0b2aad24dec17adf8268479425b81a66.ppt
- Количество слайдов: 51
 De Novo design tools for the generation of synthetically accessible ligands Peter Johnson, Krisztina Boda, Shane Weaver, Aniko Valko, Vilmos Valko
De Novo design tools for the generation of synthetically accessible ligands Peter Johnson, Krisztina Boda, Shane Weaver, Aniko Valko, Vilmos Valko
 Receptor Structure Based Drug Design Objective: To suggest potential leads that § bind strongly to a given protein because of shape and electrostatic complementarity § Are easy to synthesise Approaches: § Docking methods (preferably flexible docking) identify new lead structures by rapidly screening a database of 3 -D structures of known compounds § De novo design methods (such as SPROUT) construct a diverse set of entirely novel potential leads from scratch
Receptor Structure Based Drug Design Objective: To suggest potential leads that § bind strongly to a given protein because of shape and electrostatic complementarity § Are easy to synthesise Approaches: § Docking methods (preferably flexible docking) identify new lead structures by rapidly screening a database of 3 -D structures of known compounds § De novo design methods (such as SPROUT) construct a diverse set of entirely novel potential leads from scratch
 SPROUT Components Detects potential binding pockets of the protein structures Identifies favourable hydrogen bonding interaction sites (H-bonding, hydrophobic, covalent, metal, user defined) Docks structures to target interaction sites Generates 3 D molecular structures of novel ligands by linking the docked starting fragments together in an incremental construction scheme Scores, sorts and clusters the solutions
SPROUT Components Detects potential binding pockets of the protein structures Identifies favourable hydrogen bonding interaction sites (H-bonding, hydrophobic, covalent, metal, user defined) Docks structures to target interaction sites Generates 3 D molecular structures of novel ligands by linking the docked starting fragments together in an incremental construction scheme Scores, sorts and clusters the solutions
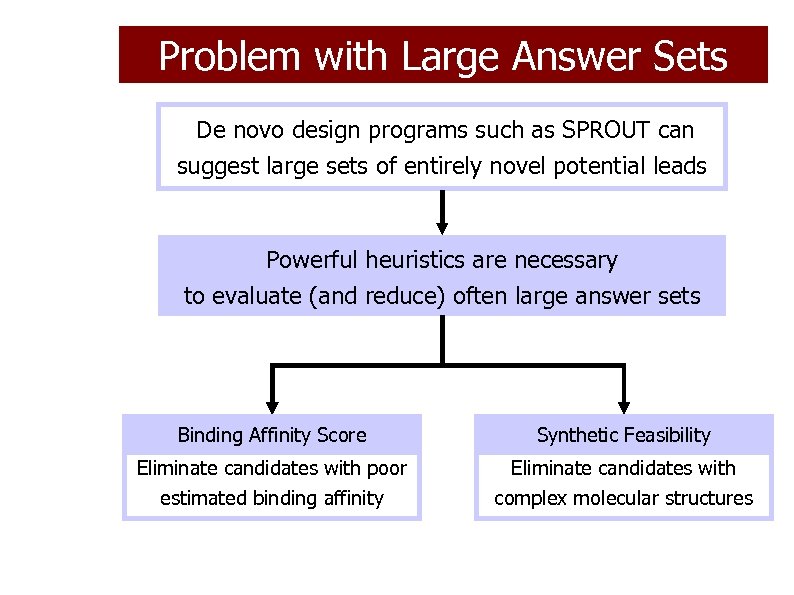 Problem with Large Answer Sets De novo design programs such as SPROUT can suggest large sets of entirely novel potential leads Powerful heuristics are necessary to evaluate (and reduce) often large answer sets Binding Affinity Score Synthetic Feasibility Eliminate candidates with poor estimated binding affinity Eliminate candidates with complex molecular structures
Problem with Large Answer Sets De novo design programs such as SPROUT can suggest large sets of entirely novel potential leads Powerful heuristics are necessary to evaluate (and reduce) often large answer sets Binding Affinity Score Synthetic Feasibility Eliminate candidates with poor estimated binding affinity Eliminate candidates with complex molecular structures
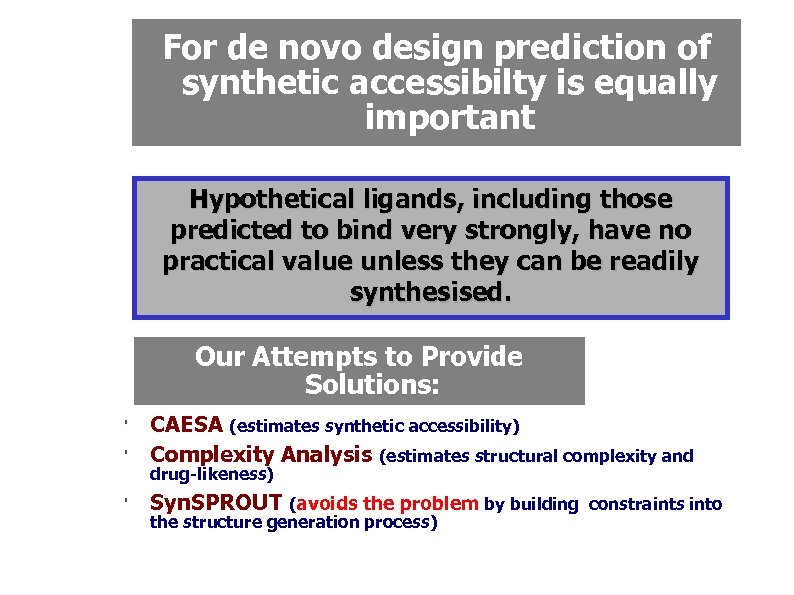 For de novo design prediction of synthetic accessibilty is equally important Hypothetical ligands, including those predicted to bind very strongly, have no practical value unless they can be readily synthesised. Our Attempts to Provide Solutions: ' CAESA (estimates synthetic accessibility) Complexity Analysis (estimates structural complexity and ' Syn. SPROUT (avoids the problem by building ' drug-likeness) the structure generation process) constraints into
For de novo design prediction of synthetic accessibilty is equally important Hypothetical ligands, including those predicted to bind very strongly, have no practical value unless they can be readily synthesised. Our Attempts to Provide Solutions: ' CAESA (estimates synthetic accessibility) Complexity Analysis (estimates structural complexity and ' Syn. SPROUT (avoids the problem by building ' drug-likeness) the structure generation process) constraints into
 CAESA Computer Assisted Estimation of Synthetic Accessibility Glenn Myatt Jon Baber
CAESA Computer Assisted Estimation of Synthetic Accessibility Glenn Myatt Jon Baber
 Goals of CAESA Project § Clear need for automated method of ranking hypothetical compounds according to perceived ease of synthesis § Good synthetic chemists can do this job themselves on small number of compounds but are unwilling to do it for hundreds or thousands of compounds § CAESA attempts to do the same job but never gets bored!
