0aff262ca18492b09b95ccb93b8c43b0.ppt
- Количество слайдов: 50
CZ 5225: Modeling and Simulation in Biology Lecture 2: Gene Expression Profiles and Microarray Data Analysis Prof. Chen Yu Zong Tel: 6874 -6877 Email: yzchen@cz 3. nus. edu. sg http: //xin. cz 3. nus. edu. sg Room 07 -24, level 7, SOC 1, NUS

Biology and Cells • All living organisms consist of cells. • Humans have trillions of cells. Yeast - one cell. • Cells are of many different types (blood, skin, nerve), but all arose from a single cell (the fertilized egg) • Each* cell contains a complete copy of the genome (the program for making the organism), encoded in DNA. 2
DNA • DNA molecules are long double-stranded chains; 4 types of bases are attached to the backbone: adenine (A), guanine (G), cytosine (C), and thymine (T). A pairs with T, C with G. • A gene is a segment of DNA that specifies how to make a protein. • Human DNA has about 25 -35 K genes; Rice about 50 -60 K but shorter genes. 3
Exons and Introns • exons are coding DNA (translated into a protein), which are only about 2% of human genome • introns are non-coding DNA, which provide structural integrity and regulatory (control) functions • exons can be thought of program data, while introns provide the program logic • Humans have much more control structure than rice 4
