are located relative to one another.
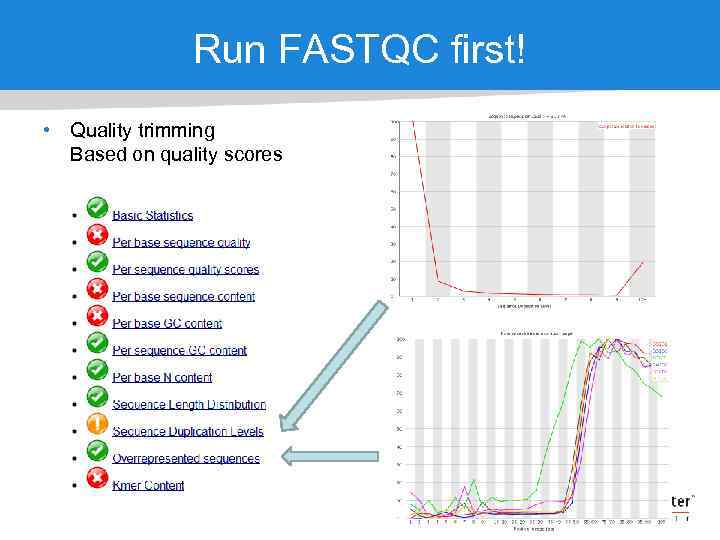
Run FASTQC first! • Quality trimming Based on quality scores

Prepare sequence reads • Quality trimming Based on quality scores • Ambiguity trimming Remove stretches of Ns • Adapter sequence trimming Remove sequence adapters • Length trimming Remove reads shorter or longer than a specified threshold

TRIMMOMATIC example $ java -cp trimmomatic-0. 15. jar org. usadellab. trimmomatic. Trimmomatic. PE s_1_1_sequence. txt. gz s_1_2_sequence. txt. gz lane 1_forward_paired. fq. gz lane 1_forward_unpaired. fq. gz lane 1_reverse_unpaired. fq. gz ILLUMINACLIP: adapters. fasta: 2: 40: 15 LEADING: 3 TRAILING: 3 SLIDINGWINDOW: 4: 15 MINLEN: 36 • • • This will perform the following: Remove adapters Remove leading low quality or N bases (below quality 3) Remove trailing low quality or N bases (below quality 3) Scan the read with a 4 -base wide sliding window, cutting when the average quality per base drops below 15 • Drop reads below the 36 bases long • Read and write files in gzipped format
2. Assembly
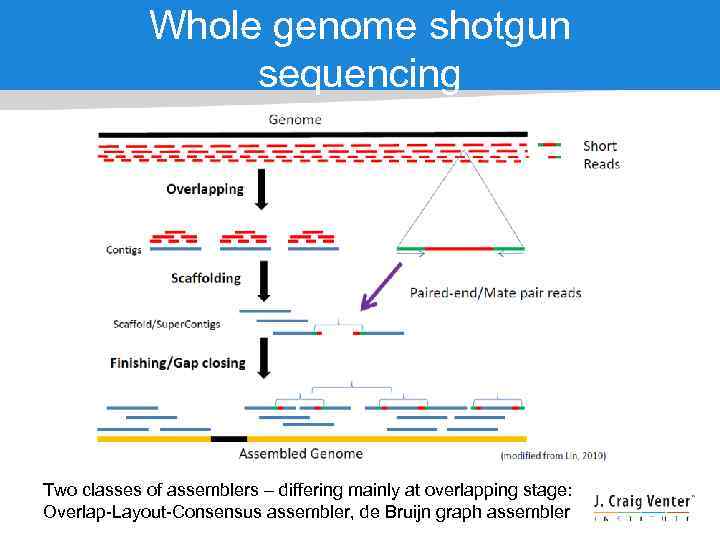