Goals of CAESA Project § Clear need for automated method of ranking hypothetical compounds according to perceived ease of synthesis § Good synthetic chemists can do this job themselves on small number of compounds but are unwilling to do it for hundreds or thousands of compounds § CAESA attempts to do the same job but never gets bored!
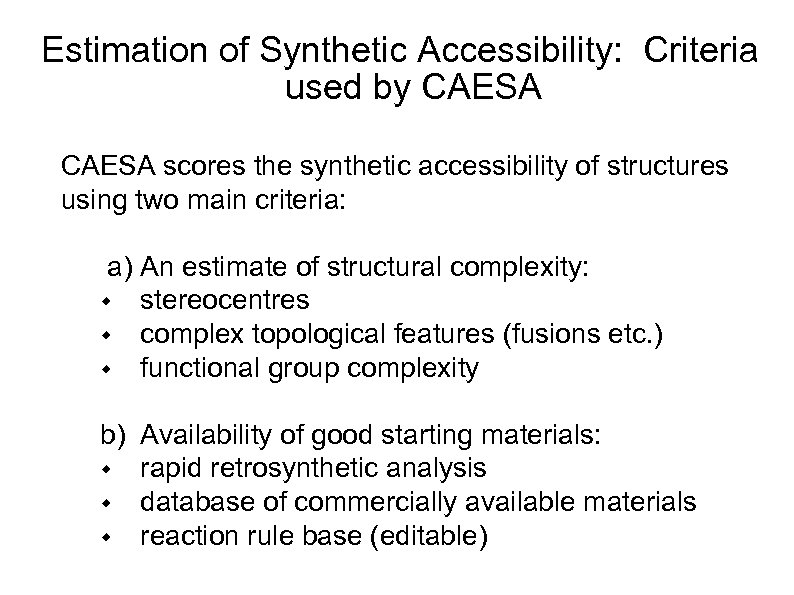 Estimation of Synthetic Accessibility: Criteria used by CAESA scores the synthetic accessibility of structures using two main criteria: a) An estimate of structural complexity: w stereocentres w complex topological features (fusions etc. ) w functional group complexity b) Availability of good starting materials: w rapid retrosynthetic analysis w database of commercially available materials w reaction rule base (editable)
Estimation of Synthetic Accessibility: Criteria used by CAESA scores the synthetic accessibility of structures using two main criteria: a) An estimate of structural complexity: w stereocentres w complex topological features (fusions etc. ) w functional group complexity b) Availability of good starting materials: w rapid retrosynthetic analysis w database of commercially available materials w reaction rule base (editable)
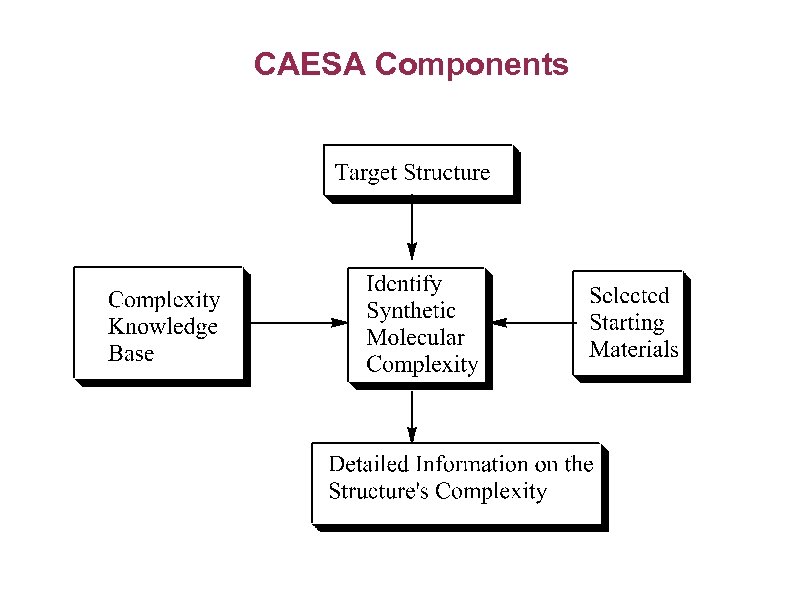 CAESA Components
CAESA Components
 Automatic Selection of Starting Materials and Synthetic Accessibility § Availability of suitable starting materials very important factor good starting materials can dramatically reduce the difficulty of synthesising a compound. § Good starting materials for part of the target molecule means the analysis of structural synthetic difficulty or complexity can be directed to just those portions of the target molecule that cannot be made from available starting materials § Finding good starting materials through retrosynthetic analysis also provides possible synthetic routes as a byproduct
Automatic Selection of Starting Materials and Synthetic Accessibility § Availability of suitable starting materials very important factor good starting materials can dramatically reduce the difficulty of synthesising a compound. § Good starting materials for part of the target molecule means the analysis of structural synthetic difficulty or complexity can be directed to just those portions of the target molecule that cannot be made from available starting materials § Finding good starting materials through retrosynthetic analysis also provides possible synthetic routes as a byproduct
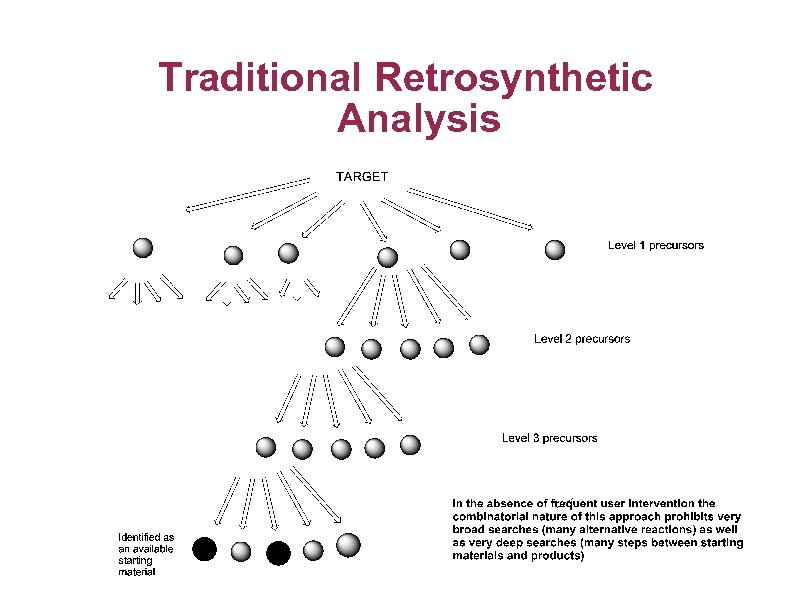 Traditional Retrosynthetic Analysis
Traditional Retrosynthetic Analysis
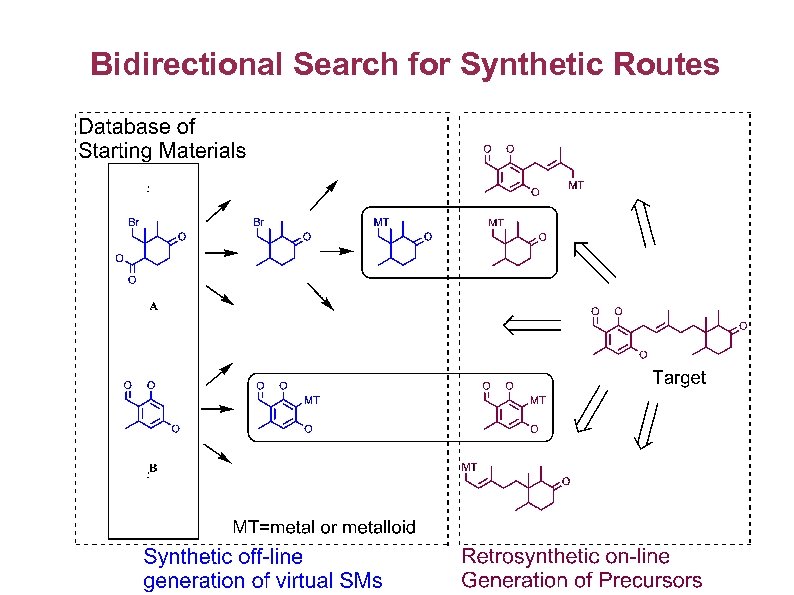 Bidirectional Search for Synthetic Routes
Bidirectional Search for Synthetic Routes
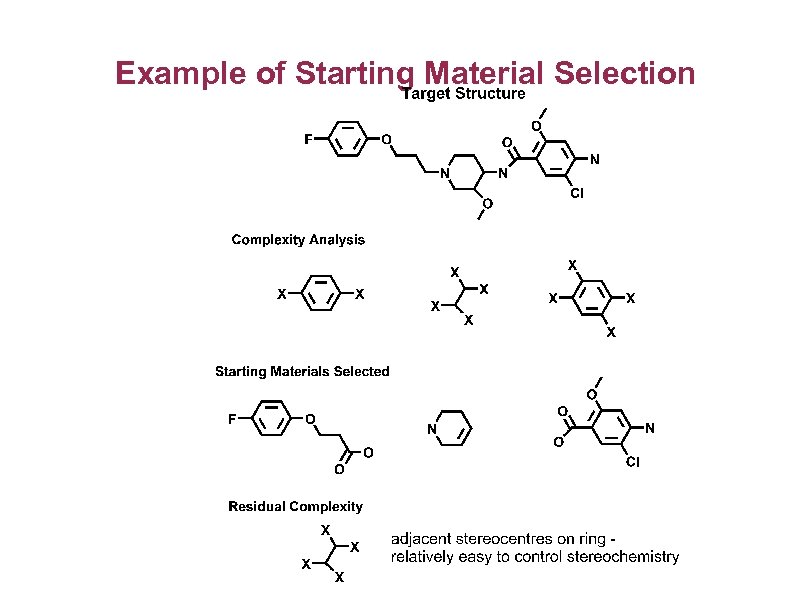 Example of Starting Material Selection
Example of Starting Material Selection
 Summary of CAESA Features § CAESA carries out a retrosynthetic analysis which terminates when a starting material from a database (such as ACD) is found § Found starting materials are scored according to length and difficulty of reaction sequence and coverage of target compound § All chemistry rules and transformations are described in editable text knowledge bases easily modified by chemists § Quality of the analysis depends on the chemistry included in the knowledge bases and the comprehensiveness of the starting material libraries § But CAESA is relatively slow and speedier methods needed for pruning of large data sets
Summary of CAESA Features § CAESA carries out a retrosynthetic analysis which terminates when a starting material from a database (such as ACD) is found § Found starting materials are scored according to length and difficulty of reaction sequence and coverage of target compound § All chemistry rules and transformations are described in editable text knowledge bases easily modified by chemists § Quality of the analysis depends on the chemistry included in the knowledge bases and the comprehensiveness of the starting material libraries § But CAESA is relatively slow and speedier methods needed for pruning of large data sets
 Alternative Approach Complexity Analysis Based on statistical distribution of various substitution patterns found in databases of existing drugs and available starting materials. Molecular Complexity Analysis of de Novo Designed Ligands Krisztina Boda and A. Peter Johnson J. Med. Chem. ; 2006; ASAP Web Release Date: 26 -Jan-2006