Gene Expression • Cells are different because of differential gene expression. • About 40% of human genes are expressed at one time. • Gene is expressed by transcribing DNA into single-stranded m. RNA • m. RNA is later translated into a protein • Microarrays measure the level of m. RNA expression 5
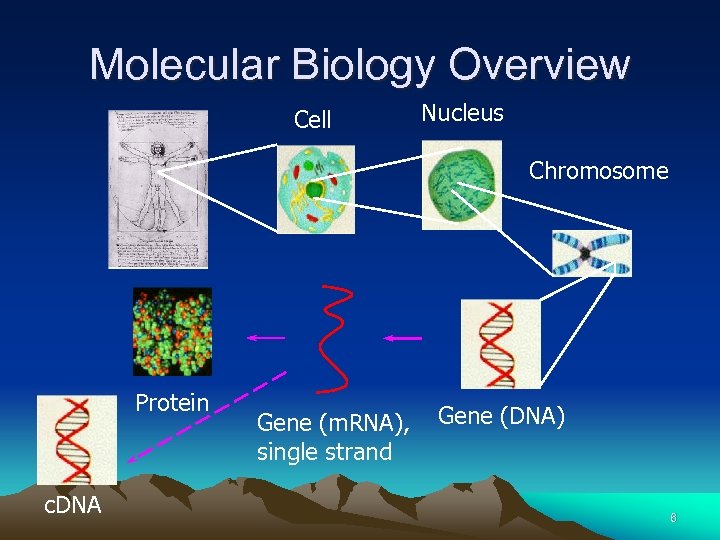
Molecular Biology Overview Cell Nucleus Chromosome Protein c. DNA Gene (m. RNA), single strand Gene (DNA) 6
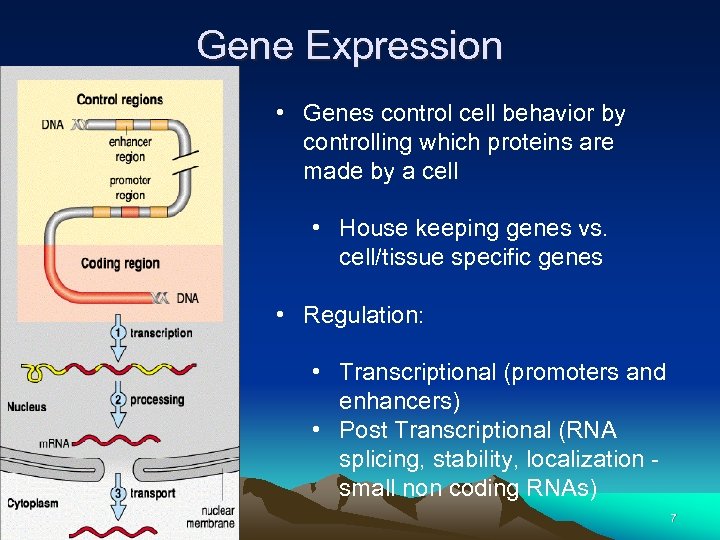
Gene Expression • Genes control cell behavior by controlling which proteins are made by a cell • House keeping genes vs. cell/tissue specific genes • Regulation: • Transcriptional (promoters and enhancers) • Post Transcriptional (RNA splicing, stability, localization small non coding RNAs) 7
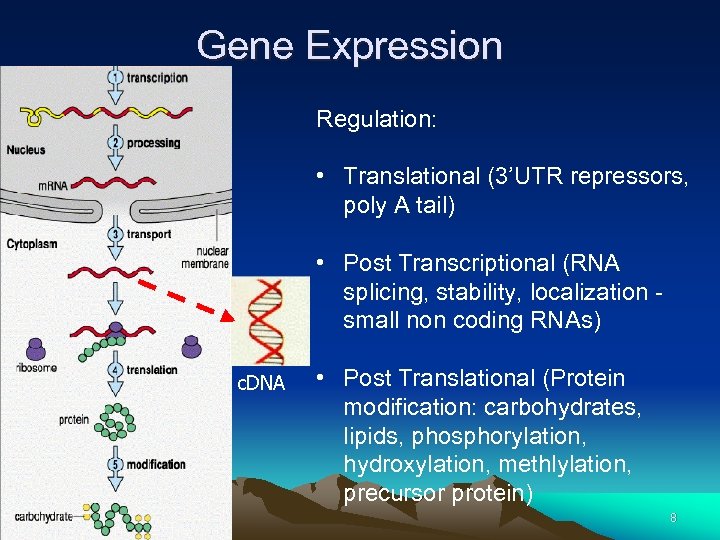
Gene Expression Regulation: • Translational (3’UTR repressors, poly A tail) • Post Transcriptional (RNA splicing, stability, localization small non coding RNAs) c. DNA • Post Translational (Protein modification: carbohydrates, lipids, phosphorylation, hydroxylation, methlylation, precursor protein) 8
Gene Expression Measurement • m. RNA expression represents dynamic aspects of cell • m. RNA expression can be measured with latest technology • m. RNA is isolated and labeled with fluorescent protein • m. RNA is hybridized to the target; level of hybridization corresponds to light emission which is measured with a laser 9
