1f0ca3f4ef4cf6d554cdd39f6cc21f31.ppt
- Количество слайдов: 63
Databases for Systems Biology Herbert M Sauro Keck Graduate Institute Claremont, CA, 91711

Systems Biology

Systems Biology • Computational Systems Biology Group (Peter Spirtes) in Pittsburgh, Pennsylvania • Biochemical Networks Modeling Group (Pedro Mendes) at the Virginia Bioinformatics Institute Computational Systems Biology Group (Reinhard Laubenbacher) at the Virginia Bioinformatics Institute • Evolution of Molecular Networks group (Andreas Wagner) at the University of New Mexico • Systems biology group (Trey Idekeker) at the Whitehead Institute for Biomedical Research, Cambridge (USA) • Computational Cell Biology (Dennis Bray) at the University of Cambridge (UK) STRC Biocomputation Group (Hamid Bolouri) at the University of Hertfordshire • Computational Molecular Biology (Ron Shamir) at the University of Tel Aviv • Complex Systems Division (Carsten Peterson) at the University of Lund • Design Principles of Protein Networks (Uri Alon) at the Weizmann Institute • Design Principles of Protein Networks (Naama Barkai) at the Weizmann Institute • Probabilistic Graphical Models (Daphne Koller) at the University of Stanford • Molecular Biology and Probabilistic Models (Nir Friedman) at the Hewbrew University of Jerusalem • Systems Optimization Group (Eckart Zitzler) at the ETH Zürich • Protein Interaction Group (Benno Schwikowski) at the Systems Biology Institute, Seattle • Systems Biology Center at TU Delft • Integrative Systems Biology at TU Denmark • U Ghent • Institute for Advanced Study, Center for Systems Biology • Ron Weiss group, Princeton University • BII Systems Biology Group (Singapore) • UC San Francisco Bio. Systems Groups World-Wide • Kitano Systems Biology Group • Davidson Lab at Caltech • Bioinformatics & Systems Biology Group at the Burnham Institute (La Jolla) • Virtual Cell Project, U Connecticut • UC Santa Barbara IGERT Program on Systems Biology • UC San Diego Bioinformatics & Systems Biology Groups • UC San Diego Systems Biodynamics Group • Integrated Systems Biology Group at Rensselaer Polytechnic Institute

Systems Biology Institutes and Larger Initiatives • Bio. SPI Project at Weizmann • Bio. SPICE • Bio. Maps Institute at Rutgers: • Institute for Systems Biology, Seattle • Bauer Center for Genomics Research (CGR) at Harvard University • Systems Biology Department at Harvard Medical School • Computational and Systems Biology Initiative at MIT • Bio-X at Stanford University • Center for Studies in Physics and Biology at The Rockefeller University • GENSCEND Initiative of the Wellcome Trust • "Genomes to Life program" (a funding initiative of the DOE) • "Cell Systems Initiative" (an initiative of the University of Washington) • "Systems of Life - System Biology" (a funding initiative of the German Ministry of Education and Research, BMBF) • SFB 618 (funded by the German Research Council DFG) • STAGSIM - Systems Biology (An Expression of Interest (Eo. I) submitted to the EU Framework Program VI) • Systems Biology in Sweden • Institute for Computational Biomedicine at the Weill Medical College of Cornell University. • Pathways/Systems Biology Working Group at I 3 C.
Systems Biology Has its Backers and Attackers Revolution or buzzword du jour, pundits ponder a pervasive term | By Mignon Fogarty Though coined 40 years ago, 1 a lot of people still ask, "What's that? " when the term systems biology comes up. "It is used in so many different contexts, nobody is really clear what you mean by it, " says John Yates III, a professor at the Scripps Research Institute in La Jolla, Calif. He's not the only one stumped by the term's meaning. David Placek, president of Sausalito, Calif. based Lexicon Branding, a company that cooks up names for pharmaceutical products such as Velcade and Meridia, says he's not so hot on the moniker. "Systems biology is just so general that it could apply to many things. When you're naming a category, the underlying principle is that if you make a statement like, 'I'm doing systems biology, ' do people know what you're talking about? '“…… Volume 17 | Issue 19 | 27 Oct. 6, 2003, The Scientist
Systems Biology? High-throughput Data?
Systems Biology? Databases? Path. DB
What is Systems Biology? Understanding the principles of how physiological/phenotypic characteristics emerge from the properties of the components. Predicting how these characteristics will change in response to alterations in the environment or system components.
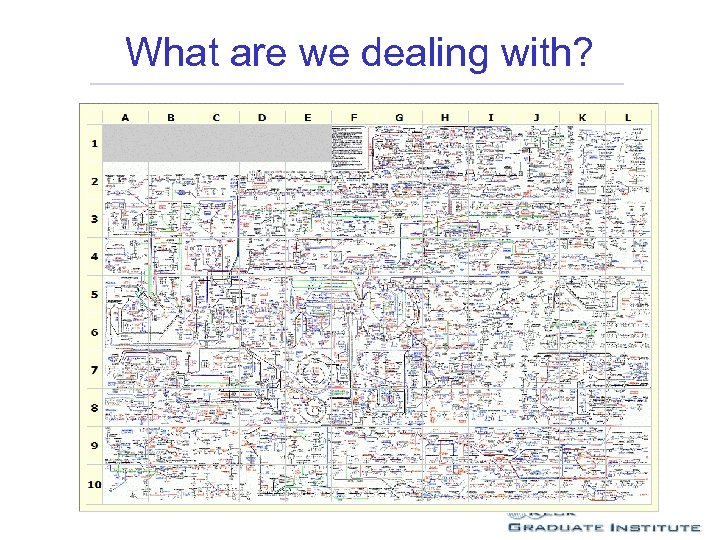
What are we dealing with?