Alternative Approach Complexity Analysis Based on statistical distribution of various substitution patterns found in databases of existing drugs and available starting materials. Molecular Complexity Analysis of de Novo Designed Ligands Krisztina Boda and A. Peter Johnson J. Med. Chem. ; 2006; ASAP Web Release Date: 26 -Jan-2006
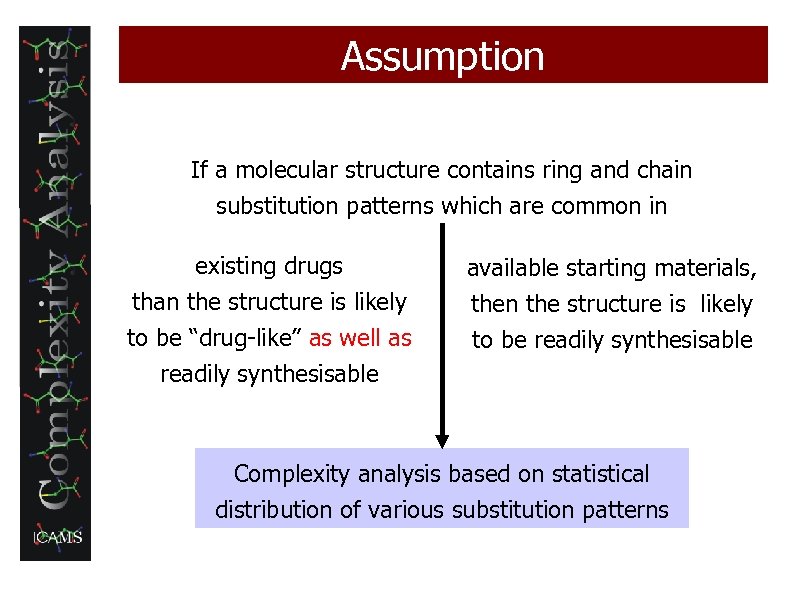 Assumption If a molecular structure contains ring and chain substitution patterns which are common in existing drugs than the structure is likely to be “drug-like” as well as available starting materials, then the structure is likely to be readily synthesisable Complexity analysis based on statistical distribution of various substitution patterns
Assumption If a molecular structure contains ring and chain substitution patterns which are common in existing drugs than the structure is likely to be “drug-like” as well as available starting materials, then the structure is likely to be readily synthesisable Complexity analysis based on statistical distribution of various substitution patterns
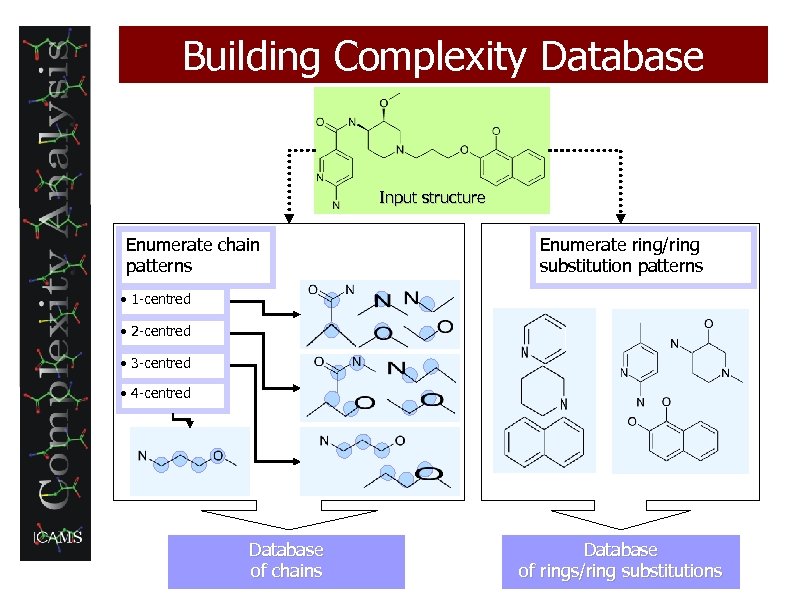 Building Complexity Database Input structure Enumerate chain patterns Enumerate ring/ring substitution patterns • 1 -centred • 2 -centred • 3 -centred • 4 -centred Database of chains Database of rings/ring substitutions
Building Complexity Database Input structure Enumerate chain patterns Enumerate ring/ring substitution patterns • 1 -centred • 2 -centred • 3 -centred • 4 -centred Database of chains Database of rings/ring substitutions
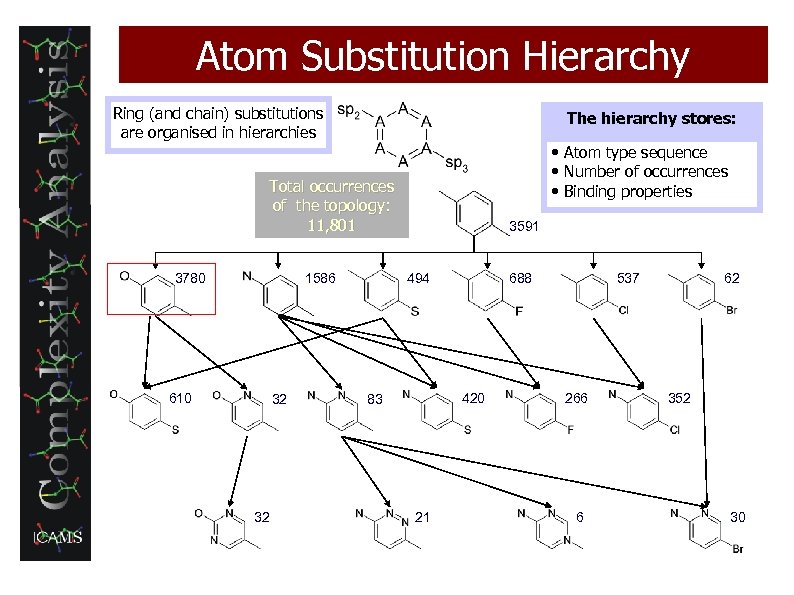 Atom Substitution Hierarchy Ring (and chain) substitutions are organised in hierarchies The hierarchy stores: • Atom type sequence • Number of occurrences • Binding properties Total occurrences of the topology: 11, 801 3780 1586 610 32 32 3591 494 688 420 83 21 537 266 6 62 352 30
Atom Substitution Hierarchy Ring (and chain) substitutions are organised in hierarchies The hierarchy stores: • Atom type sequence • Number of occurrences • Binding properties Total occurrences of the topology: 11, 801 3780 1586 610 32 32 3591 494 688 420 83 21 537 266 6 62 352 30
![Ligand Complexity Analysis 1. Enumerate ring and chain patterns [More Patterns] Canonical name : Ligand Complexity Analysis 1. Enumerate ring and chain patterns [More Patterns] Canonical name :](https://present5.com/presentation/0b2aad24dec17adf8268479425b81a66/image-19.jpg) Ligand Complexity Analysis 1. Enumerate ring and chain patterns [More Patterns] Canonical name : A DATABASE of hierarchies + frequency of occurrences Canonical name : B Canonical name : C 3. Match canonical name against the hierarchy roots of the database 2. Generate canonical names for each atom pattern 5. Rank structures by complexity score 4. Retrieval of frequency of occurrences → Calculate score Speed of Complexity Analysis ~ 1000 -1200 structures / minute on Linux PC (3 GHz)
Ligand Complexity Analysis 1. Enumerate ring and chain patterns [More Patterns] Canonical name : A DATABASE of hierarchies + frequency of occurrences Canonical name : B Canonical name : C 3. Match canonical name against the hierarchy roots of the database 2. Generate canonical names for each atom pattern 5. Rank structures by complexity score 4. Retrieval of frequency of occurrences → Calculate score Speed of Complexity Analysis ~ 1000 -1200 structures / minute on Linux PC (3 GHz)
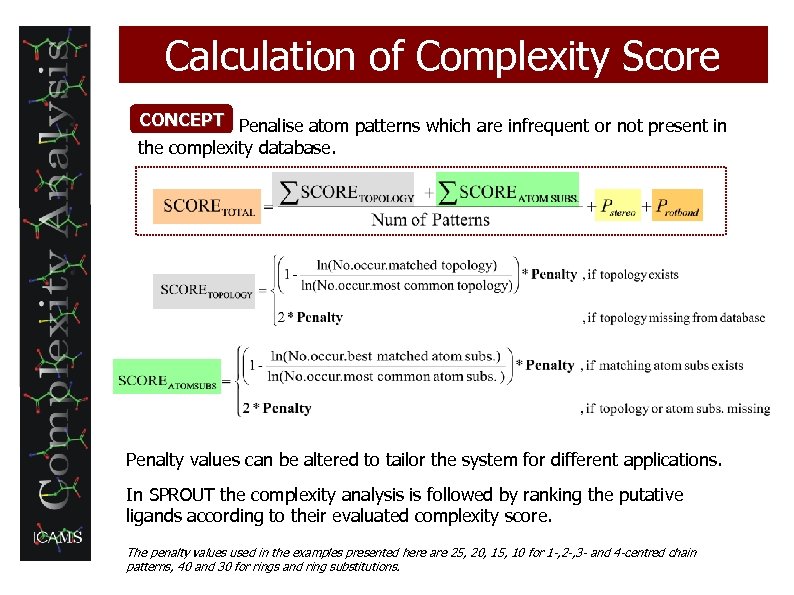 Calculation of Complexity Score CONCEPT Penalise atom patterns which are infrequent or not present in the complexity database. Penalty values can be altered to tailor the system for different applications. In SPROUT the complexity analysis is followed by ranking the putative ligands according to their evaluated complexity score. The penalty values used in the examples presented here are 25, 20, 15, 10 for 1 -, 2 -, 3 - and 4 -centred chain patterns, 40 and 30 for rings and ring substitutions.
Calculation of Complexity Score CONCEPT Penalise atom patterns which are infrequent or not present in the complexity database. Penalty values can be altered to tailor the system for different applications. In SPROUT the complexity analysis is followed by ranking the putative ligands according to their evaluated complexity score. The penalty values used in the examples presented here are 25, 20, 15, 10 for 1 -, 2 -, 3 - and 4 -centred chain patterns, 40 and 30 for rings and ring substitutions.