Traditional Methods • Northern Blotting – Single RNA isolated – Probed with labeled c. DNA • RT-PCR – Primers amplify specific c. DNA transcripts 10

Microarray Technology • Microarray: – New Technology (first paper: 1995) • Allows study of thousands of genes at same time – Glass slide of DNA molecules • Molecule: string of bases (25 bp – 500 bp) • uniquely identifies gene or unit to be studied 11
Gene Expression Microarrays The main types of gene expression microarrays: • Short oligonucleotide arrays (Affymetrix) • c. DNA or spotted arrays (Brown/Botstein). • Long oligonucleotide arrays (Agilent Inkjet); • Fiber-optic arrays • . . . 12
Fabrications of Microarrays • Size of a microscope slide Images: http: //www. affymetrix. com/ 13
Differing Conditions • Ultimate Goal: – Understand expression level of genes under different conditions • Helps to: – Determine genes involved in a disease – Pathways to a disease – Used as a screening tool 14
Gene Conditions • • • Cell types (brain vs. liver) Developmental (fetal vs. adult) Response to stimulus Gene activity (wild vs. mutant) Disease states (healthy vs. diseased) 15
Expressed Genes • Genes under a given condition – m. RNA extracted from cells – m. RNA labeled – Labeled m. RNA is m. RNA present in a given condition – Labeled m. RNA will hybridize (base pair) with corresponding sequence on slide 16