Whole genome shotgun sequencing Two classes of assemblers – differing mainly at overlapping stage: Overlap-Layout-Consensus assembler, de Bruijn graph assembler
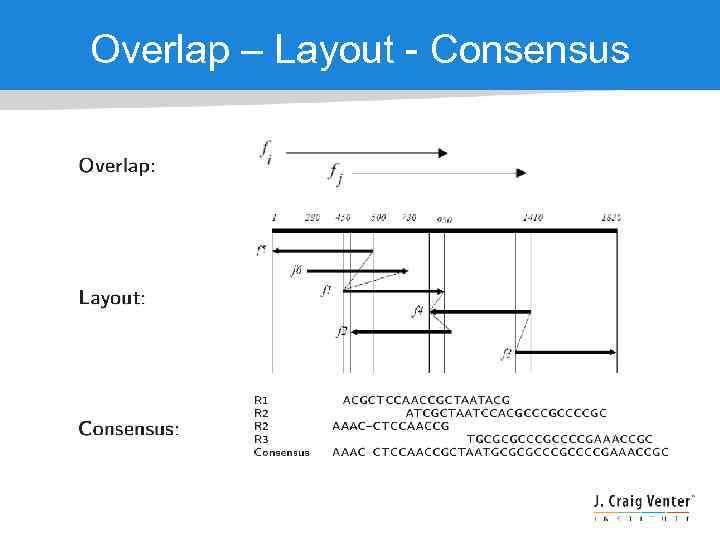
Overlap – Layout - Consensus
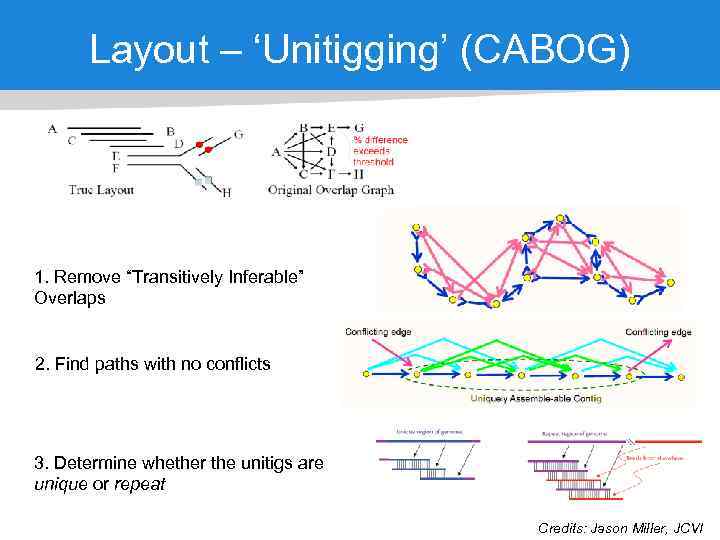
Layout – ‘Unitigging’ (CABOG) 1. Remove “Transitively Inferable” Overlaps 2. Find paths with no conflicts 3. Determine whether the unitigs are unique or repeat Credits: Jason Miller, JCVI
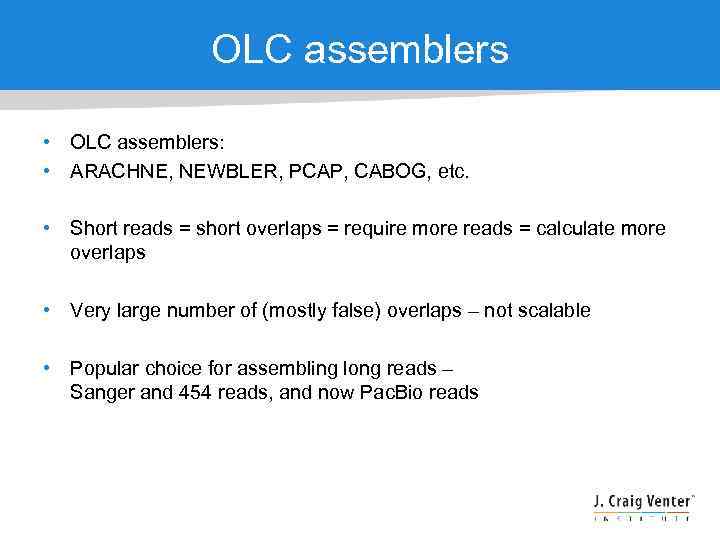
OLC assemblers • OLC assemblers: • ARACHNE, NEWBLER, PCAP, CABOG, etc. • Short reads = short overlaps = require more reads = calculate more overlaps • Very large number of (mostly false) overlaps – not scalable • Popular choice for assembling long reads – Sanger and 454 reads, and now Pac. Bio reads
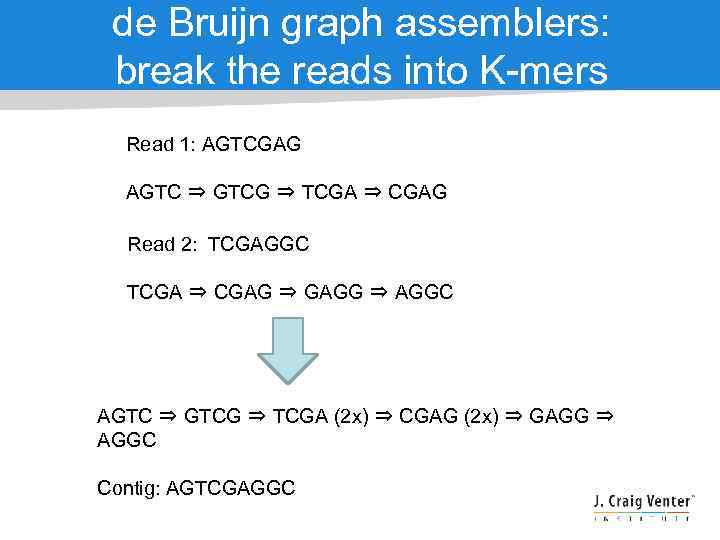
de Bruijn graph assemblers: break the reads into K-mers Read 1: AGTCGAG AGTC ⇒ GTCG ⇒ TCGA ⇒ CGAG Read 2: TCGAGGC TCGA ⇒ CGAG ⇒ GAGG ⇒ AGGC AGTC ⇒ GTCG ⇒ TCGA (2 x) ⇒ CGAG (2 x) ⇒ GAGG ⇒ AGGC Contig: AGTCGAGGC
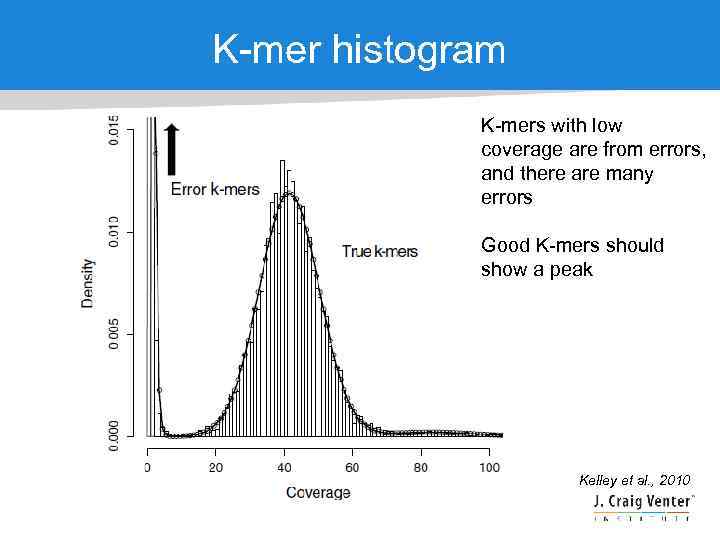