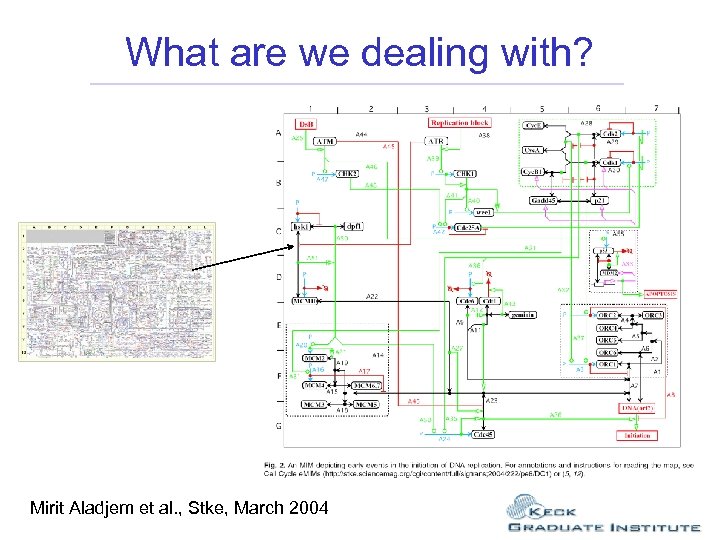
What are we dealing with? Mirit Aladjem et al. , Stke, March 2004
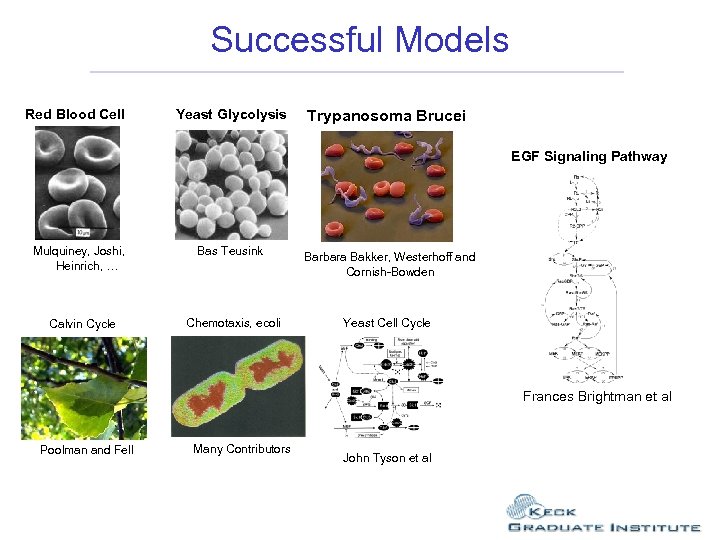
Successful Models Red Blood Cell Yeast Glycolysis Trypanosoma Brucei EGF Signaling Pathway Mulquiney, Joshi, Heinrich, … Bas Teusink Calvin Cycle Chemotaxis, ecoli Barbara Bakker, Westerhoff and Cornish-Bowden Yeast Cell Cycle Frances Brightman et al Poolman and Fell Many Contributors John Tyson et al
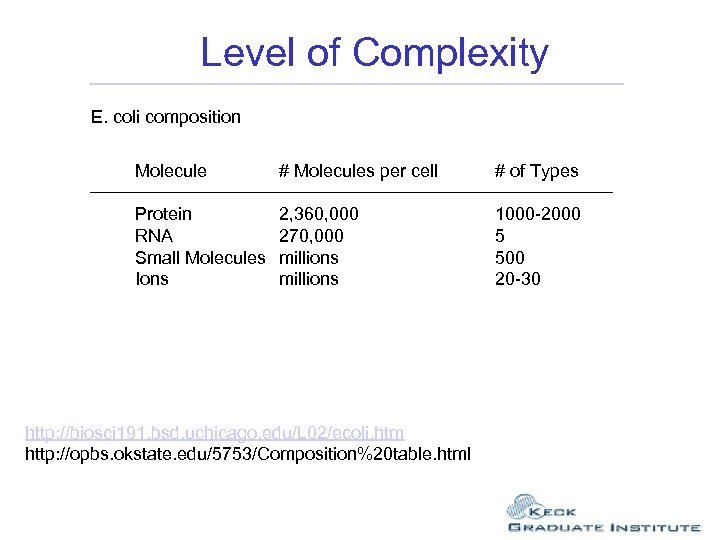
Level of Complexity E. coli composition Molecule # Molecules per cell # of Types Protein RNA Small Molecules Ions 2, 360, 000 270, 000 millions 1000 -2000 5 500 20 -30 http: //biosci 191. bsd. uchicago. edu/L 02/ecoli. htm http: //opbs. okstate. edu/5753/Composition%20 table. html
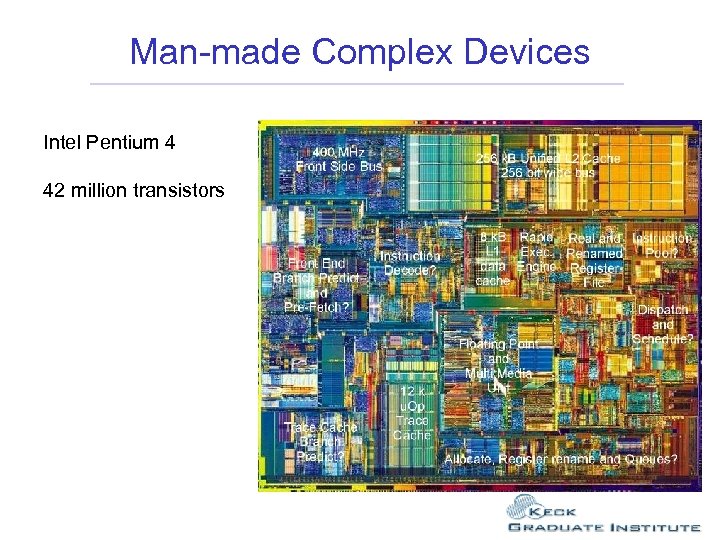
Man-made Complex Devices Intel Pentium 4 42 million transistors
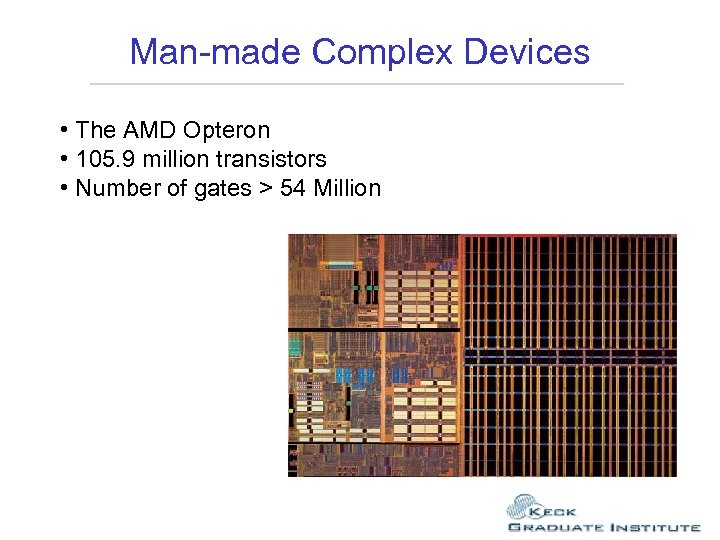
Man-made Complex Devices • The AMD Opteron • 105. 9 million transistors • Number of gates > 54 Million
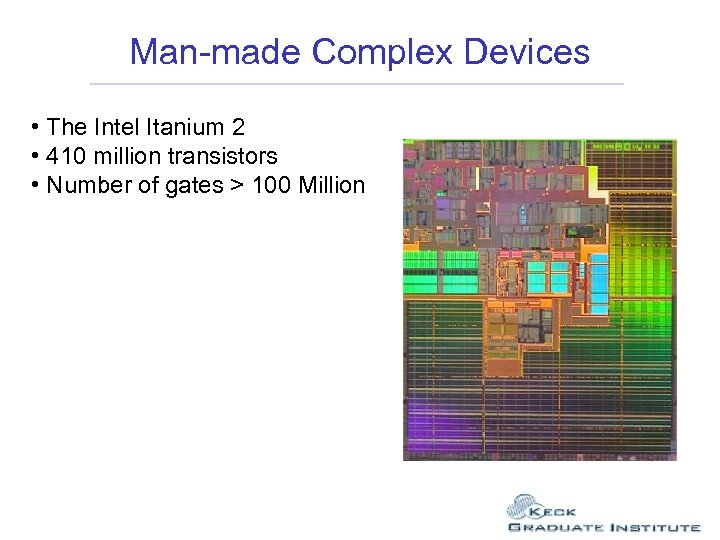
Man-made Complex Devices • The Intel Itanium 2 • 410 million transistors • Number of gates > 100 Million
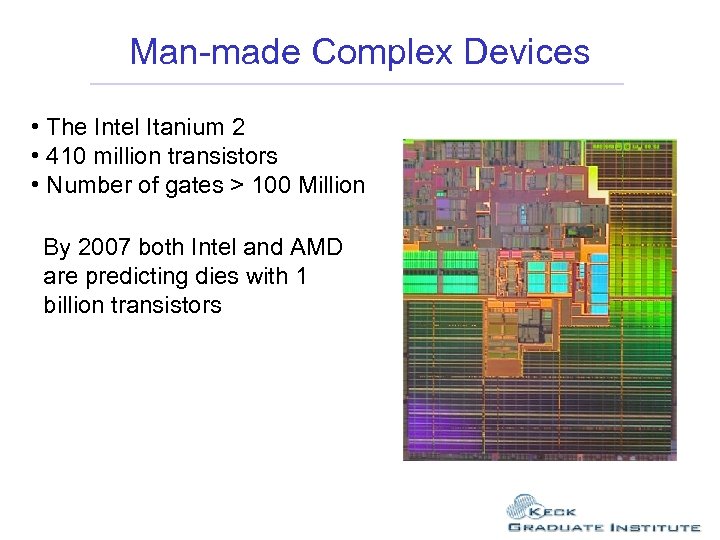