Two Different Types of Microarrays • Custom spotted arrays (up to 20, 000 sequences) – c. DNA – Oligonucleotide • High-density (up to 100, 000 sequences) synthetic oligonucleotide arrays – Affymetrix (25 bases) – SHOW AFFYMETRIX LAYOUT 17
Custom Arrays • Mostly c. DNA arrays • 2 -dye (2 -channel) – RNA from two sources (c. DNA created) • Source 1: labeled with red dye • Source 2: labeled with green dye 18
Two Channel Microarrays • Microarrays measure gene expression • Two different samples: – Control (green label) – Sample (red label) • Both are washed over the microarray – Hybridization occurs – Each spot is one of 4 colors 19
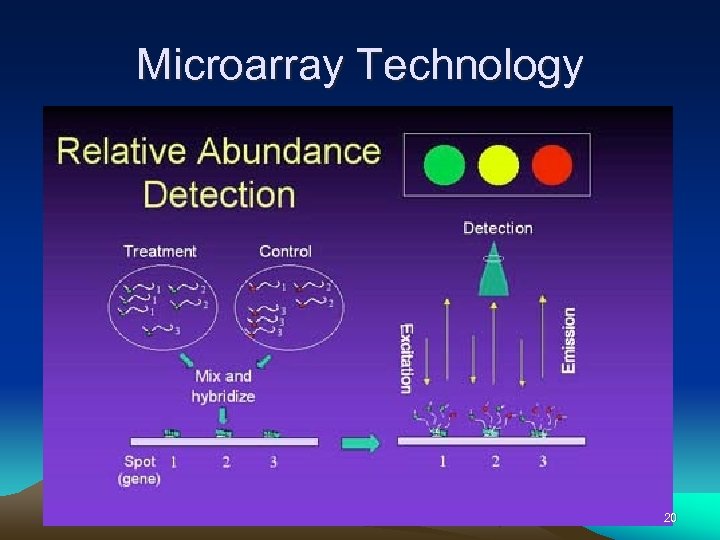
Microarray Technology 20
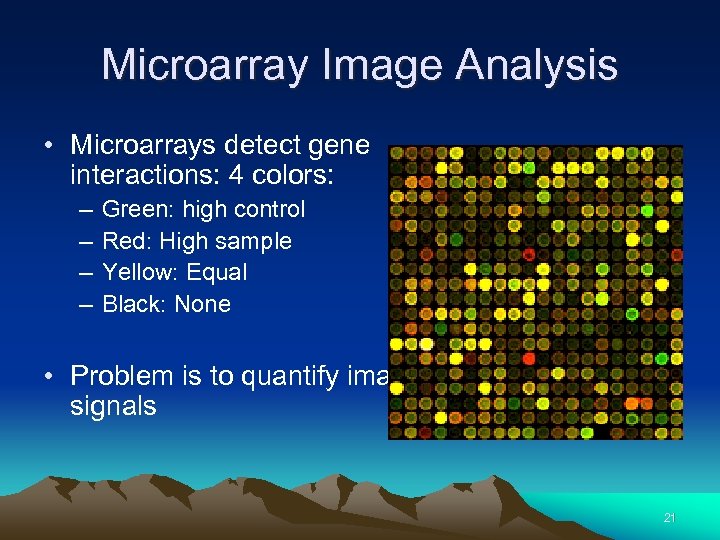
Microarray Image Analysis • Microarrays detect gene interactions: 4 colors: – – Green: high control Red: High sample Yellow: Equal Black: None • Problem is to quantify image signals 21
Single Color Microarrays • Prefabricated – Affymetrix (25 mers) • Custom – c. DNA (500 bases or so) – Spotted oligos (70 -80 bases) 22

Microarray Animations • Davidson University: • http: //www. bio. davidson. edu/courses/genomics/chip. html • Imagecyte: • http: //www. imagecyte. com/array 2. html 23
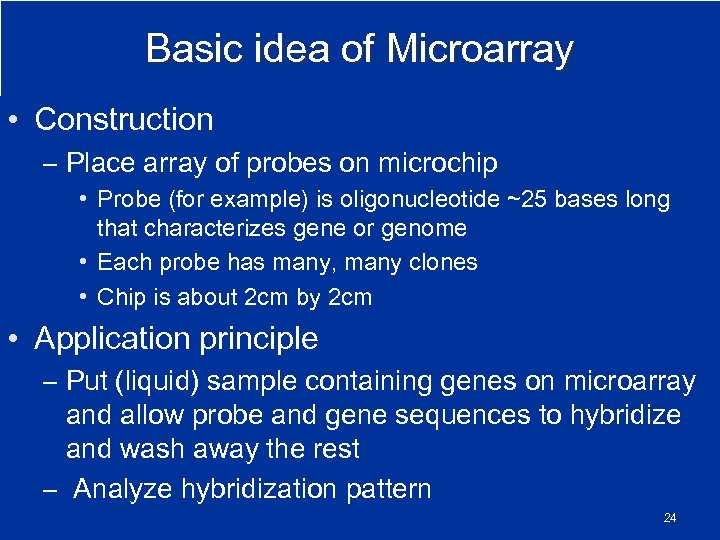