K-mer histogram K-mers with low coverage are from errors, and there are many errors Good K-mers should show a peak Kelley et al. , 2010
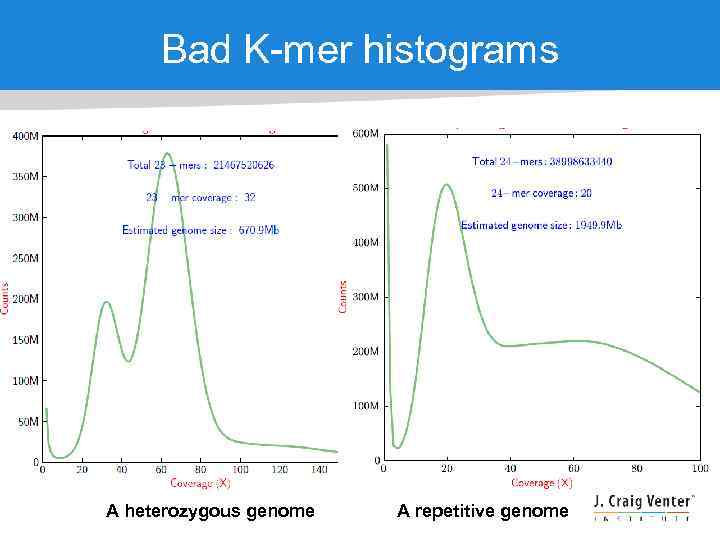
Bad K-mer histograms A heterozygous genome A repetitive genome
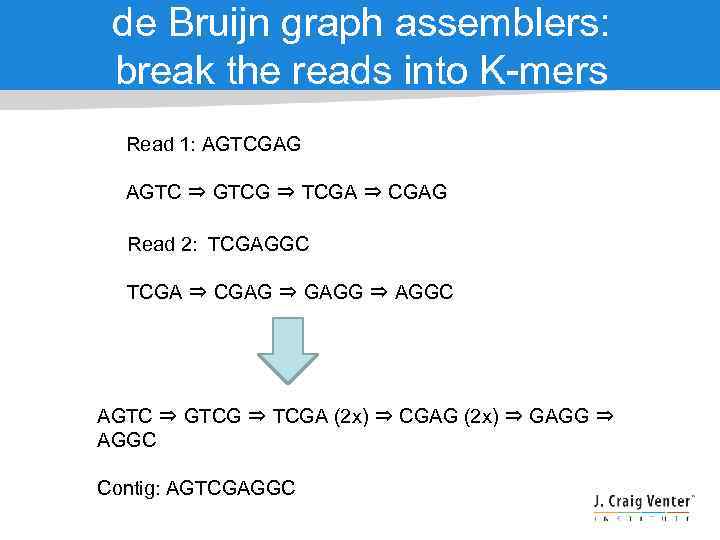
de Bruijn graph assemblers: break the reads into K-mers Read 1: AGTCGAG AGTC ⇒ GTCG ⇒ TCGA ⇒ CGAG Read 2: TCGAGGC TCGA ⇒ CGAG ⇒ GAGG ⇒ AGGC AGTC ⇒ GTCG ⇒ TCGA (2 x) ⇒ CGAG (2 x) ⇒ GAGG ⇒ AGGC Contig: AGTCGAGGC
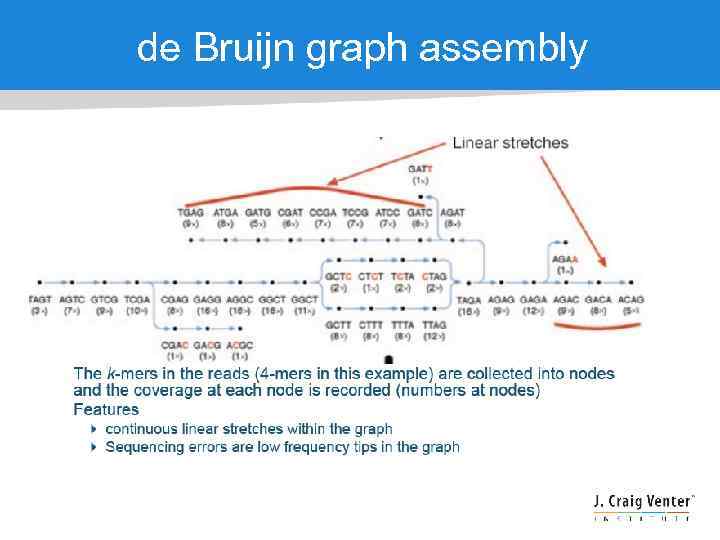
de Bruijn graph assembly
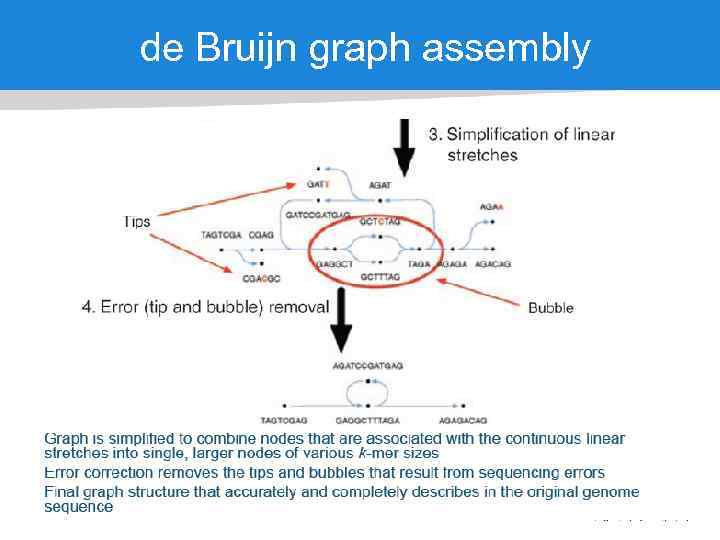
de Bruijn graph assembly
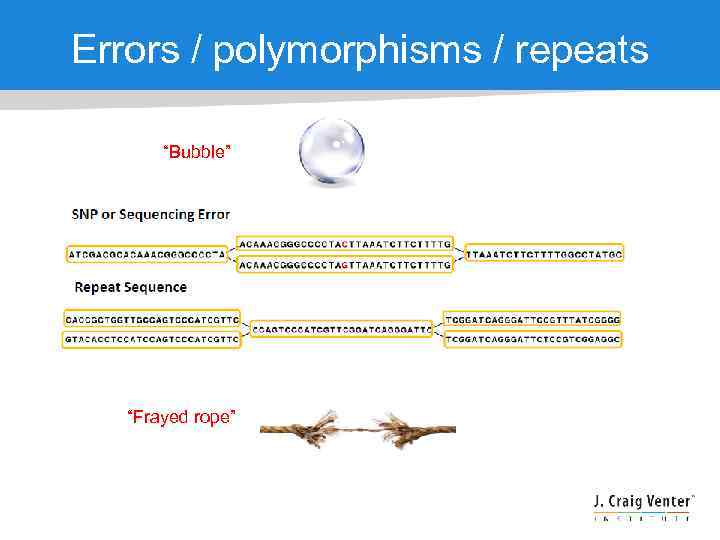
Errors / polymorphisms / repeats “Bubble” “Frayed rope”

de Bruijn graph assemblers • No need for all-against-all overlap discovery – fast (but more RAM) • More sensitive to repeats and sequencing errors • de Bruijn graph assemblers: • SOAPdenovo, ALLPATHS, Abyss, Velvet, SGA, CLC Bio, etc. • We will use ALLPATHS, SOAP, Abyss in today’s exercises • It’s always a good idea to run different assemblers, with different options (K-mer size, etc. ) and COMPARE!