 Validation Experiment Comparison with CAESA Both methods used to estimate synthetic accessibility for the same set of 50 top selling drugs
Validation Experiment Comparison with CAESA Both methods used to estimate synthetic accessibility for the same set of 50 top selling drugs
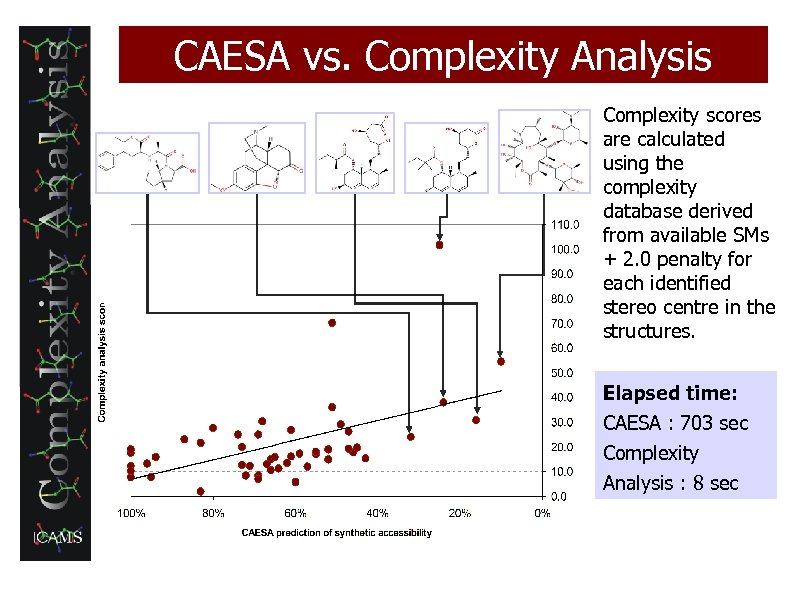 CAESA vs. Complexity Analysis Complexity scores are calculated using the complexity database derived from available SMs + 2. 0 penalty for each identified stereo centre in the structures. Elapsed time: CAESA : 703 sec Complexity Analysis : 8 sec
CAESA vs. Complexity Analysis Complexity scores are calculated using the complexity database derived from available SMs + 2. 0 penalty for each identified stereo centre in the structures. Elapsed time: CAESA : 703 sec Complexity Analysis : 8 sec
 Complexity Analysis vs CAESA § More suitable for prioritization of thousands of structures within a reasonable time frame. § Provides acceptable compromise between the speed of the analysis and the accuracy of calculated scores. § Because this approach is based on characteristics of existing readily available compounds, simple but novel structural features may be wrongly identified as complex
Complexity Analysis vs CAESA § More suitable for prioritization of thousands of structures within a reasonable time frame. § Provides acceptable compromise between the speed of the analysis and the accuracy of calculated scores. § Because this approach is based on characteristics of existing readily available compounds, simple but novel structural features may be wrongly identified as complex
 Yet another alternative approach Build synthetic feasibility into the structure generation process ~
Yet another alternative approach Build synthetic feasibility into the structure generation process ~
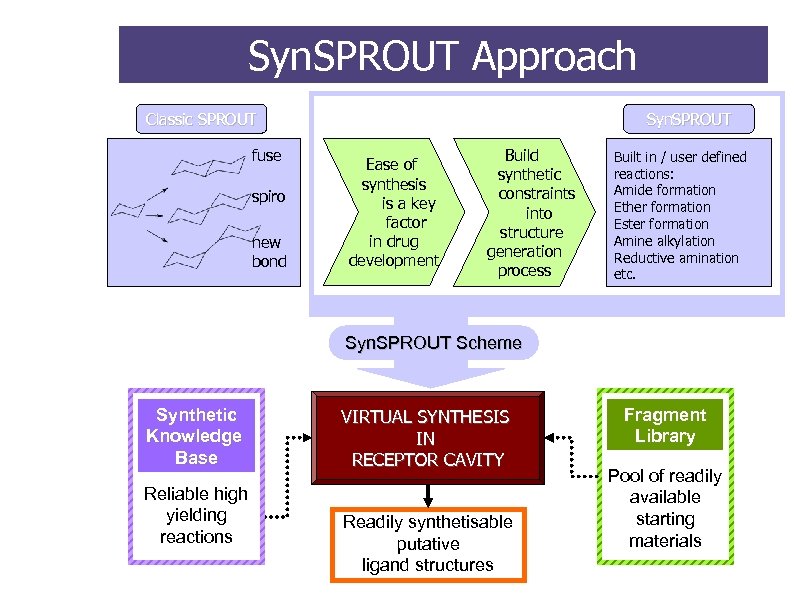 Syn. SPROUT Approach Classic SPROUT fuse spiro new bond Syn. SPROUT Ease of synthesis is a key factor in drug development Build synthetic constraints into structure generation process Built in / user defined reactions: Amide formation Ether formation Ester formation Amine alkylation Reductive amination etc. Syn. SPROUT Scheme Synthetic Knowledge Base Reliable high yielding reactions VIRTUAL SYNTHESIS IN RECEPTOR CAVITY Readily synthetisable putative ligand structures Fragment Library Pool of readily available starting materials
Syn. SPROUT Approach Classic SPROUT fuse spiro new bond Syn. SPROUT Ease of synthesis is a key factor in drug development Build synthetic constraints into structure generation process Built in / user defined reactions: Amide formation Ether formation Ester formation Amine alkylation Reductive amination etc. Syn. SPROUT Scheme Synthetic Knowledge Base Reliable high yielding reactions VIRTUAL SYNTHESIS IN RECEPTOR CAVITY Readily synthetisable putative ligand structures Fragment Library Pool of readily available starting materials
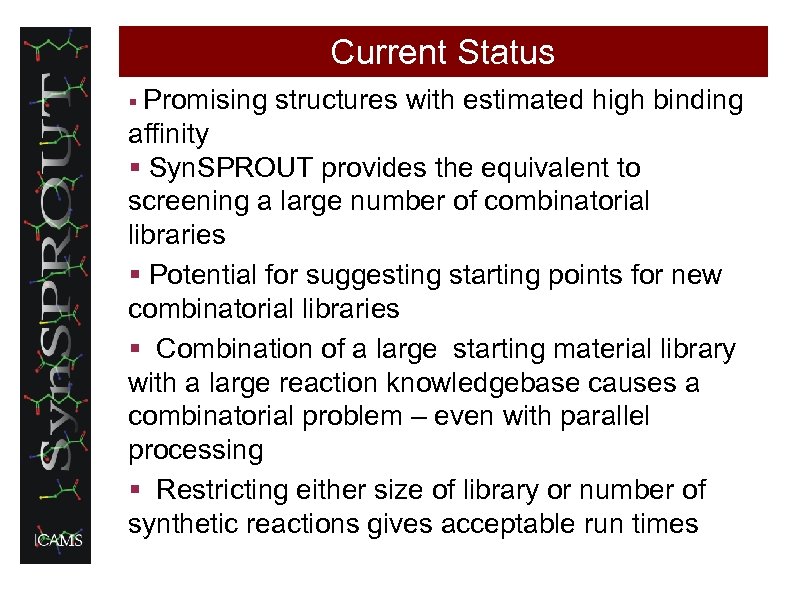 Current Status § Promising structures with estimated high binding affinity § Syn. SPROUT provides the equivalent to screening a large number of combinatorial libraries § Potential for suggesting starting points for new combinatorial libraries § Combination of a large starting material library with a large reaction knowledgebase causes a combinatorial problem – even with parallel processing § Restricting either size of library or number of synthetic reactions gives acceptable run times
Current Status § Promising structures with estimated high binding affinity § Syn. SPROUT provides the equivalent to screening a large number of combinatorial libraries § Potential for suggesting starting points for new combinatorial libraries § Combination of a large starting material library with a large reaction knowledgebase causes a combinatorial problem – even with parallel processing § Restricting either size of library or number of synthetic reactions gives acceptable run times