Man-made Complex Devices • The Intel Itanium 2 • 410 million transistors • Number of gates > 100 Million By 2007 both Intel and AMD are predicting dies with 1 billion transistors
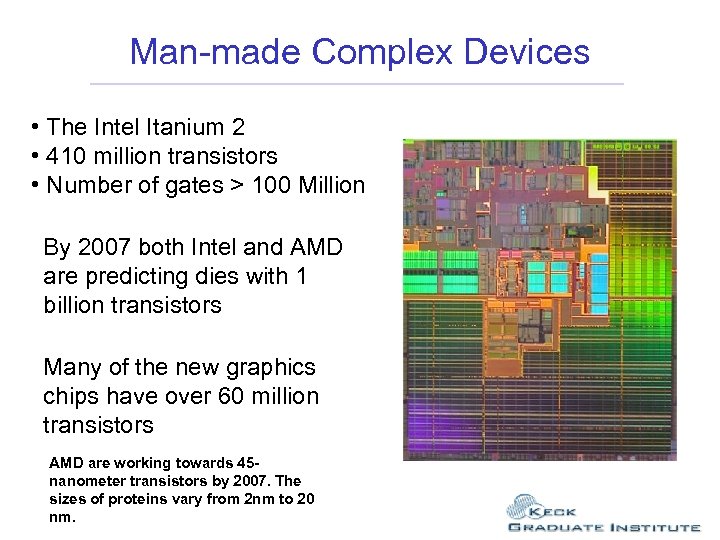
Man-made Complex Devices • The Intel Itanium 2 • 410 million transistors • Number of gates > 100 Million By 2007 both Intel and AMD are predicting dies with 1 billion transistors Many of the new graphics chips have over 60 million transistors AMD are working towards 45 nanometer transistors by 2007. The sizes of proteins vary from 2 nm to 20 nm.
Man-made Complex Devices Probably by 2010, man-made devices will have comparable complexity to bacterial cells if not greater.
Cellular Models Building computational models of cells seems more and more like a viable project. Such a project would bring a much clearer understanding of how cellular systems are controlled and ultimately it should bring unprecedented predictive power.

Are Biologists Ready? Xo S 1 S 2 S 3 v S 4 S 5 S 6 X 1 Xo and X 1 fixed, all reactions reversible, assume stable steady state.
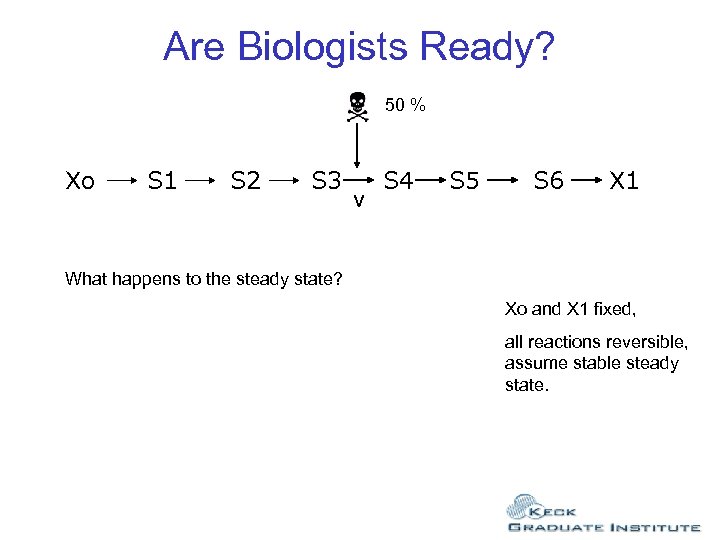
Are Biologists Ready? 50 % Xo S 1 S 2 S 3 v S 4 S 5 S 6 X 1 What happens to the steady state? Xo and X 1 fixed, all reactions reversible, assume stable steady state.