3. Assembly QC
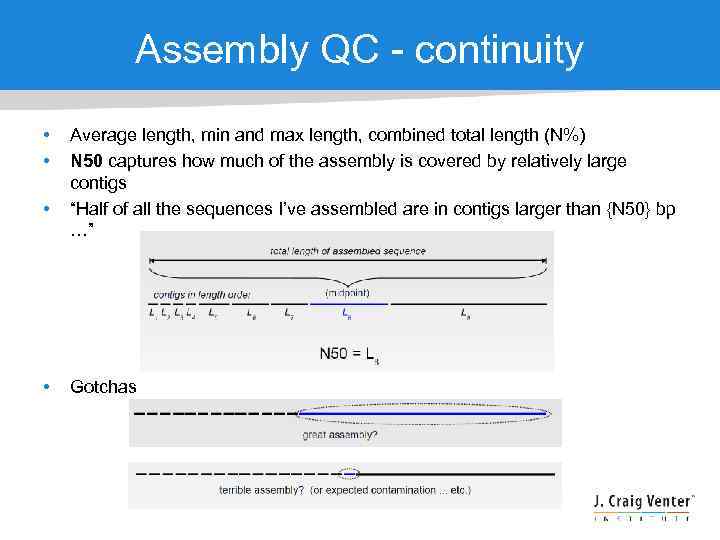
Assembly QC - continuity • • Average length, min and max length, combined total length (N%) N 50 captures how much of the assembly is covered by relatively large contigs “Half of all the sequences I’ve assembled are in contigs larger than {N 50} bp …” Gotchas
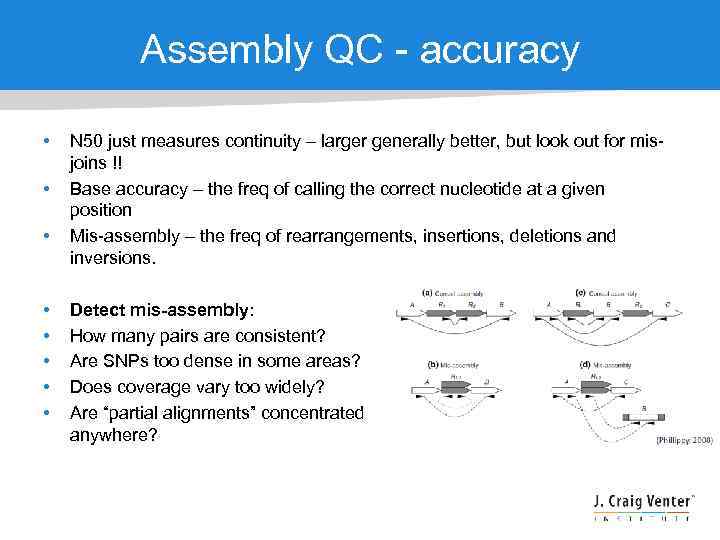
Assembly QC - accuracy • • N 50 just measures continuity – larger generally better, but look out for misjoins !! Base accuracy – the freq of calling the correct nucleotide at a given position Mis-assembly – the freq of rearrangements, insertions, deletions and inversions. Detect mis-assembly: How many pairs are consistent? Are SNPs too dense in some areas? Does coverage vary too widely? Are “partial alignments” concentrated anywhere?
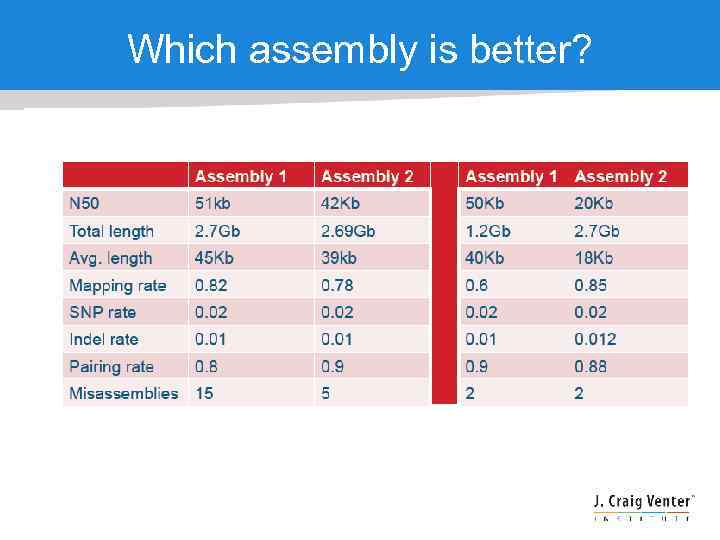
Which assembly is better?