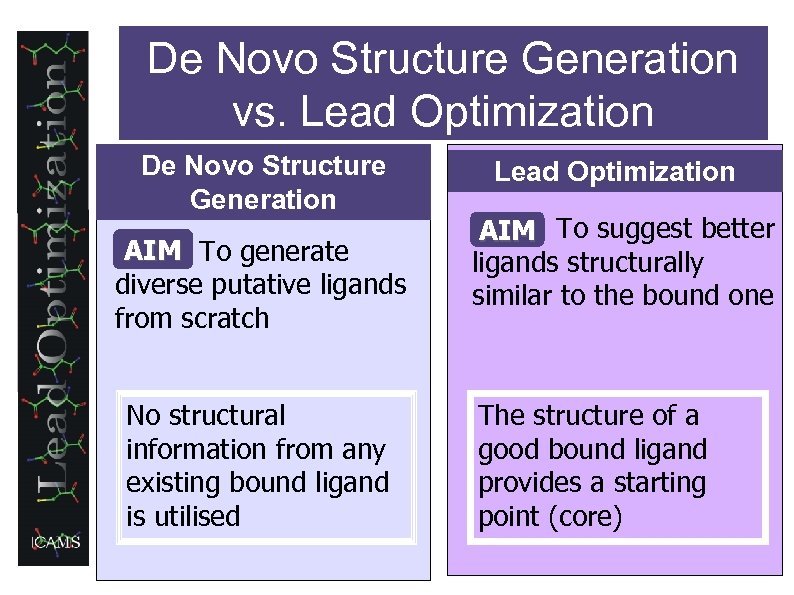 De Novo Structure Generation vs. Lead Optimization De Novo Structure Generation AIM To generate diverse putative ligands from scratch No structural information from any existing bound ligand is utilised Lead Optimization AIM To suggest better ligands structurally similar to the bound one The structure of a good bound ligand provides a starting point (core)
De Novo Structure Generation vs. Lead Optimization De Novo Structure Generation AIM To generate diverse putative ligands from scratch No structural information from any existing bound ligand is utilised Lead Optimization AIM To suggest better ligands structurally similar to the bound one The structure of a good bound ligand provides a starting point (core)
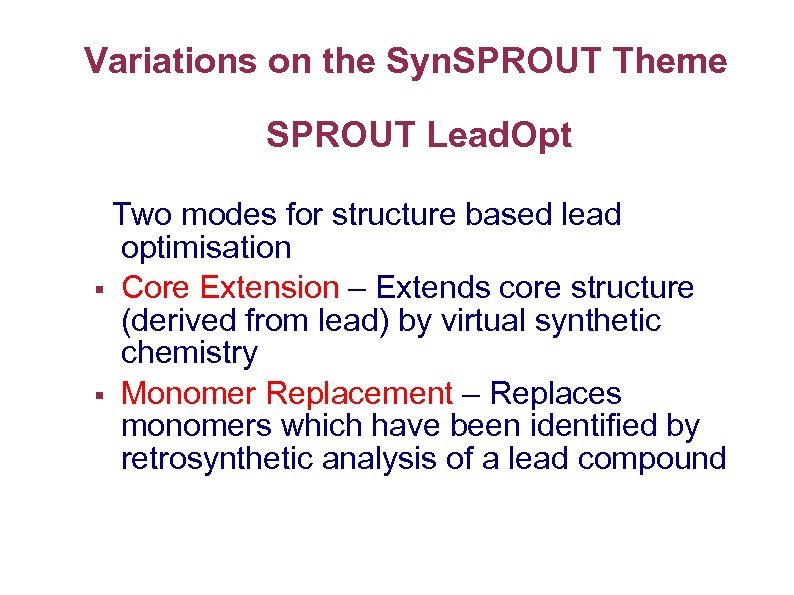 Variations on the Syn. SPROUT Theme SPROUT Lead. Opt Two modes for structure based lead optimisation § Core Extension – Extends core structure (derived from lead) by virtual synthetic chemistry § Monomer Replacement – Replaces monomers which have been identified by retrosynthetic analysis of a lead compound
Variations on the Syn. SPROUT Theme SPROUT Lead. Opt Two modes for structure based lead optimisation § Core Extension – Extends core structure (derived from lead) by virtual synthetic chemistry § Monomer Replacement – Replaces monomers which have been identified by retrosynthetic analysis of a lead compound
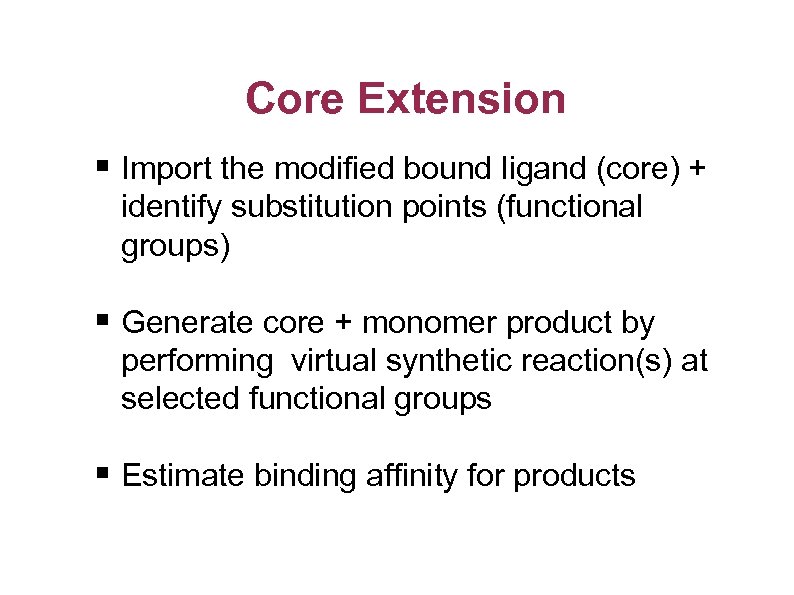 Core Extension § Import the modified bound ligand (core) + identify substitution points (functional groups) § Generate core + monomer product by performing virtual synthetic reaction(s) at selected functional groups § Estimate binding affinity for products
Core Extension § Import the modified bound ligand (core) + identify substitution points (functional groups) § Generate core + monomer product by performing virtual synthetic reaction(s) at selected functional groups § Estimate binding affinity for products
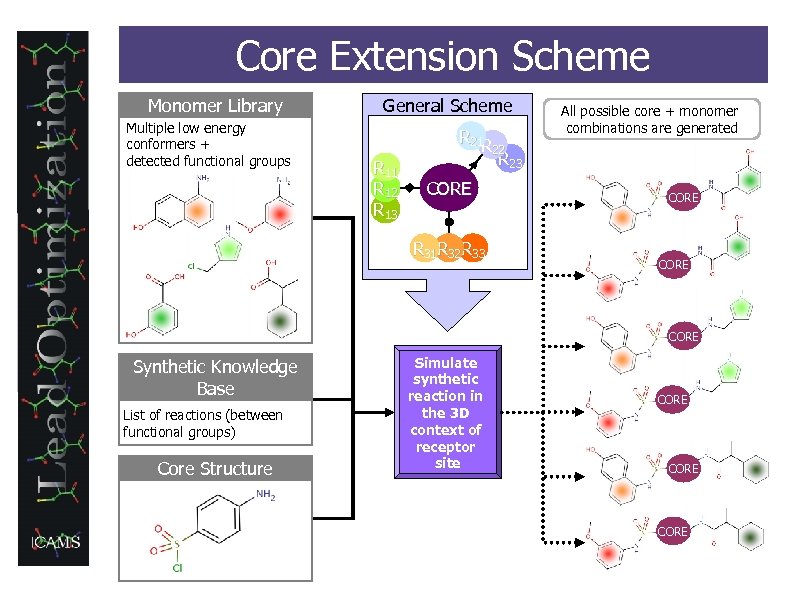 Core Extension Scheme Monomer Library Multiple low energy conformers + detected functional groups General Scheme R 21 R R 11 R 12 R 13 All possible core + monomer combinations are generated 22 CORE R 31 R 32 R 33 R 23 CORE Synthetic Knowledge Base List of reactions (between functional groups) Core Structure Simulate synthetic reaction in the 3 D context of receptor site CORE
Core Extension Scheme Monomer Library Multiple low energy conformers + detected functional groups General Scheme R 21 R R 11 R 12 R 13 All possible core + monomer combinations are generated 22 CORE R 31 R 32 R 33 R 23 CORE Synthetic Knowledge Base List of reactions (between functional groups) Core Structure Simulate synthetic reaction in the 3 D context of receptor site CORE
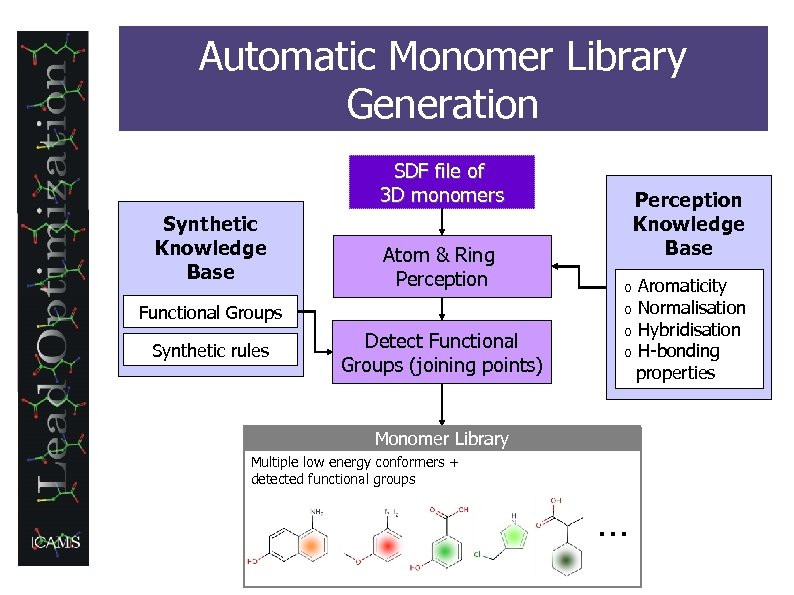 Automatic Monomer Library Generation SDF file of 3 D monomers Synthetic Knowledge Base Atom & Ring Perception Functional Groups Synthetic rules Detect Functional Groups (joining points) Perception Knowledge Base o o Monomer Library Multiple low energy conformers + detected functional groups … Aromaticity Normalisation Hybridisation H-bonding properties
Automatic Monomer Library Generation SDF file of 3 D monomers Synthetic Knowledge Base Atom & Ring Perception Functional Groups Synthetic rules Detect Functional Groups (joining points) Perception Knowledge Base o o Monomer Library Multiple low energy conformers + detected functional groups … Aromaticity Normalisation Hybridisation H-bonding properties