Are Biologists Ready? 50 % Xo S 1 S 2 S 3 v S 4 S 5 Students reply: 1. Nothing happens. 2. Nothing happens unless it is the rate-limiting step. 3. The rate v goes down, but that’s all. 4. S 3 goes up. 5. S 4 goes down. 6. Species downstream of v go up. 7. Steady State flow changes but species levels don’t. 8. Xo and X 1 change S 6 X 1
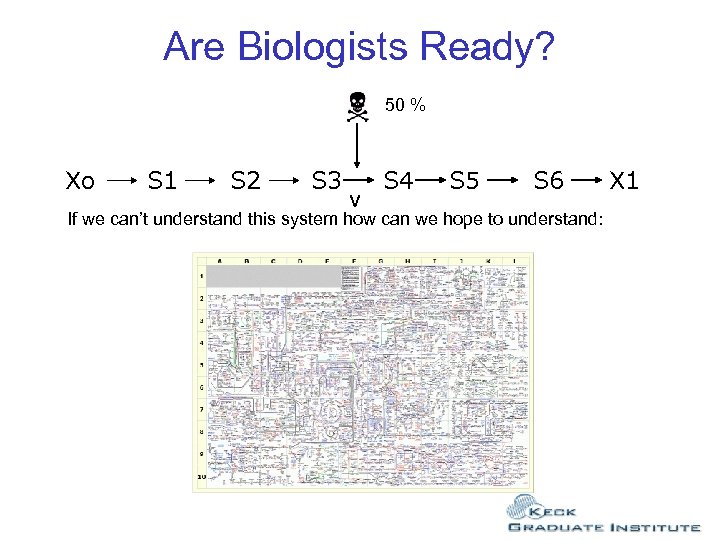
Are Biologists Ready? 50 % Xo S 1 S 2 S 3 v S 4 S 5 S 6 If we can’t understand this system how can we hope to understand: X 1
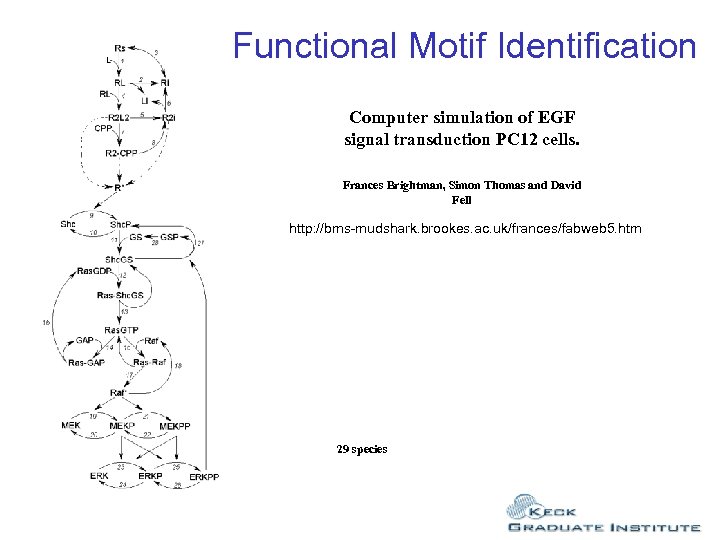
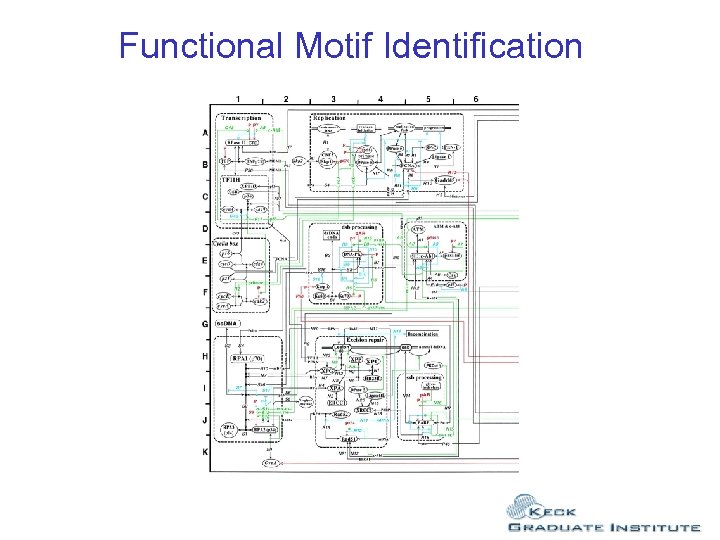
Functional Motif Identification Computer simulation of EGF signal transduction PC 12 cells. Frances Brightman, Simon Thomas and David Fell http: //bms-mudshark. brookes. ac. uk/frances/fabweb 5. htm 29 species
Functional Motif Identification Computer simulation of EGF signal transduction PC 12 cells. Frances Brightman, Simon Thomas and David Fell http: //bms-mudshark. brookes. ac. uk/frances/fabweb 5. htm
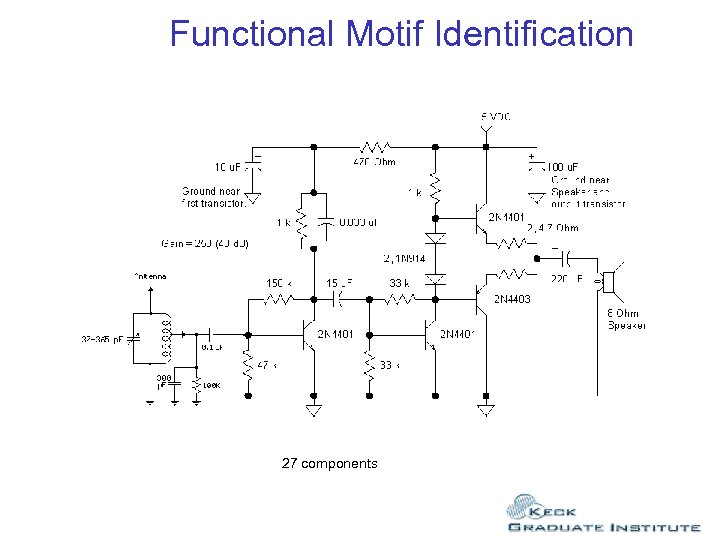
Functional Motif Identification 27 components
Functional Motif Identification
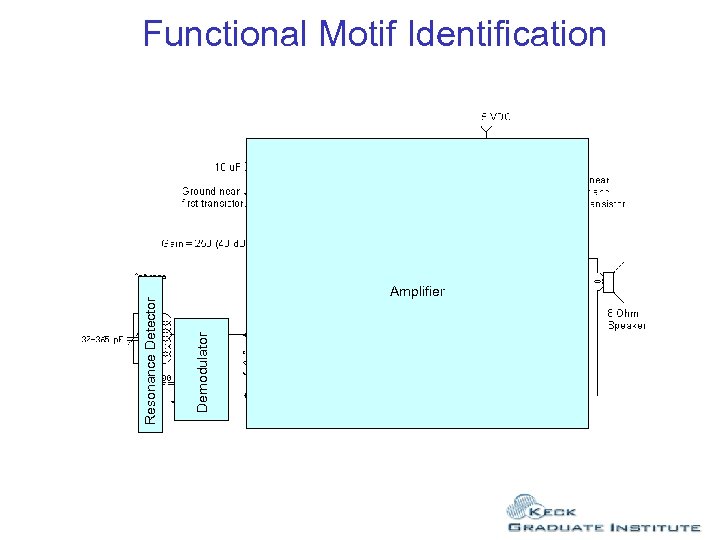
Amplifier Demodulator Resonance Detector Functional Motif Identification
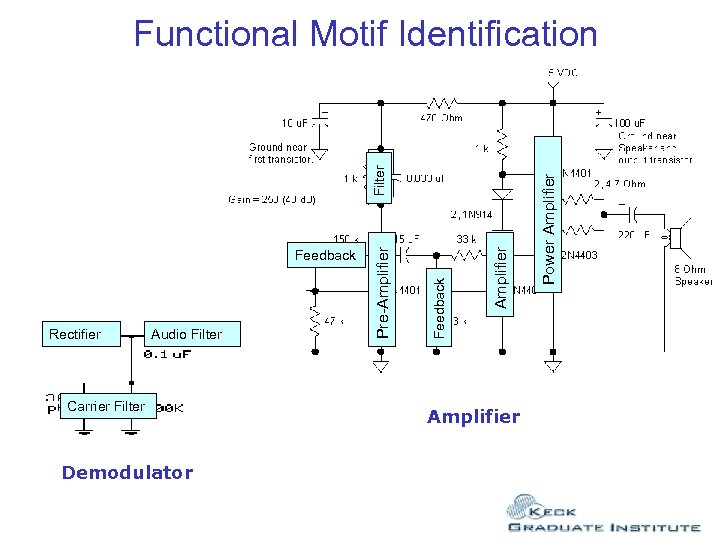