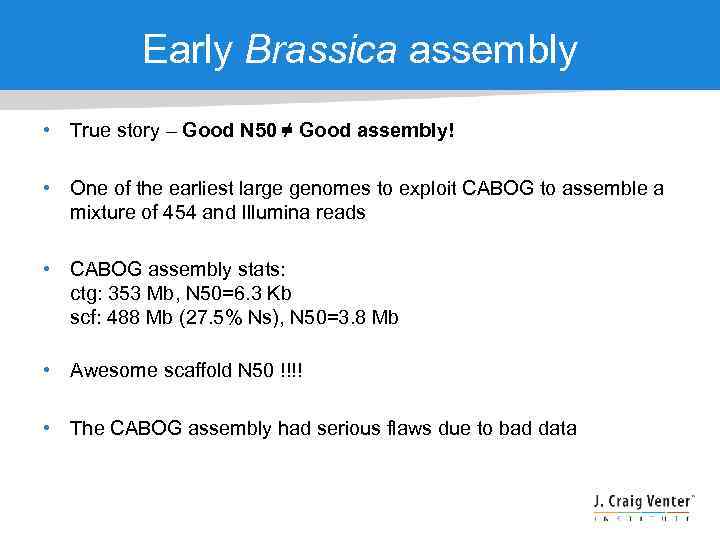
Early Brassica assembly • True story – Good N 50 ≠ Good assembly! • One of the earliest large genomes to exploit CABOG to assemble a mixture of 454 and Illumina reads • CABOG assembly stats: ctg: 353 Mb, N 50=6. 3 Kb scf: 488 Mb (27. 5% Ns), N 50=3. 8 Mb • Awesome scaffold N 50 !!!! • The CABOG assembly had serious flaws due to bad data
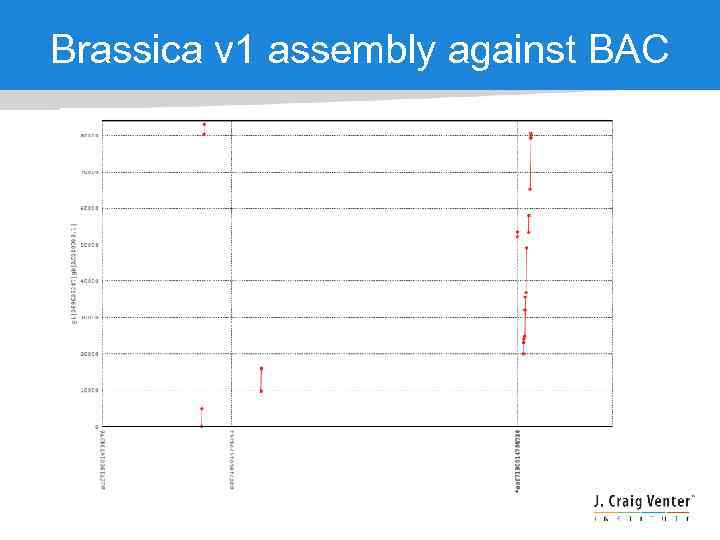
Brassica v 1 assembly against BAC
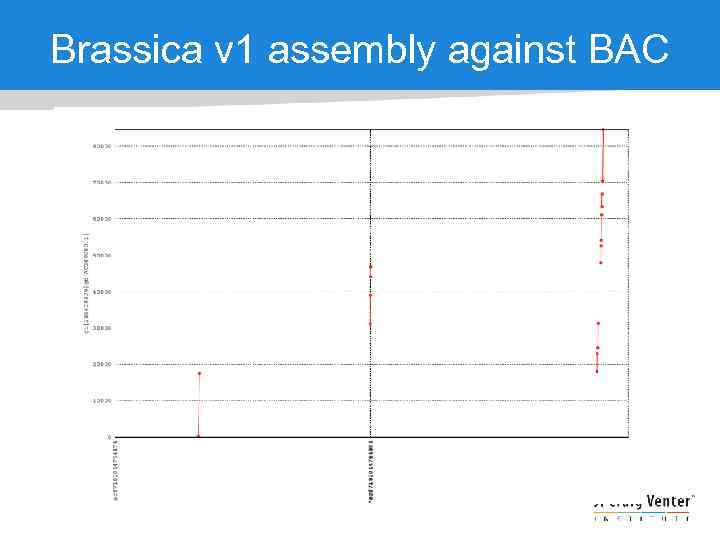
Brassica v 1 assembly against BAC
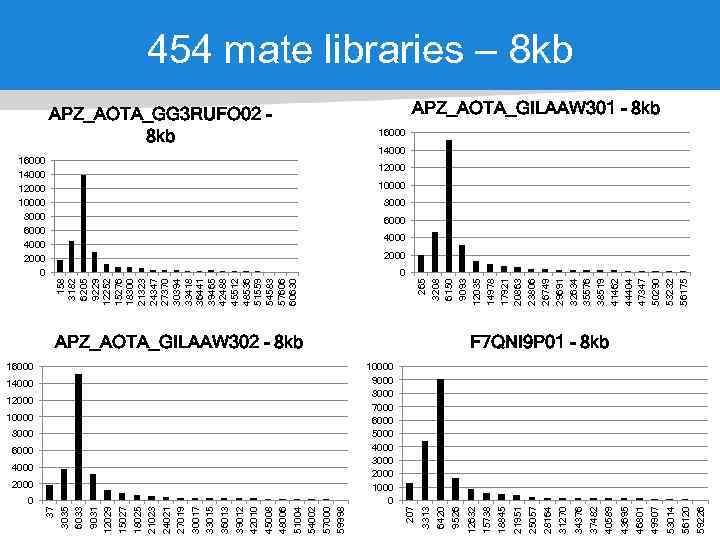
0 37 3035 6033 9031 12029 15027 18025 21023 24021 27019 30017 33015 36013 39012 42010 45008 48006 51004 54002 57000 59998 16000 14000 12000 10000 8000 6000 4000 2000 10000 9000 8000 7000 6000 5000 4000 3000 2000 1000 0 21951 18845 15738 39 59226 56120 53014 49907 46801 43695 40589 37482 34376 31270 28164 25057 APZ_AOTA_GILAAW 302 - 8 kb 12632 9526 6420 265 3208 6150 9093 12035 14978 17921 20863 23806 26749 29691 32634 35576 38519 41462 44404 47347 50290 53232 56175 0 3313 APZ_AOTA_GG 3 RUFO 02 8 kb 207 16000 14000 12000 10000 8000 6000 4000 2000 0 