-O[HS=1] CHEMICAL-LABEL <Primary Amine> C-N[HS=2]; [CONNECTION=1] EXPLANATION Amide Synthetic Knowledge Base CHEMICAL-LABEL <Carboxylic Acid> C[SPCENTRE=2](=O)-O[HS=1] CHEMICAL-LABEL <Primary Amine> C-N[HS=2]; [CONNECTION=1] EXPLANATION Amide](https://present5.com/presentation/0b2aad24dec17adf8268479425b81a66/image-32.jpg) Synthetic Knowledge Base CHEMICAL-LABEL
Synthetic Knowledge Base CHEMICAL-LABEL
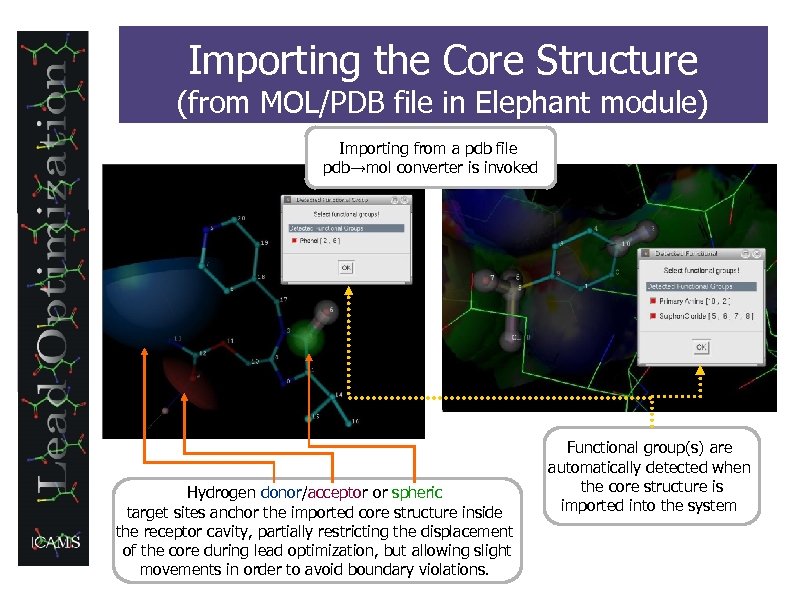 Importing the Core Structure (from MOL/PDB file in Elephant module) Importing from a pdb file pdb→mol converter is invoked Hydrogen donor/acceptor or spheric target sites anchor the imported core structure inside the receptor cavity, partially restricting the displacement of the core during lead optimization, but allowing slight movements in order to avoid boundary violations. Functional group(s) are automatically detected when the core structure is imported into the system
Importing the Core Structure (from MOL/PDB file in Elephant module) Importing from a pdb file pdb→mol converter is invoked Hydrogen donor/acceptor or spheric target sites anchor the imported core structure inside the receptor cavity, partially restricting the displacement of the core during lead optimization, but allowing slight movements in order to avoid boundary violations. Functional group(s) are automatically detected when the core structure is imported into the system
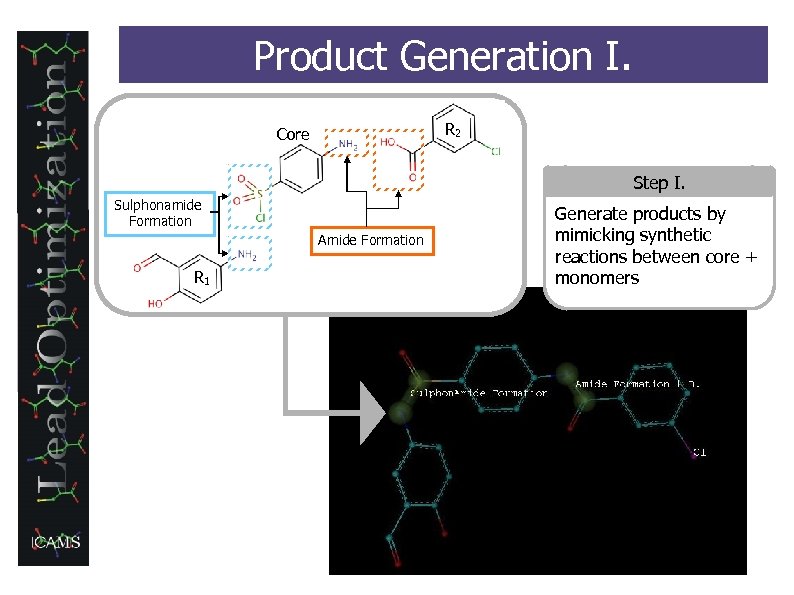 Product Generation I. R 2 Core Step I. Sulphonamide Formation Amide Formation R 1 Generate products by mimicking synthetic reactions between core + monomers
Product Generation I. R 2 Core Step I. Sulphonamide Formation Amide Formation R 1 Generate products by mimicking synthetic reactions between core + monomers
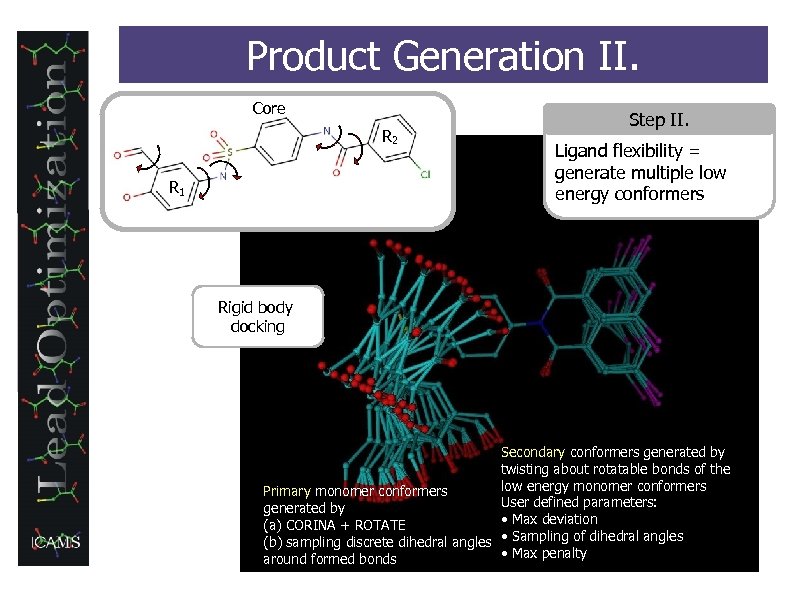 Product Generation II. Core R 2 R 1 Step II. Ligand flexibility = generate multiple low energy conformers Rigid body docking Secondary conformers generated by twisting about rotatable bonds of the low energy monomer conformers Primary monomer conformers User defined parameters: generated by • Max deviation (a) CORINA + ROTATE (b) sampling discrete dihedral angles • Sampling of dihedral angles • Max penalty around formed bonds
Product Generation II. Core R 2 R 1 Step II. Ligand flexibility = generate multiple low energy conformers Rigid body docking Secondary conformers generated by twisting about rotatable bonds of the low energy monomer conformers Primary monomer conformers User defined parameters: generated by • Max deviation (a) CORINA + ROTATE (b) sampling discrete dihedral angles • Sampling of dihedral angles • Max penalty around formed bonds
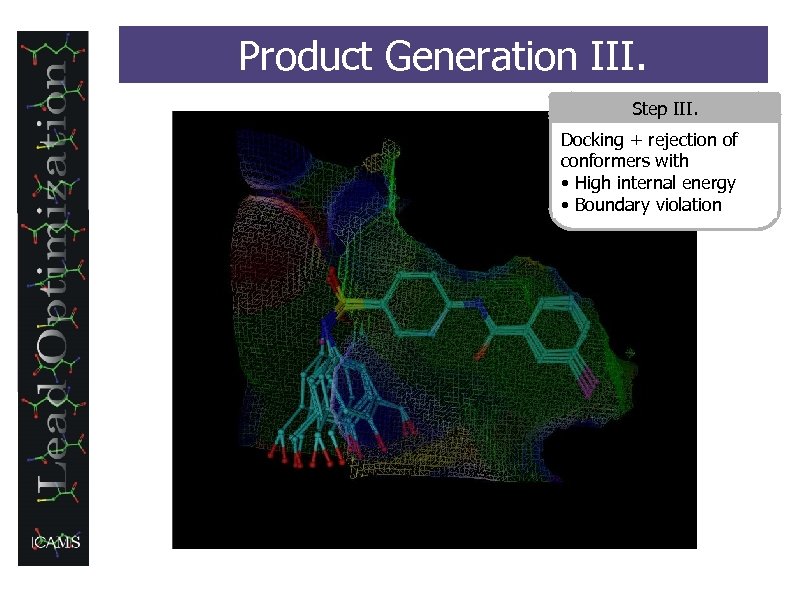 Product Generation III. Step III. Docking + rejection of conformers with • High internal energy • Boundary violation
Product Generation III. Step III. Docking + rejection of conformers with • High internal energy • Boundary violation
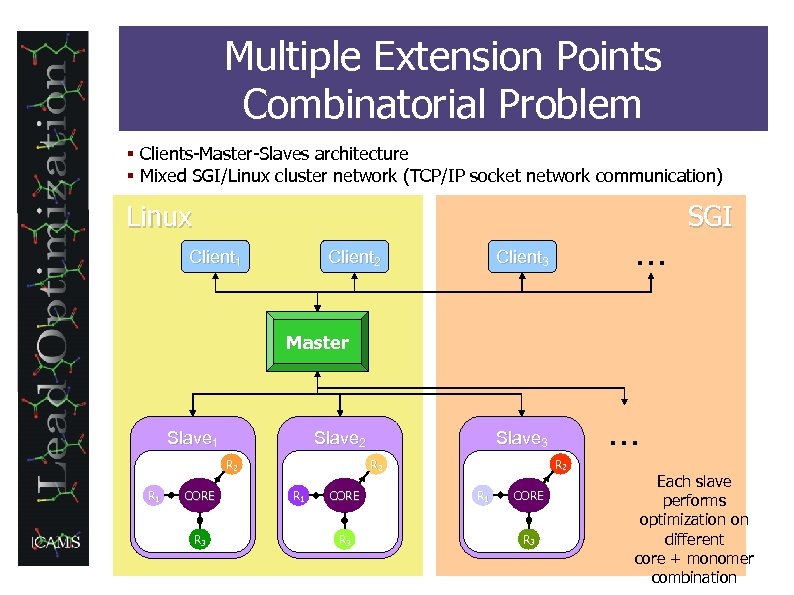 Multiple Extension Points Combinatorial Problem § Clients-Master-Slaves architecture § Mixed SGI/Linux cluster network (TCP/IP socket network communication) Linux Client 1 Client 2 … Client 3 SGI Master Slave 2 Slave 1 R 2 R 1 CORE R 3 … Slave 3 R 1 CORE R 3 Each slave performs optimization on different core + monomer combination
Multiple Extension Points Combinatorial Problem § Clients-Master-Slaves architecture § Mixed SGI/Linux cluster network (TCP/IP socket network communication) Linux Client 1 Client 2 … Client 3 SGI Master Slave 2 Slave 1 R 2 R 1 CORE R 3 … Slave 3 R 1 CORE R 3 Each slave performs optimization on different core + monomer combination
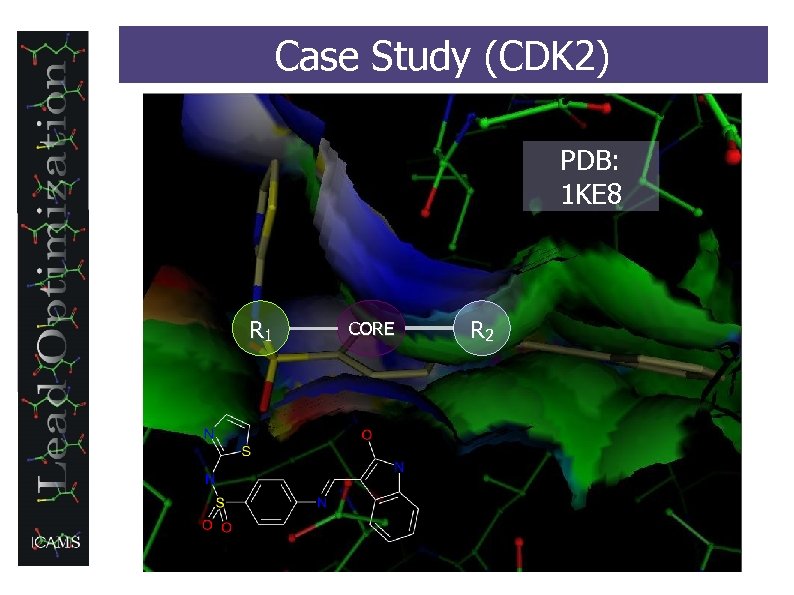 Case Study (CDK 2) PDB: 1 KE 8 R 1 CORE R 2
Case Study (CDK 2) PDB: 1 KE 8 R 1 CORE R 2