Basic idea of Microarray • Construction – Place array of probes on microchip • Probe (for example) is oligonucleotide ~25 bases long that characterizes gene or genome • Each probe has many, many clones • Chip is about 2 cm by 2 cm • Application principle – Put (liquid) sample containing genes on microarray and allow probe and gene sequences to hybridize and wash away the rest – Analyze hybridization pattern 24
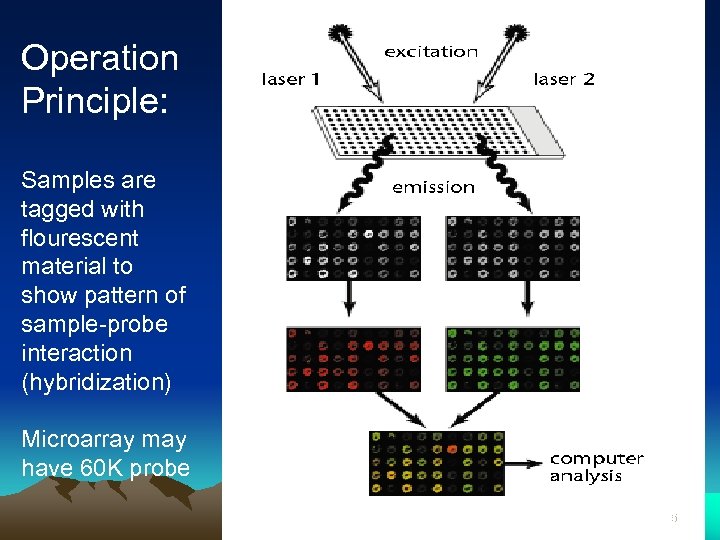
Operation Principle: Samples are tagged with flourescent material to show pattern of sample-probe interaction (hybridization) Microarray analysis Microarray may have 60 K probe 25
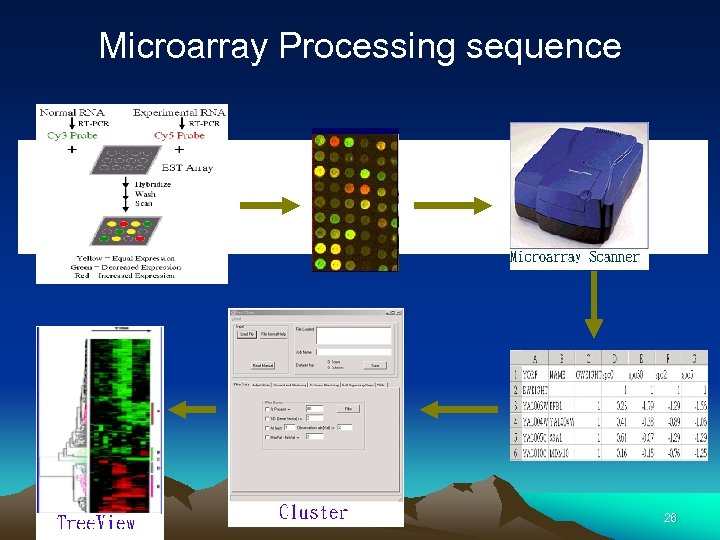
Microarray Processing sequence 26
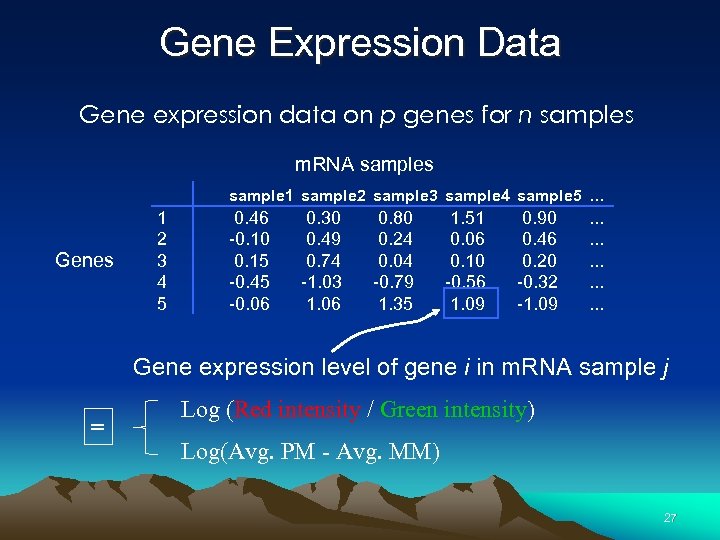
Gene Expression Data Gene expression data on p genes for n samples m. RNA samples sample 1 sample 2 sample 3 sample 4 sample 5 … Genes 1 2 3 4 5 0. 46 -0. 10 0. 15 -0. 45 -0. 06 0. 30 0. 49 0. 74 -1. 03 1. 06 0. 80 0. 24 0. 04 -0. 79 1. 35 1. 51 0. 06 0. 10 -0. 56 1. 09 0. 90 0. 46 0. 20 -0. 32 -1. 09 . . . . Gene expression level of gene i in m. RNA sample j = Log (Red intensity / Green intensity) Log(Avg. PM - Avg. MM) 27
Some possible applications • Sample from specific organ to show which genes are expressed • Compare samples from healthy and sick host to find gene-disease connection • Probes are sets of human pathogens for disease detection 28