158 3182 6205 9229 12252 15276 18300 21323 24347 27370 30394 33418 36441 39465 42488 45512 48536 51559 54583 57606 60630 454 mate libraries – 8 kb APZ_AOTA_GILAAW 301 - 8 kb 16000 14000 12000 10000 8000 6000 4000 2000 F 7 QNI 9 P 01 - 8 kb
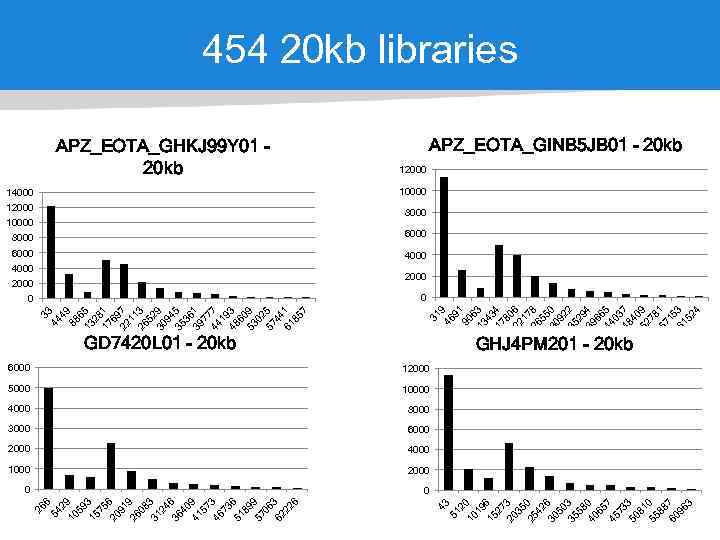
10000 12000 8000 6000 4000 2000 0 6000 12000 5000 10000 4000 8000 3000 6000 2000 4000 1000 2000 0 2 10 0 19 15 6 27 20 3 35 25 0 42 30 6 50 35 3 58 40 0 65 45 7 73 50 3 81 55 0 88 60 7 96 3 14000 51 10000 31 9 46 91 90 6 13 3 43 17 4 80 22 6 17 26 8 55 30 0 92 35 2 29 39 4 66 44 5 03 48 7 40 52 9 78 57 1 15 61 3 52 4 33 44 49 88 6 13 5 28 17 1 69 22 7 11 26 3 52 30 9 94 35 5 36 39 1 77 44 7 19 48 3 60 53 9 02 57 5 44 61 1 85 7 APZ_EOTA_GHKJ 99 Y 01 20 kb 43 6 54 29 10 59 15 3 75 20 6 91 26 9 08 31 3 24 36 6 40 41 9 57 46 3 73 51 6 89 57 9 06 62 3 22 6 26 454 20 kb libraries APZ_EOTA_GINB 5 JB 01 - 20 kb 12000 8000 4000 2000 0 GD 7420 L 01 - 20 kb GHJ 4 PM 201 - 20 kb 0 40
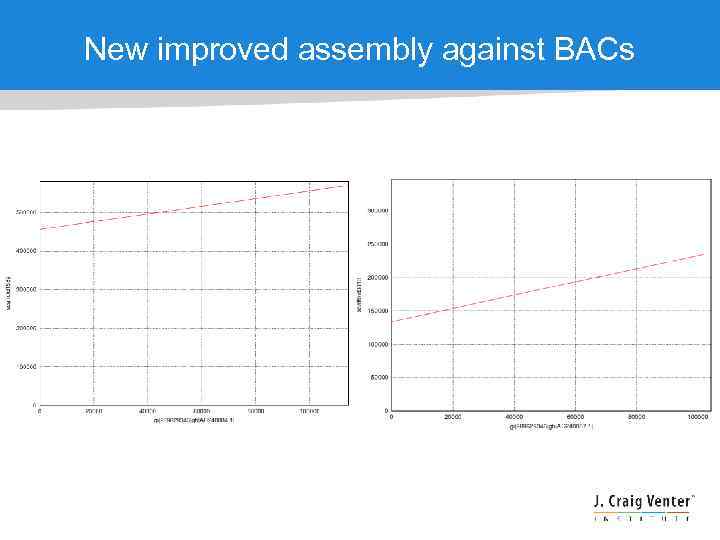
New improved assembly against BACs