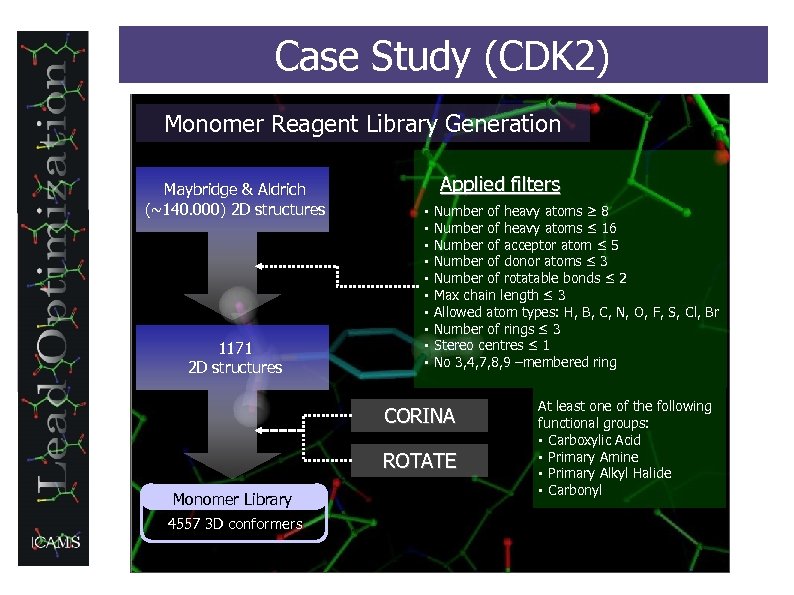 Case Study (CDK 2) Monomer Reagent Library Generation Maybridge & Aldrich (~140. 000) 2 D structures 1171 2 D structures Applied filters § § § § § Number of heavy atoms ≥ 8 Number of heavy atoms ≤ 16 Number of acceptor atom ≤ 5 Number of donor atoms ≤ 3 Number of rotatable bonds ≤ 2 Max chain length ≤ 3 Allowed atom types: H, B, C, N, O, F, S, Cl, Br Number of rings ≤ 3 Stereo centres ≤ 1 No 3, 4, 7, 8, 9 –membered ring CORINA ROTATE Monomer Library 4557 3 D conformers At least one of the following functional groups: § Carboxylic Acid § Primary Amine § Primary Alkyl Halide § Carbonyl
Case Study (CDK 2) Monomer Reagent Library Generation Maybridge & Aldrich (~140. 000) 2 D structures 1171 2 D structures Applied filters § § § § § Number of heavy atoms ≥ 8 Number of heavy atoms ≤ 16 Number of acceptor atom ≤ 5 Number of donor atoms ≤ 3 Number of rotatable bonds ≤ 2 Max chain length ≤ 3 Allowed atom types: H, B, C, N, O, F, S, Cl, Br Number of rings ≤ 3 Stereo centres ≤ 1 No 3, 4, 7, 8, 9 –membered ring CORINA ROTATE Monomer Library 4557 3 D conformers At least one of the following functional groups: § Carboxylic Acid § Primary Amine § Primary Alkyl Halide § Carbonyl
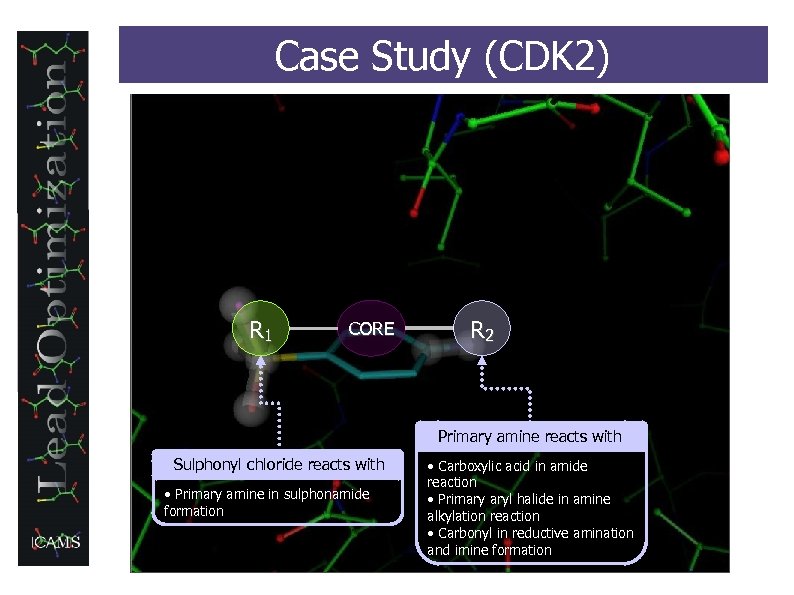 Case Study (CDK 2) R 1 CORE R 2 Primary amine reacts with Sulphonyl chloride reacts with • Primary amine in sulphonamide formation • Carboxylic acid in amide reaction • Primary aryl halide in amine alkylation reaction • Carbonyl in reductive amination and imine formation
Case Study (CDK 2) R 1 CORE R 2 Primary amine reacts with Sulphonyl chloride reacts with • Primary amine in sulphonamide formation • Carboxylic acid in amide reaction • Primary aryl halide in amine alkylation reaction • Carbonyl in reductive amination and imine formation
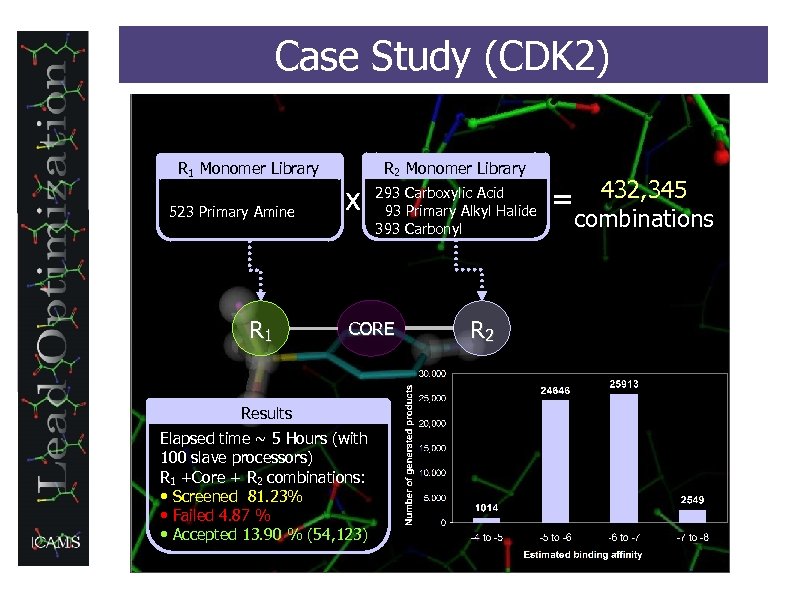 Case Study (CDK 2) R 1 Monomer Library 523 Primary Amine R 1 x R 2 Monomer Library 293 Carboxylic Acid 93 Primary Alkyl Halide 393 Carbonyl CORE Results Elapsed time ~ 5 Hours (with 100 slave processors) R 1 +Core + R 2 combinations: • Screened 81. 23% • Failed 4. 87 % • Accepted 13. 90 % (54, 123) R 2 432, 345 combinations =
Case Study (CDK 2) R 1 Monomer Library 523 Primary Amine R 1 x R 2 Monomer Library 293 Carboxylic Acid 93 Primary Alkyl Halide 393 Carbonyl CORE Results Elapsed time ~ 5 Hours (with 100 slave processors) R 1 +Core + R 2 combinations: • Screened 81. 23% • Failed 4. 87 % • Accepted 13. 90 % (54, 123) R 2 432, 345 combinations =
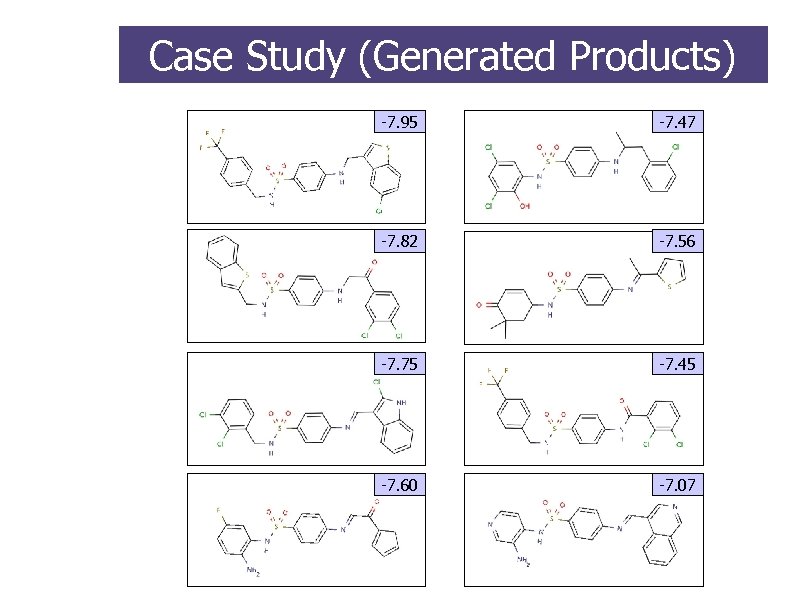 Case Study (Generated Products) -7. 95 -7. 47 -7. 82 -7. 56 -7. 75 -7. 45 -7. 60 -7. 07
Case Study (Generated Products) -7. 95 -7. 47 -7. 82 -7. 56 -7. 75 -7. 45 -7. 60 -7. 07
 Monomer Replacement • Many lead compounds are composed of readily available starting materials (monomers) linked by reliable high yielding reactions • Retrosynthetic analysis can be used to identify the monomers • Structurally related analogues could be generated by exhaustive monomer replacement • Considerable efficiency gains if monomer library is arranged in a hierarchy based on substructural relationships
Monomer Replacement • Many lead compounds are composed of readily available starting materials (monomers) linked by reliable high yielding reactions • Retrosynthetic analysis can be used to identify the monomers • Structurally related analogues could be generated by exhaustive monomer replacement • Considerable efficiency gains if monomer library is arranged in a hierarchy based on substructural relationships
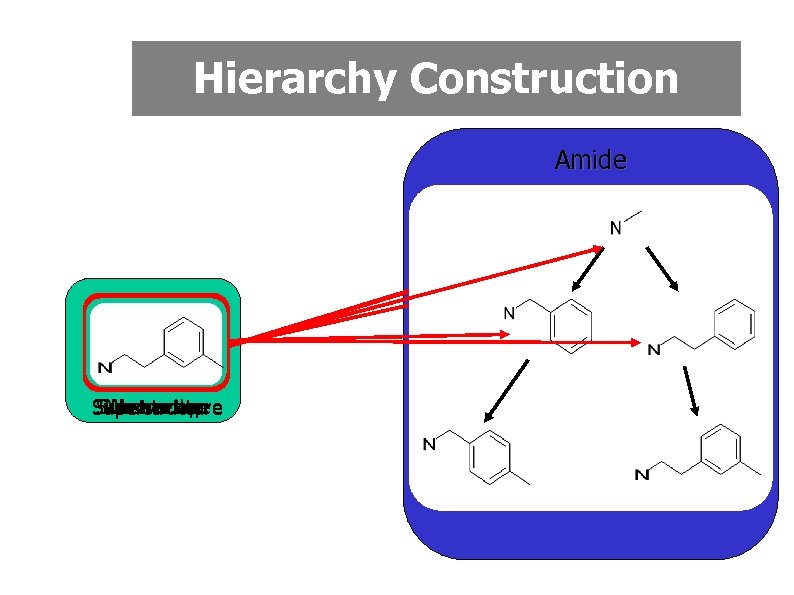 Hierarchy Construction Amide No overlap Substructure Superstructure
Hierarchy Construction Amide No overlap Substructure Superstructure
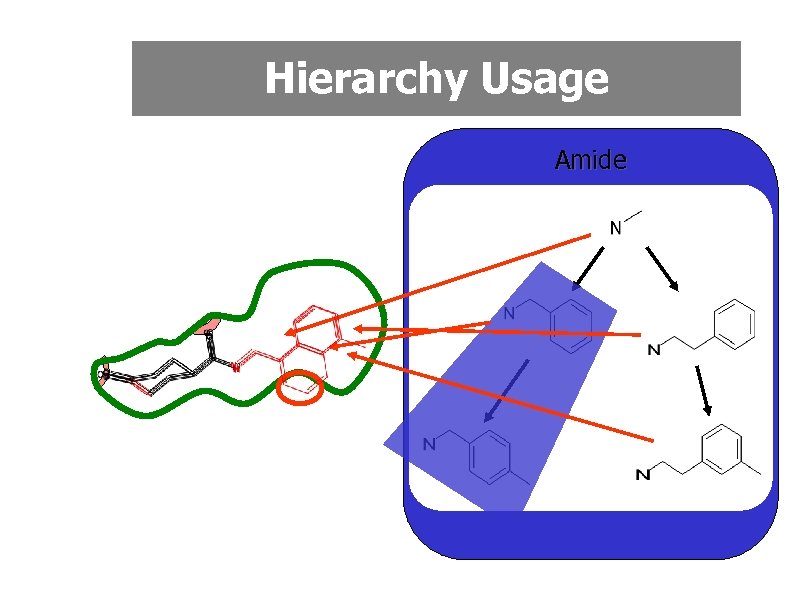 Hierarchy Usage Amide
Hierarchy Usage Amide
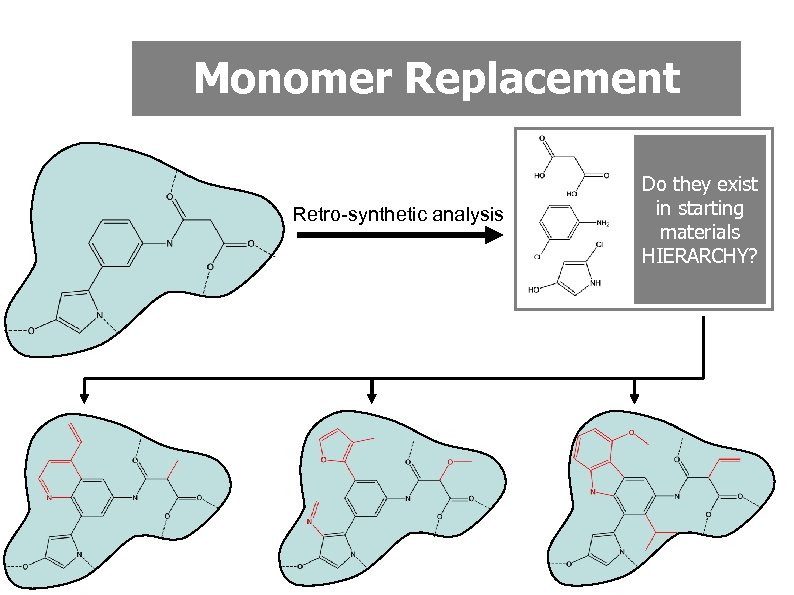 Monomer Replacement Retro-synthetic analysis Do they exist in starting materials HIERARCHY?