Huge amount of data from single microarray • If just two color, then amount of data on array with N probes is 2 N • Cannot analyze pixel by pixel • Analyze by pattern – cluster analysis 29
Major Data Mining Techniques • Link Analysis – Associations Discovery – Sequential Pattern Discovery – Similar Time Series Discovery • Predictive Modeling – Classification – Clustering 30
Cluster Analysis: Grouping Similarly Expressed Genes, Cell Samples, or Both • Strengthens signal when averages are taken within clusters of genes (Eisen) • Useful (essential ? ) when seeking new subclasses of cells, tumours, etc. • Leads to readily interpreted figures 31
Some clustering methods and software • Partitioning:K-Means, K-Medoids, PAM, CLARA … • Hierarchical:Cluster, HAC、BIRCH、CURE、 ROCK • Density-based: CAST, DBSCAN、OPTICS、 CLIQUE… • Grid-based:STING、CLIQUE、Wave. Cluster… • Model-based:SOM (self-organized map)、 COBWEB、CLASSIT、Auto. Class… • Two-way Clustering • Block clustering 32
Assessment of various methods • Algorithmic Approaches to Clustering Gene Expression Data, Ron Shamir School of Computer Science, Tel-Aviv University Tel-Aviv – http: //citeseer. nj. nec. com/shamir 01 algorithmic. html • Conclusion: hierarchical clustering exceptional 33
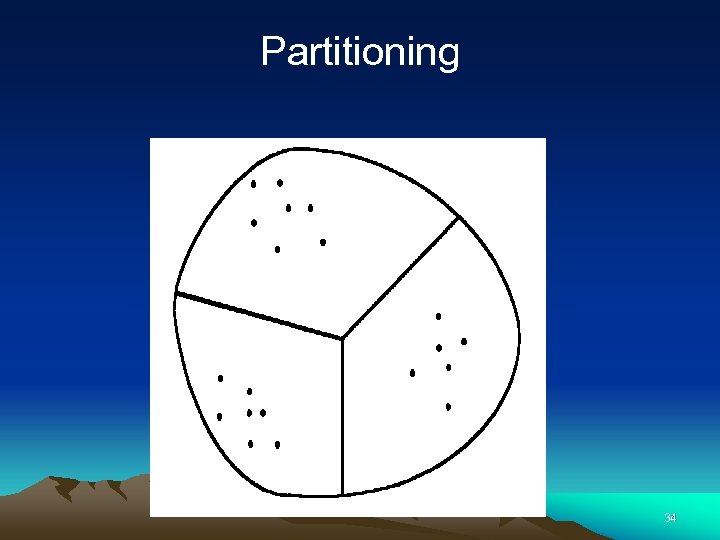
Partitioning 34
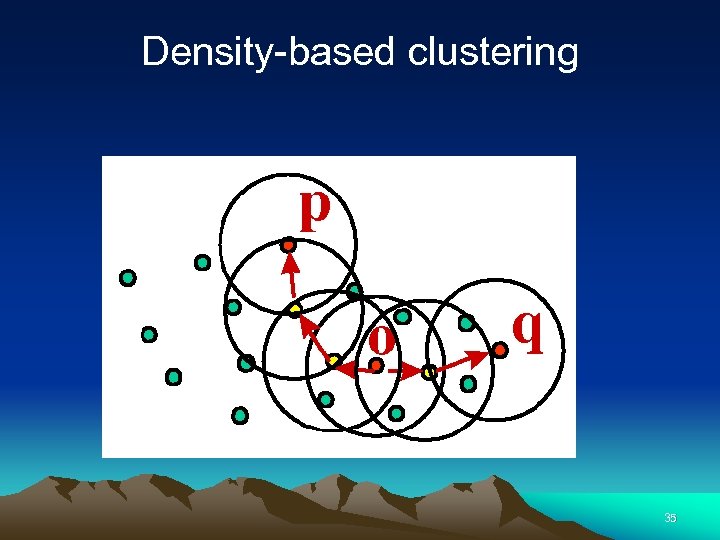