Rectifier Audio Filter Carrier Filter Demodulator Amplifier Power Amplifier Feedback Pre-Amplifier Filter Functional Motif Identification
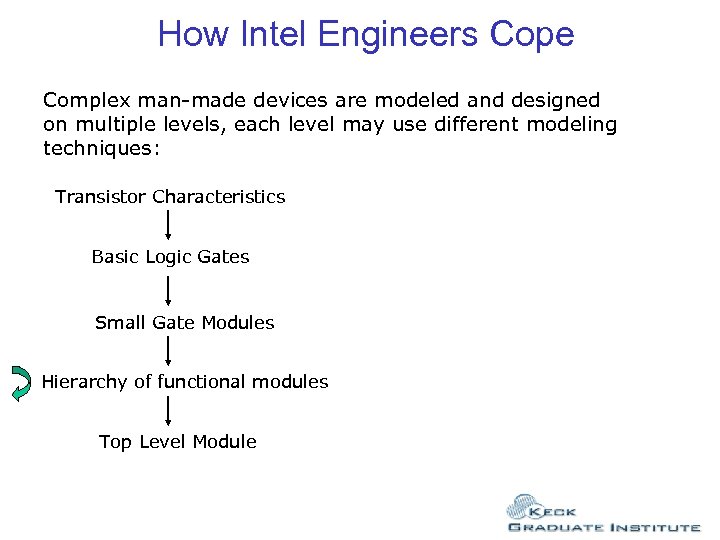
How Intel Engineers Cope Complex man-made devices are modeled and designed on multiple levels, each level may use different modeling techniques: Transistor Characteristics Basic Logic Gates Small Gate Modules Hierarchy of functional modules Top Level Module
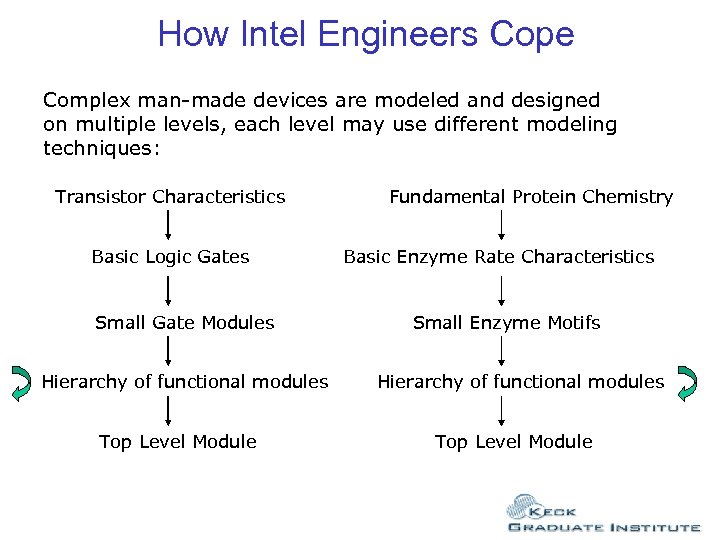
How Intel Engineers Cope Complex man-made devices are modeled and designed on multiple levels, each level may use different modeling techniques: Transistor Characteristics Basic Logic Gates Small Gate Modules Hierarchy of functional modules Top Level Module Fundamental Protein Chemistry Basic Enzyme Rate Characteristics Small Enzyme Motifs Hierarchy of functional modules Top Level Module
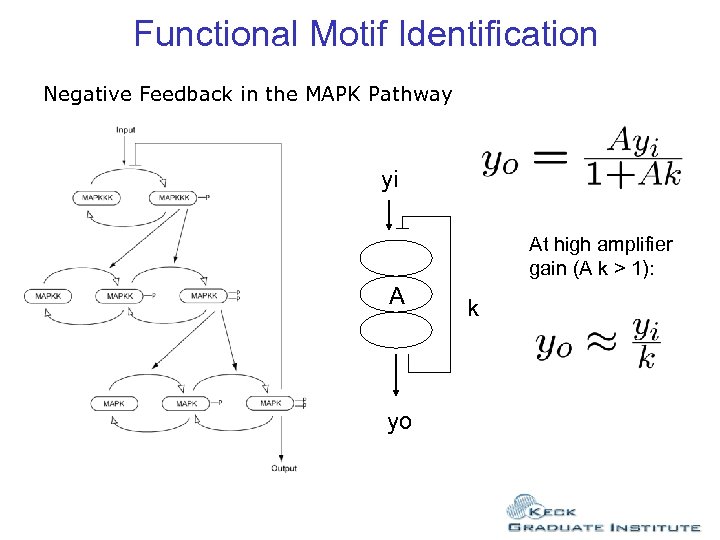
Functional Motif Identification Negative Feedback in the MAPK Pathway yi At high amplifier gain (A k > 1): A yo k
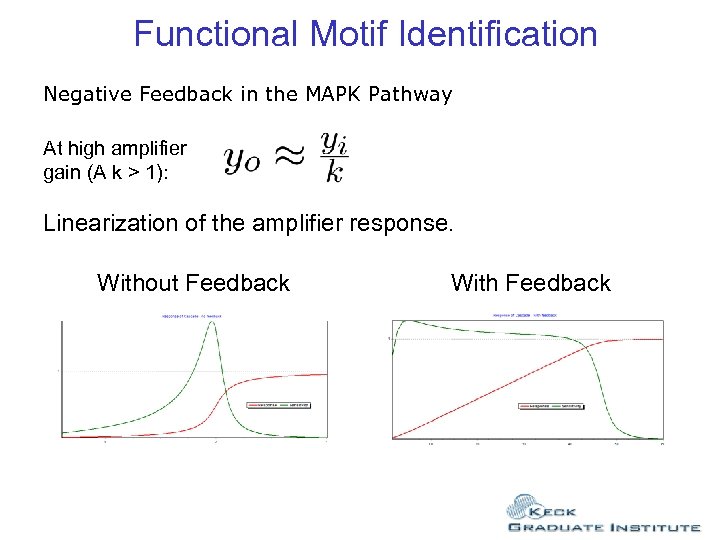
Functional Motif Identification Negative Feedback in the MAPK Pathway At high amplifier gain (A k > 1): Linearization of the amplifier response. Without Feedback With Feedback
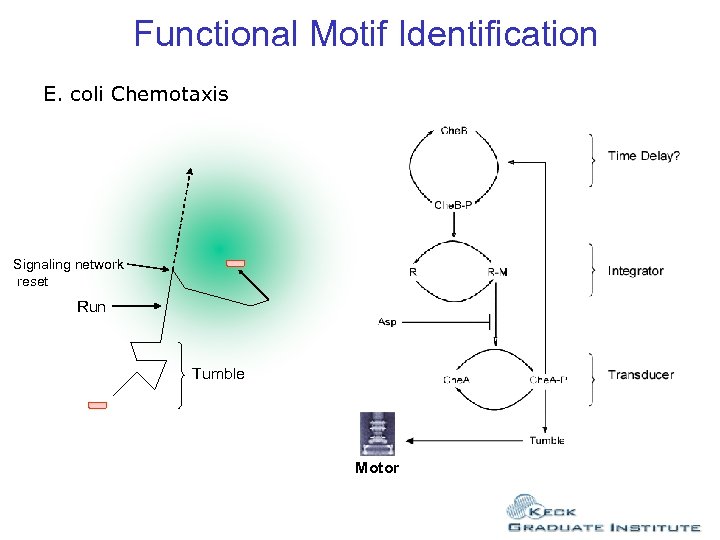
Functional Motif Identification E. coli Chemotaxis Signaling network reset Run Tumble Motor