Is my assembly complete? • CEGMA evaluates the assembly against 248 ultra-conserved proteins across eukaryotes • Map the transcriptome set (ESTs or RNASeq assemblies) to the genome assembly to see how much of expressed genes are covered
4. Assembly Curation

Assembly curation • What to do after you’ve got an assembly? • • Large insert mate pair library (fosmids, BACs) Optical (restriction) map Genetic map Synteny • Two goals: • Fix chimeric (mis-joined) scaffolds • Build larger scaffolds towards chromosomes
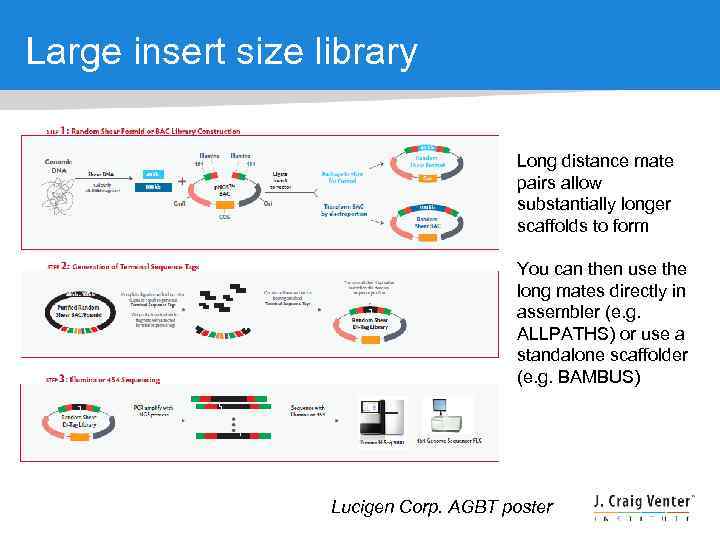
Large insert size library Long distance mate pairs allow substantially longer scaffolds to form You can then use the long mates directly in assembler (e. g. ALLPATHS) or use a standalone scaffolder (e. g. BAMBUS) Lucigen Corp. AGBT poster
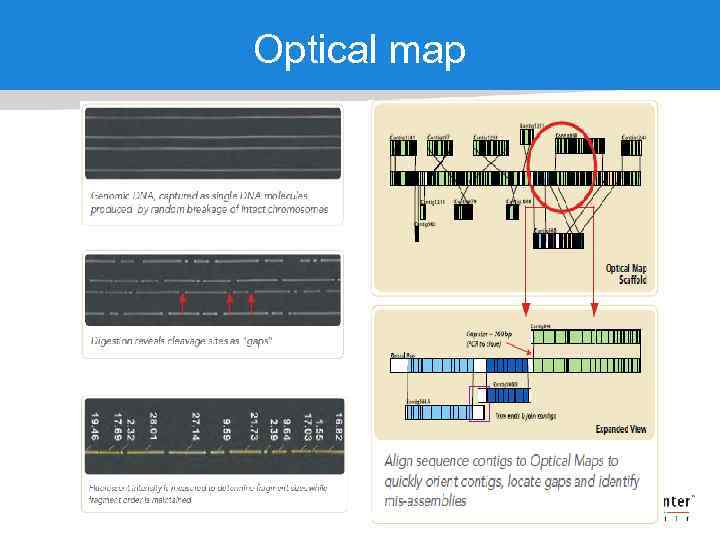
Optical map
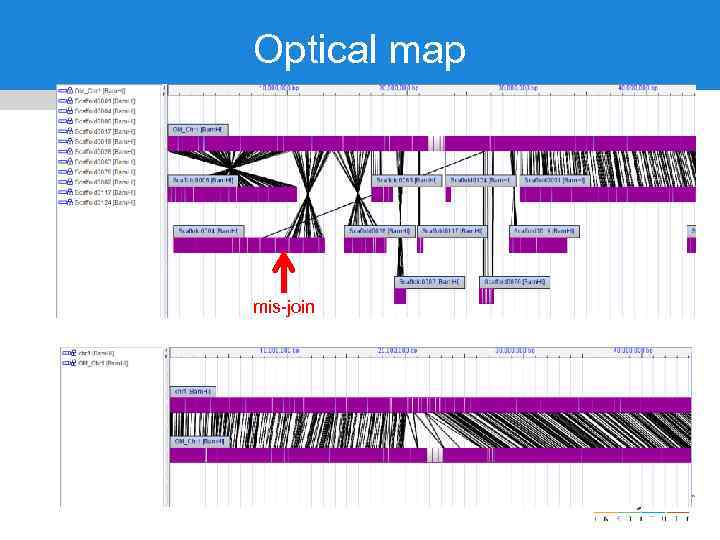
Optical map mis-join
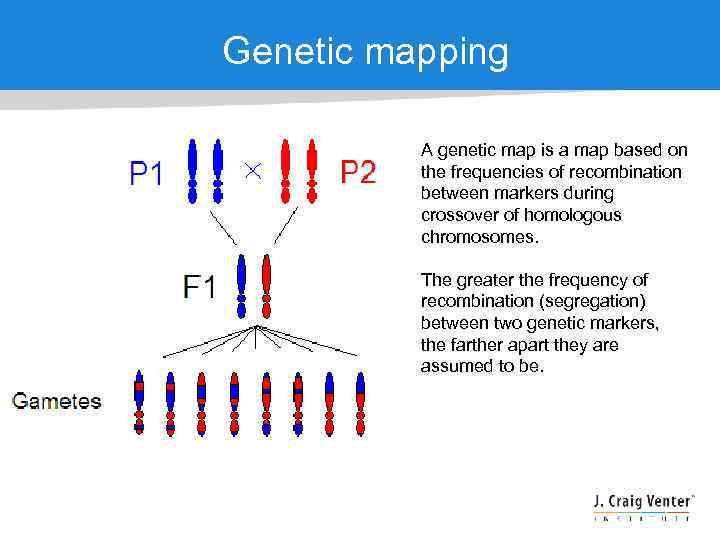
Genetic mapping A genetic map is a map based on the frequencies of recombination between markers during crossover of homologous chromosomes. The greater the frequency of recombination (segregation) between two genetic markers, the farther apart they are assumed to be.