Density-based clustering 35
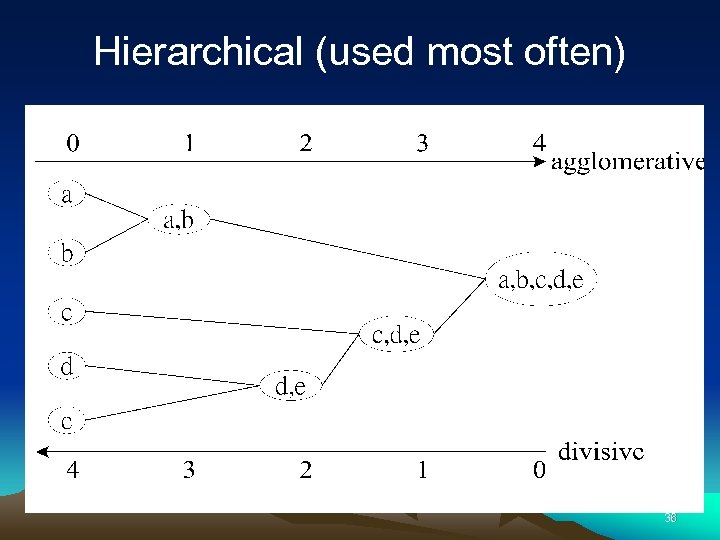
Hierarchical (used most often) 36
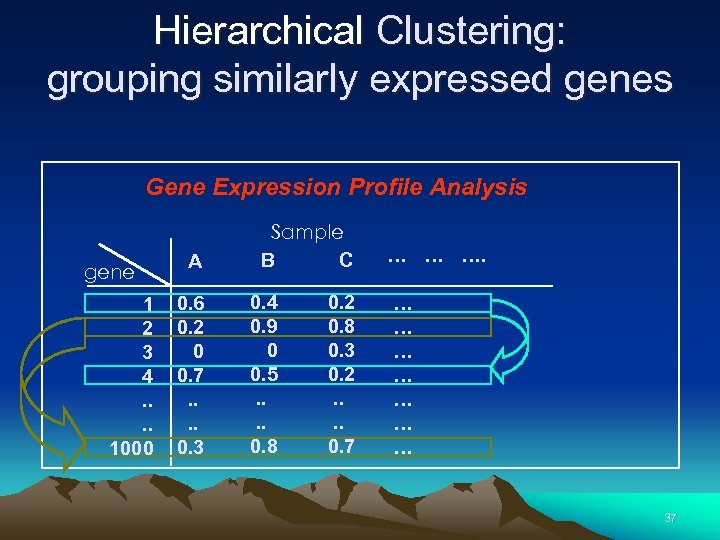
Hierarchical Clustering: grouping similarly expressed genes Gene Expression Profile Analysis gene 1 2 3 4. . 1000 A 0. 6 0. 2 0 0. 7. . 0. 3 Sample B C 0. 4 0. 9 0 0. 5. . 0. 8 0. 2 0. 8 0. 3 0. 2. . 0. 7 … … … … 37
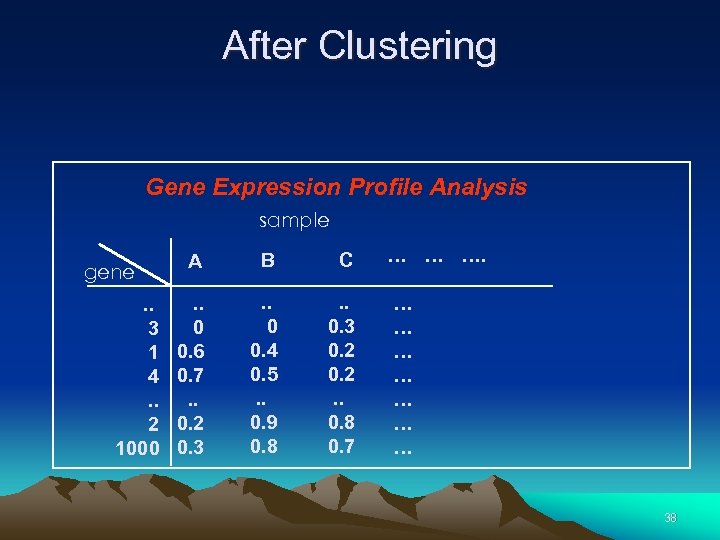
After Clustering Gene Expression Profile Analysis sample gene. . 3 1 4. . 2 1000 A B C . . 0 0. 6 0. 7. . 0. 2 0. 3 . . 0 0. 4 0. 5. . 0. 9 0. 8 . . 0. 3 0. 2. . 0. 8 0. 7 … … … … 38
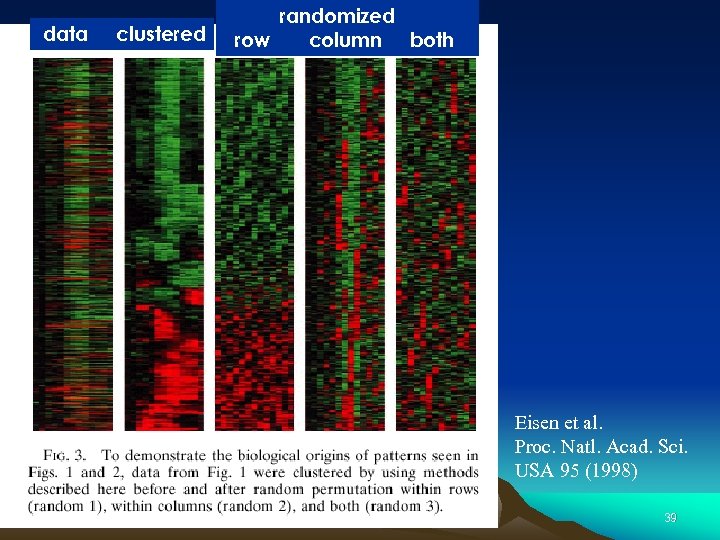
data clustered time randomized row column both Eisen et al. Proc. Natl. Acad. Sci. USA 95 (1998) 39
Types of Similarity Measurements • Distance measurements • Correlation coefficients • Association coefficients • Probabilistic similarity coefficients 40
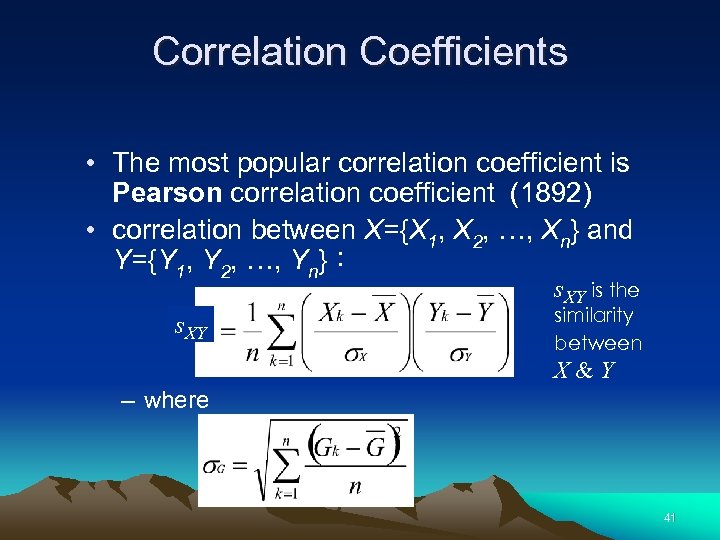
Correlation Coefficients • The most popular correlation coefficient is Pearson correlation coefficient (1892) • correlation between X={X 1, X 2, …, Xn} and Y={Y 1, Y 2, …, Yn}: s. XY is the s. XY similarity between X&Y – where 41
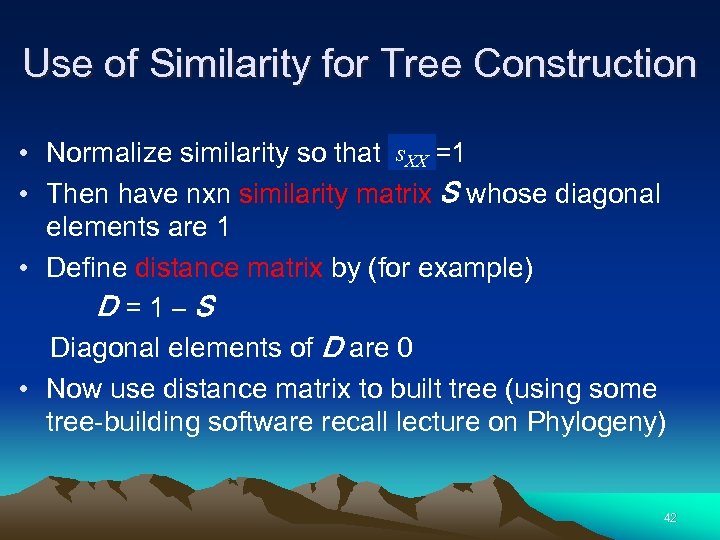
Use of Similarity for Tree Construction • Normalize similarity so that s. XX =1 • Then have nxn similarity matrix S whose diagonal elements are 1 • Define distance matrix by (for example) D=1–S Diagonal elements of D are 0 • Now use distance matrix to built tree (using some tree-building software recall lecture on Phylogeny) 42