Software Tools and Resources: Software Infrastructure Interchange Formats Analysis Algorithms Model Editors Visualization Model Databases Theoretical Foundation
Databases for Systems Biology • Kinetic Data • Network Information
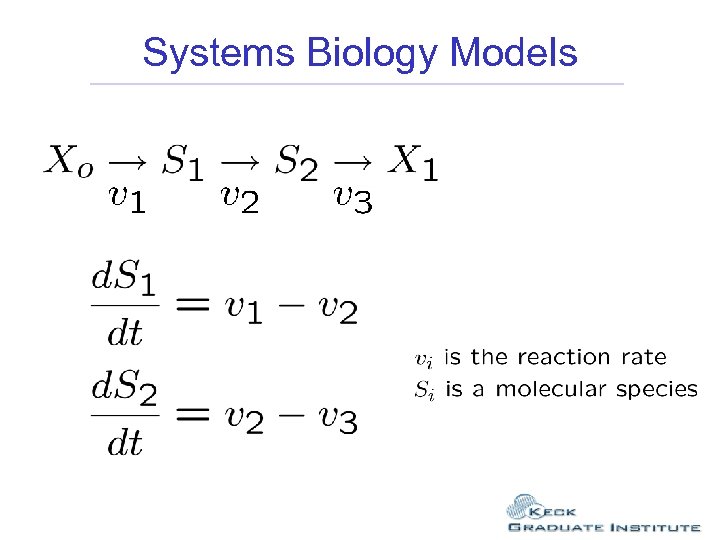
Systems Biology Models
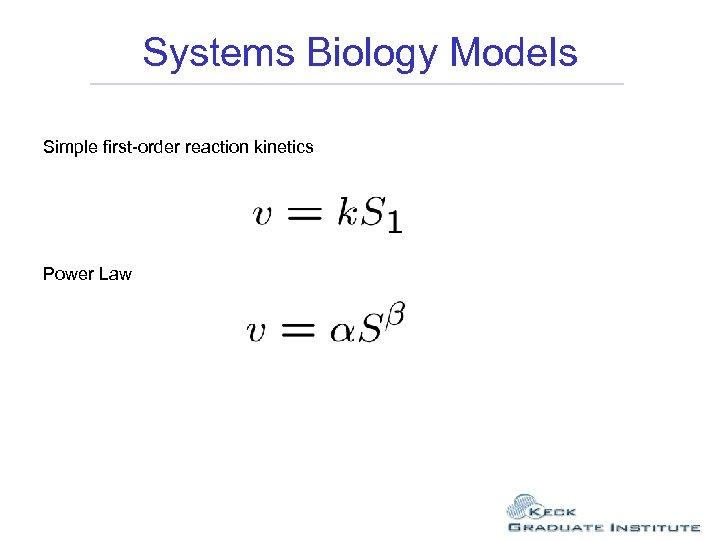
Systems Biology Models Simple first-order reaction kinetics Power Law
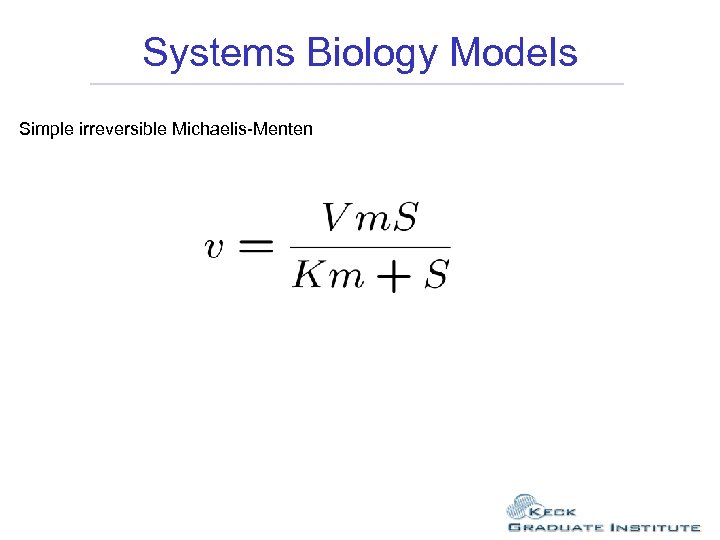
Systems Biology Models Simple irreversible Michaelis-Menten
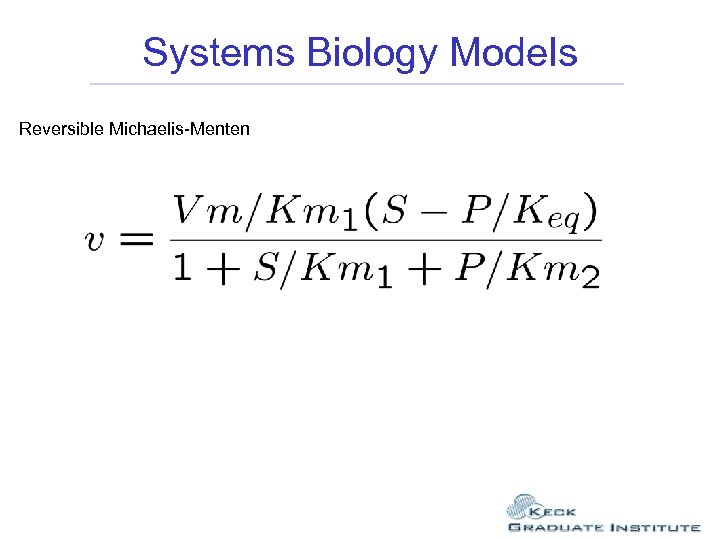
Systems Biology Models Reversible Michaelis-Menten
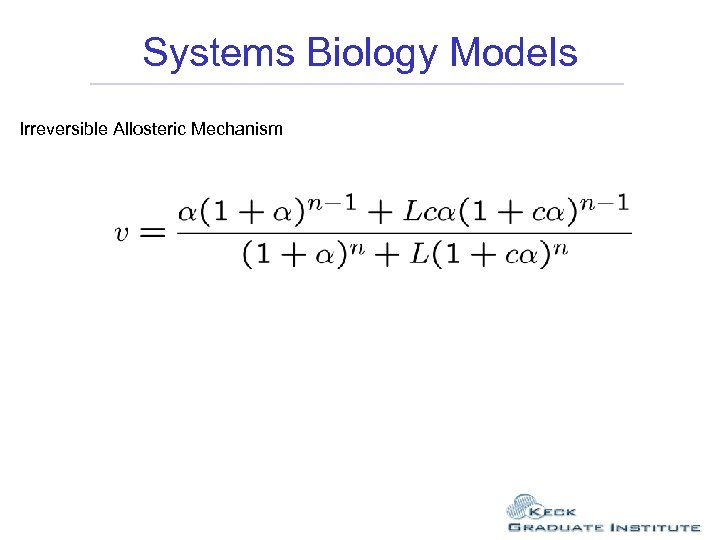
Systems Biology Models Irreversible Allosteric Mechanism
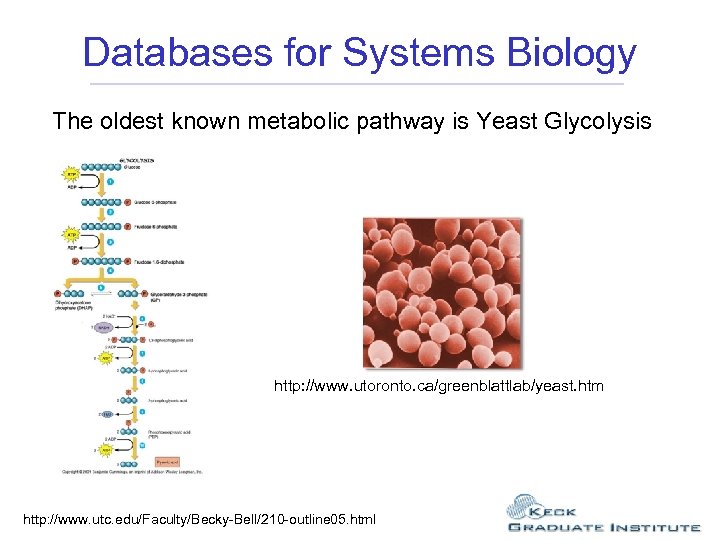
Databases for Systems Biology The oldest known metabolic pathway is Yeast Glycolysis http: //www. utoronto. ca/greenblattlab/yeast. htm http: //www. utc. edu/Faculty/Becky-Bell/210 -outline 05. html

Databases for Systems Biology Hexokinase 2. 7. 1. 1
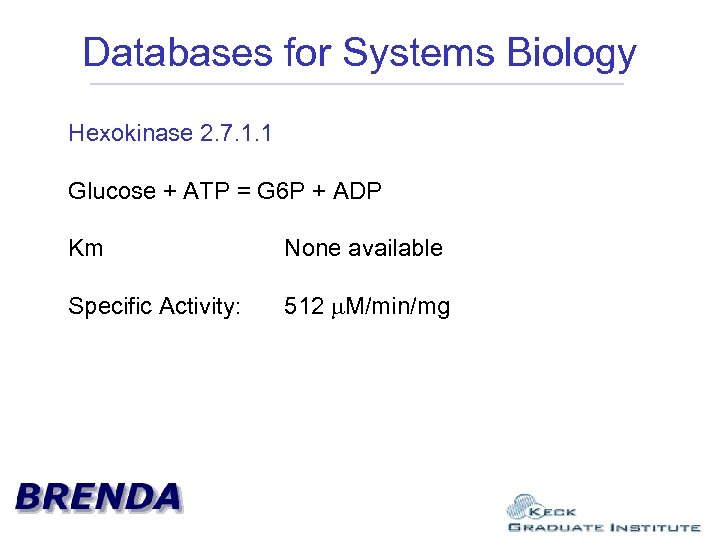
Databases for Systems Biology Hexokinase 2. 7. 1. 1 Glucose + ATP = G 6 P + ADP Km None available Specific Activity: 512 M/min/mg

Databases for Systems Biology Phosphofructokinase 2. 7. 1. 11