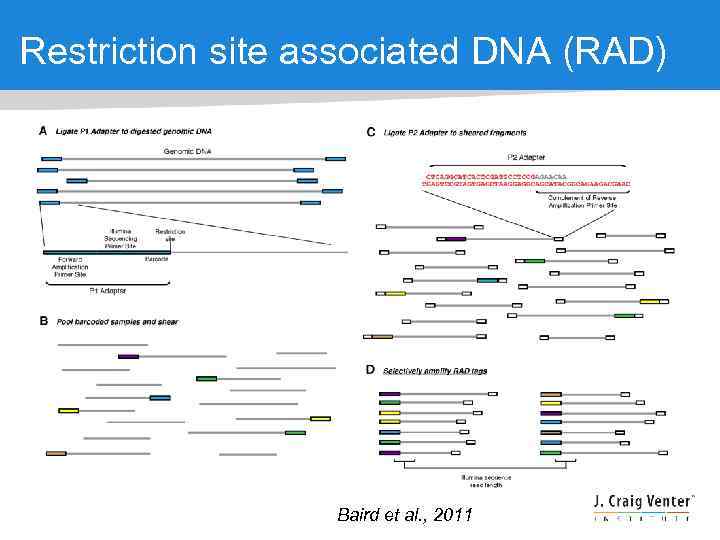
Restriction site associated DNA (RAD) Baird et al. , 2011
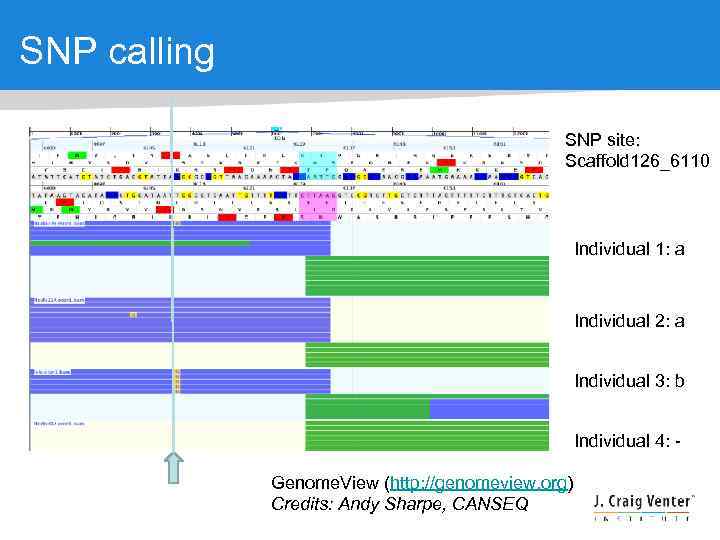
SNP calling SNP site: Scaffold 126_6110 Individual 1: a Individual 2: a Individual 3: b Individual 4: Genome. View (http: //genomeview. org) Credits: Andy Sharpe, CANSEQ

RAD segregation data a=ref allele, b=alt allele, - = missing 1 al ual 2 u … vidivid i d Ind. In SNP sites MSTmap (http: //alumni. cs. ucr. edu/~yonghui/mstmap. html)
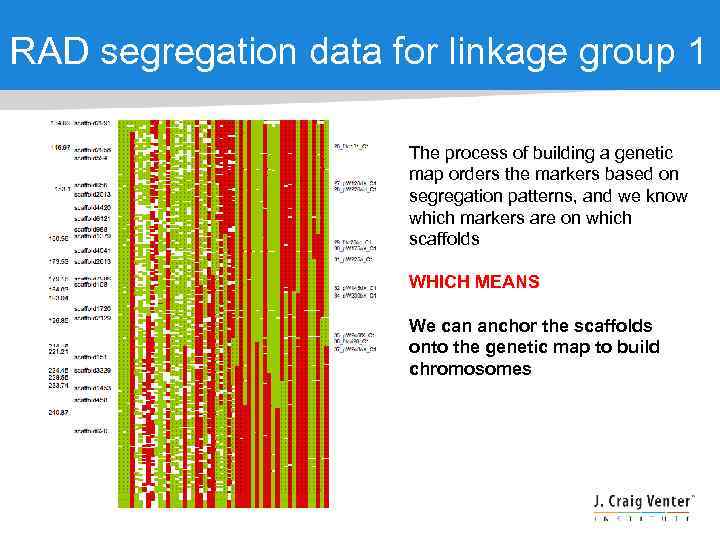
RAD segregation data for linkage group 1 The process of building a genetic map orders the markers based on segregation patterns, and we know which markers are on which scaffolds WHICH MEANS We can anchor the scaffolds onto the genetic map to build chromosomes
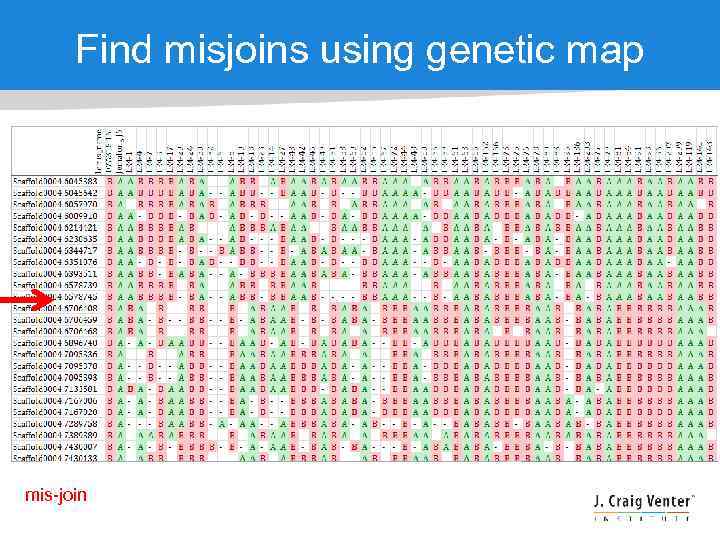
Find misjoins using genetic map mis-join

Talk summary • Good genome assembly is dependent on good preparation of data • Don’t rely on the results of your assembler, check adequately and check again using whatever references you can find • External scaffolding using maps (genetic map, physical map, optical map) allow repair of chimeric scaffolds, and anchor onto chromosomes • FIN
Genome-Assembly-Haibao.pptx