1d0f874378e1b0bb99bdfe92cb942de1.ppt
- Количество слайдов: 13
Database Population and Curation Michael J. Donoghue, Yale University William H. Piel, University at Buffalo
Data Entry/Populating • • • Manual Entry -- students and staff Data Migration -- from Tree. BASE to ITR Value Added Data -- federating data Submission System -- burden on user Sustainability -- beyond ITR

Manual Entry • Advantages – Selective coverage – Full control of quality and depth – Good source for student training/outreach • Disadvantages – Work intensive, seemingly endless – Not all data can be digitized – Analyses not as accurate as user-entered
Manual Entry • PIs build Endnote database of desired studies: – Author names – Citation – Abstract • Students prepare datasets – Scan and OCR characters and character descriptions from the paper or PDF or download sequence data from Genbank; Google for data; seek data from authors – Use regular expressions in BBEdit (or equivalent) so that data is ready for Mac. Clade – Recreate trees with PAUP and Mac. Clade as needed – Verify Parameters (e. g. tree lengths) – Examine paper for basic outline of analyses – Enter data into Tree. BASE; later into ITR product
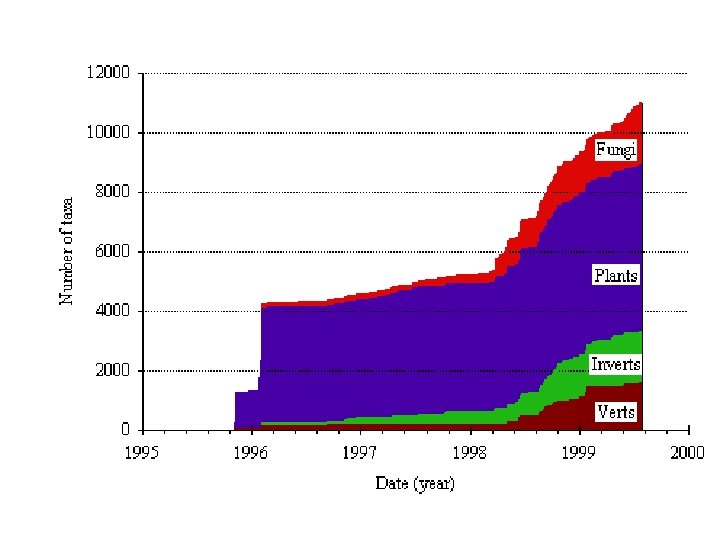
Data Migration • Currently: – 1, 526 authors – 847 studies – 2, 273 trees – 32, 490 taxa. • Modest but, with about 80% connectivity • Connectivity will increase after solving the nomenclature Pandora's box

Data Migration • Design an export format – XML? – NEXUS with proprietary block? – Diacritical translation/ASCII Character Sets – Preserve Matrix IDs and Study IDs? – Resolve nomenclature in Tree. BASE or ITR?
Data Migration • Tree. BASE's "Shadow Database" – For submissions "in progress" (~ 500) – Uses slightly different data schema – Uses slightly different IDs (positive integers) – Treatment depends on ITR data model

Value Added Data • Automated vs. Manually Curated VA – Do we upgrade existing and new datasets? – Identify taxa using SOAP with ITIS/Genbank? – Identify genes based on automated BLASTs? – Rank trees per study: identify "the" tree? – Automate some tree parameters? – Others? • GIS for phylogeography • Culture numbers and other IDs
Other Changes to the Data Model • Expand data types (distance, genomic, etc) • Adapt to "Electronic Notebook"model – Much more complex analysis description • Separate "real data" from benchmark/simulated • Separate "published data" from data under active research

Submission System • Tree. BASE Submission will continue – Demands constant editorial effort • New Submission System GUI: – Must maximize burden on the user – But cannot be excessively arduous – Must incorporate quality control flags – Best to use solid, client-side helper applications

Sustainability • Consider strategies for sustainability – Lobby societies and journals • Require Author Submission • Pass on modest Submission Charges? – Establish a Tree-of-Life electronic journal • Designed to publish massive trees – Mission-Oriented Funding Sources • NIH • Foundations
1d0f874378e1b0bb99bdfe92cb942de1.ppt