Monomer Replacement Retro-synthetic analysis Do they exist in starting materials HIERARCHY?
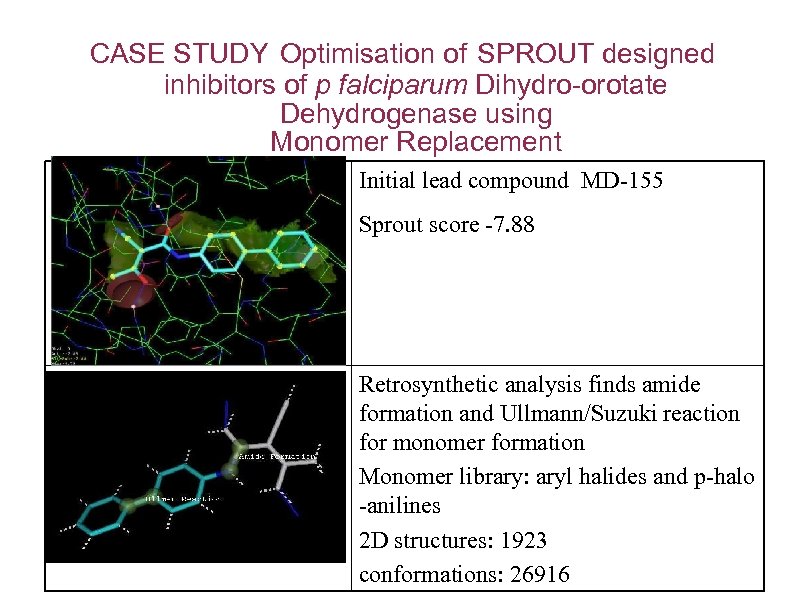 CASE STUDY Optimisation of SPROUT designed inhibitors of p falciparum Dihydro-orotate Dehydrogenase using Monomer Replacement Initial lead compound MD-155 Sprout score -7. 88 Retrosynthetic analysis finds amide formation and Ullmann/Suzuki reaction for monomer formation Monomer library: aryl halides and p-halo -anilines 2 D structures: 1923 conformations: 26916
CASE STUDY Optimisation of SPROUT designed inhibitors of p falciparum Dihydro-orotate Dehydrogenase using Monomer Replacement Initial lead compound MD-155 Sprout score -7. 88 Retrosynthetic analysis finds amide formation and Ullmann/Suzuki reaction for monomer formation Monomer library: aryl halides and p-halo -anilines 2 D structures: 1923 conformations: 26916
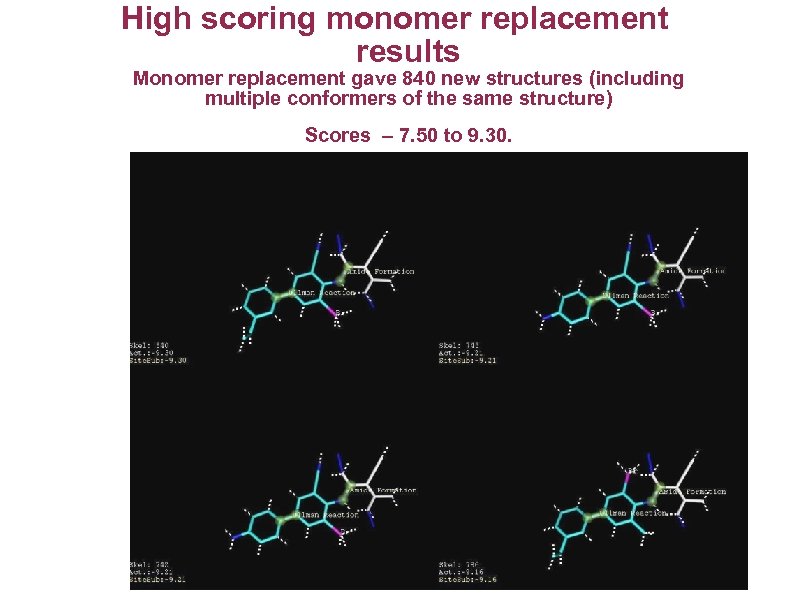 High scoring monomer replacement results Monomer replacement gave 840 new structures (including multiple conformers of the same structure) Scores – 7. 50 to 9. 30.
High scoring monomer replacement results Monomer replacement gave 840 new structures (including multiple conformers of the same structure) Scores – 7. 50 to 9. 30.
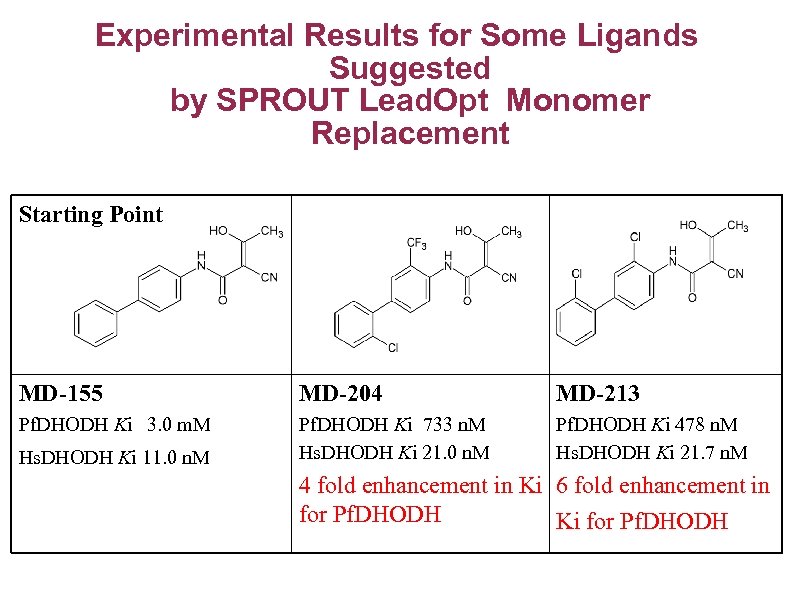 Experimental Results for Some Ligands Suggested by SPROUT Lead. Opt Monomer Replacement Starting Point MD-155 MD-204 MD-213 Pf. DHODH Ki 3. 0 m. M Pf. DHODH Ki 733 n. M Hs. DHODH Ki 21. 0 n. M Pf. DHODH Ki 478 n. M Hs. DHODH Ki 21. 7 n. M Hs. DHODH Ki 11. 0 n. M 4 fold enhancement in Ki 6 fold enhancement in for Pf. DHODH Ki for Pf. DHODH
Experimental Results for Some Ligands Suggested by SPROUT Lead. Opt Monomer Replacement Starting Point MD-155 MD-204 MD-213 Pf. DHODH Ki 3. 0 m. M Pf. DHODH Ki 733 n. M Hs. DHODH Ki 21. 0 n. M Pf. DHODH Ki 478 n. M Hs. DHODH Ki 21. 7 n. M Hs. DHODH Ki 11. 0 n. M 4 fold enhancement in Ki 6 fold enhancement in for Pf. DHODH Ki for Pf. DHODH
 Conclusions § Scoring functions for assessment of binding affinity of the hypothetical compounds produced by de novo design are far from perfect § Hence only readily synthesisable putative ligands will undergo experimental evaluation by medicinal chemists § Assessment of synthetic feasibility is a tractable problem
Conclusions § Scoring functions for assessment of binding affinity of the hypothetical compounds produced by de novo design are far from perfect § Hence only readily synthesisable putative ligands will undergo experimental evaluation by medicinal chemists § Assessment of synthetic feasibility is a tractable problem
 Acknowledgements § Matt Davies, Phil Bone and Timo Heikkala for experimental work § Molecular Networks Gmb. H for providing CORINA & ROTATE § MDL for providing MDDR, one of the databases used in the complexity analysis project § for sponsoring the lead optimization project
Acknowledgements § Matt Davies, Phil Bone and Timo Heikkala for experimental work § Molecular Networks Gmb. H for providing CORINA & ROTATE § MDL for providing MDDR, one of the databases used in the complexity analysis project § for sponsoring the lead optimization project