Databases for Systems Biology Phosphofructokinase 2. 7. 1. 11 ATP + F 6 P = ADP + FBP Km None available Specific Activity: 180 M/min/mg 148 M/min/mg 114 M/min/mg

Databases for Systems Biology Pyruvate Kinase 2. 7. 1. 40
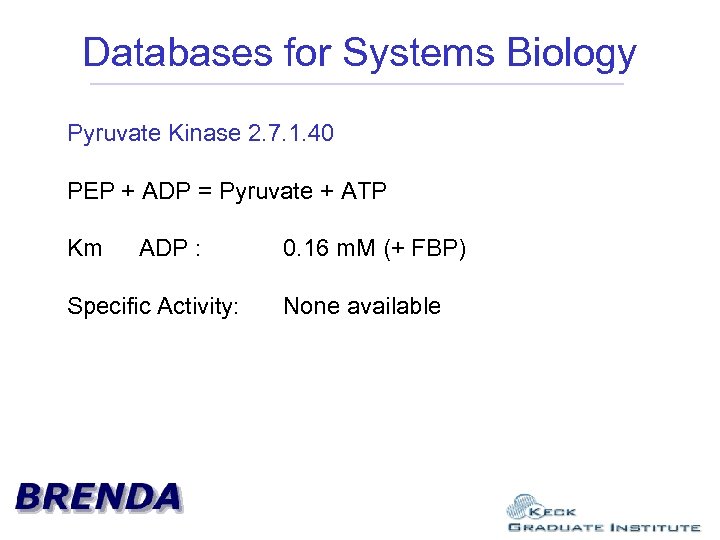
Databases for Systems Biology Pyruvate Kinase 2. 7. 1. 40 PEP + ADP = Pyruvate + ATP Km ADP : Specific Activity: 0. 16 m. M (+ FBP) None available
Databases for Systems Biology 1. Kinetic equations 2. Values for kinetic constants plus standard errors 3. Conditions under which enzyme was characterized
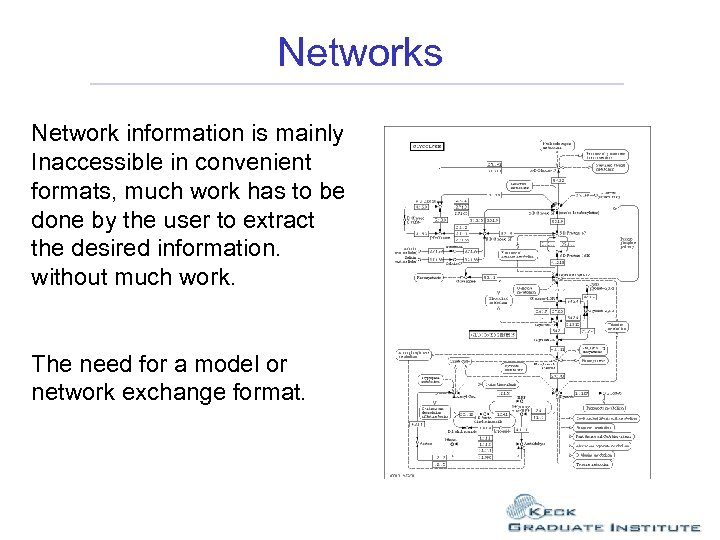
Networks Network information is mainly Inaccessible in convenient formats, much work has to be done by the user to extract the desired information. without much work. The need for a model or network exchange format.
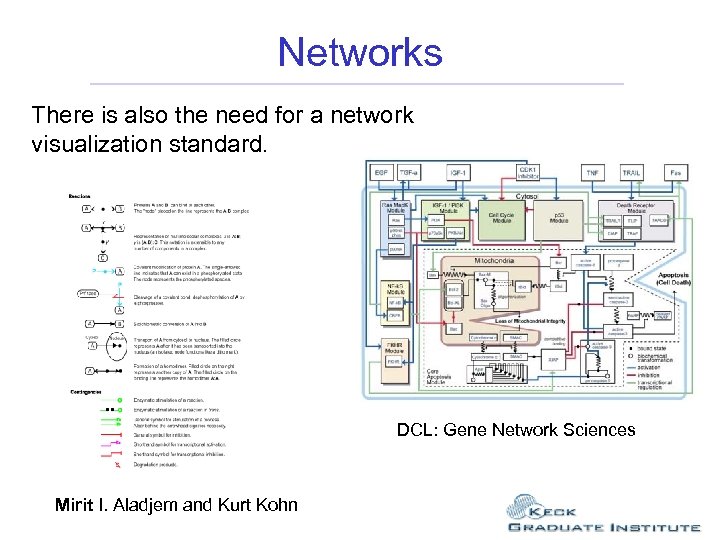
Networks There is also the need for a network visualization standard. DCL: Gene Network Sciences Mirit I. Aladjem and Kurt Kohn
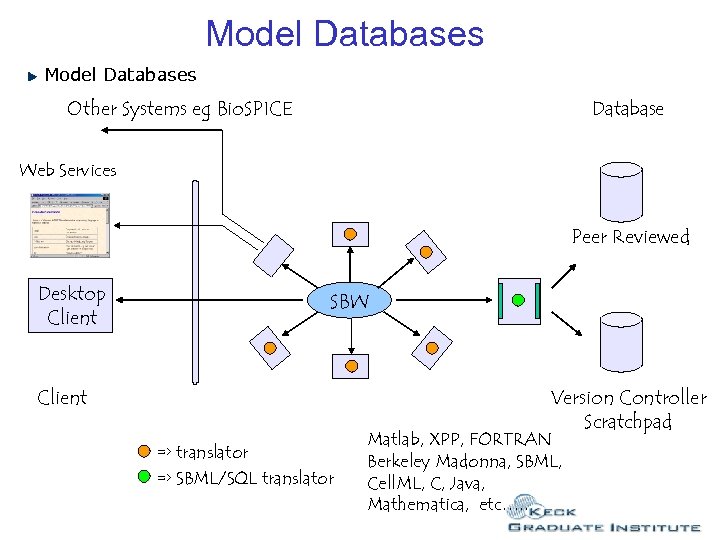
Model Databases Other Systems eg Bio. SPICE Database Web Services Peer Reviewed Desktop Client SBW Client Version Controller Scratchpad => translator => SBML/SQL translator Matlab, XPP, FORTRAN Berkeley Madonna, SBML, Cell. ML, C, Java, Mathematica, etc…. .
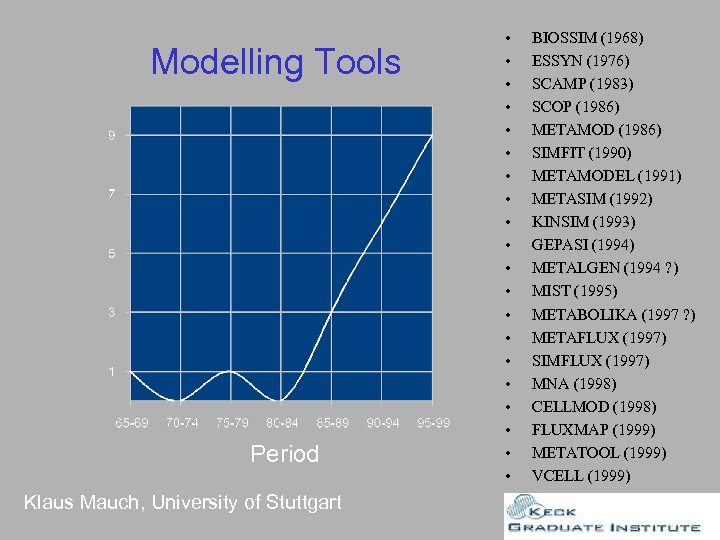
Modelling Tools Period Klaus Mauch, University of Stuttgart • • • • • BIOSSIM (1968) ESSYN (1976) SCAMP (1983) SCOP (1986) METAMOD (1986) SIMFIT (1990) METAMODEL (1991) METASIM (1992) KINSIM (1993) GEPASI (1994) METALGEN (1994 ? ) MIST (1995) METABOLIKA (1997 ? ) METAFLUX (1997) SIMFLUX (1997) MNA (1998) CELLMOD (1998) FLUXMAP (1999) METATOOL (1999) VCELL (1999)
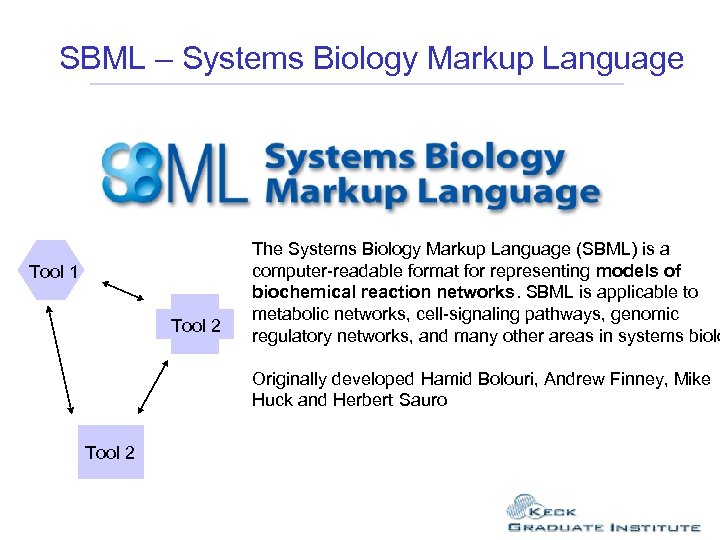
SBML – Systems Biology Markup Language The Systems Biology Markup Language (SBML) is a computer-readable format for representing models of biochemical reaction networks. SBML is applicable to metabolic networks, cell-signaling pathways, genomic regulatory networks, and many other areas in systems biology. Tool 1 Tool 2 The Systems Biology Markup Language (SBML) is a computer-readable format for representing models of biochemical reaction networks. SBML is applicable to metabolic networks, cell-signaling pathways, genomic regulatory networks, and many other areas in systems biolo Originally developed Hamid Bolouri, Andrew Finney, Mike Huck and Herbert Sauro Tool 2
SBML – Systems Biology Markup Language XML based Standard • Simple Compartments (well stirred reactor) • Internal/External Species • Reaction Schemes • Global Parameters • Arbitrary Rate Laws • DAEs (ODE + Algebraic functions, Constraints) • Physical Units/Model Notes • Annotation – extension capability

SBML – Systems Biology Markup Language What is XML? <? xml version="1. 0" ? > <note> <to> Hobbit </to> <from> Orc </from> <heading> Note to Frodo </heading> <body> I want to eat you </body> </note>

SBML – Systems Biology Markup Language XML has a hierarchical structure <root> <child> <subchild>. . . </subchild> </root> Each node can also have optional attributes, eg <child name = “john”>

<? xml version="1. 0" encoding="UTF-8"? > <!-- Created by XMLPretty. Printer on 11/14/2002 --> <sbml level = "1" version = "1" xmlns = "http: //www. sbml. org/sbml/level 1"> <!---> <!-- Model Starts Here --> <!---> <model name = "untitled"> <list. Of. Compartments> <compartment name = "u. Vol" volume = "1"/> </list. Of. Compartments> <list. Of. Species> <specie boundary. Condition = "false" compartment = "u. Vol" initial. Amount = "0" name ="Node 0"/> <specie boundary. Condition = "false" compartment = "u. Vol" initial. Amount = "0" name = "Node 1"/> <specie boundary. Condition = "false" compartment = "u. Vol" initial. Amount = "0" name = "Node 2"/> </list. Of. Species> SBML – Example <list. Of. Reactions> <reaction name = "J 0" reversible = "false"> <list. Of. Reactants> <specie. Reference specie = "Node 0" stoichiometry = "1"/> </list. Of. Reactants> <list. Of. Products> <specie. Reference specie = "Node 1" stoichiometry = "1"/> </list. Of. Products> <kinetic. Law formula = "v"> </kinetic. Law> </reaction> <reaction name = "J 1" reversible = "false"> <list. Of. Reactants> <specie. Reference specie = "Node 1" stoichiometry = "1"/> </list. Of. Reactants> <list. Of. Products> <specie. Reference specie = "Node 2" stoichiometry = "1"/> </list. Of. Products> <kinetic. Law formula = "v"> </kinetic. Law> </reaction> </list. Of. Reactions> </model> </sbml>
Other Related Efforts - Cell. ML is a more comprehensive attempt at developing an exchange standard, also defined in terms of XML. However, it is much more complex and the designers of Cell. ML have not provided software support in the form of tools and software libraries.
Data Formats One other area which is even more difficult to resolve is experimental data formats, microarray, proteomic, metabolmic, basically all the omics. Two projects are attempting to put some order in the data format area, bio. SPICE and particularly the DOE GTL project.
The Future There is obviously a long way to go. Kinetic data must be more carefully curated. Standards for exchanging data, models, including visualization notations need to be developed further.

What are we dealing with? Reaction systems working on multiple time scales: 1. Discrete deterministic events 2. Fast reactions 3. Continuous variables (modeled by ODES) 4. Continuous variables with additive and multiplicative noise 5. Stochastic discrete systems (Gillespie type)
Functional Motif Identification
1f0ca3f4ef4cf6d554cdd39f6cc21f31.ppt