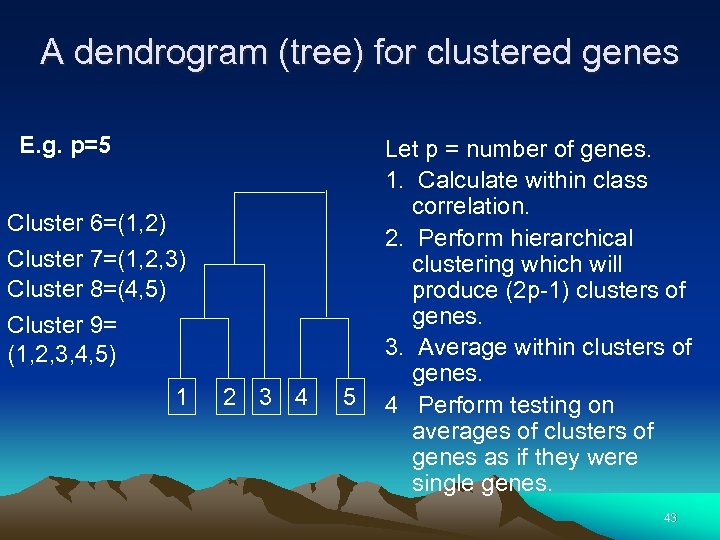
A dendrogram (tree) for clustered genes E. g. p=5 Cluster 6=(1, 2) Cluster 7=(1, 2, 3) Cluster 8=(4, 5) Cluster 9= (1, 2, 3, 4, 5) 1 2 3 4 5 Let p = number of genes. 1. Calculate within class correlation. 2. Perform hierarchical clustering which will produce (2 p-1) clusters of genes. 3. Average within clusters of genes. 4 Perform testing on averages of clusters of genes as if they were single genes. 43
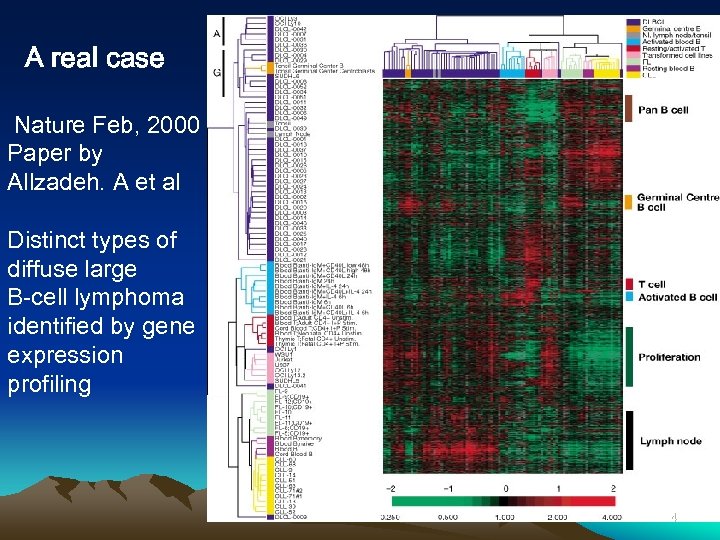
A real case Nature Feb, 2000 Paper by Allzadeh. A et al Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling 44
![Validation Techniques: Hubert’s Γ Statistics • X=[X(i, j)] and Y=[Y(i, j)] are two n Validation Techniques: Hubert’s Γ Statistics • X=[X(i, j)] and Y=[Y(i, j)] are two n](https://present5.com/presentation/0aff262ca18492b09b95ccb93b8c43b0/image-45.jpg)
Validation Techniques: Hubert’s Γ Statistics • X=[X(i, j)] and Y=[Y(i, j)] are two n × n matrix – X(i, j): similarity of gene i and gene j if genes i and j are in same cluster, otherwise – Hubert’s Γ statistic represents the point serial correlation: • where M = n (n - 1) / 2 – A higher value of Γ represents the better clustering quality. 45
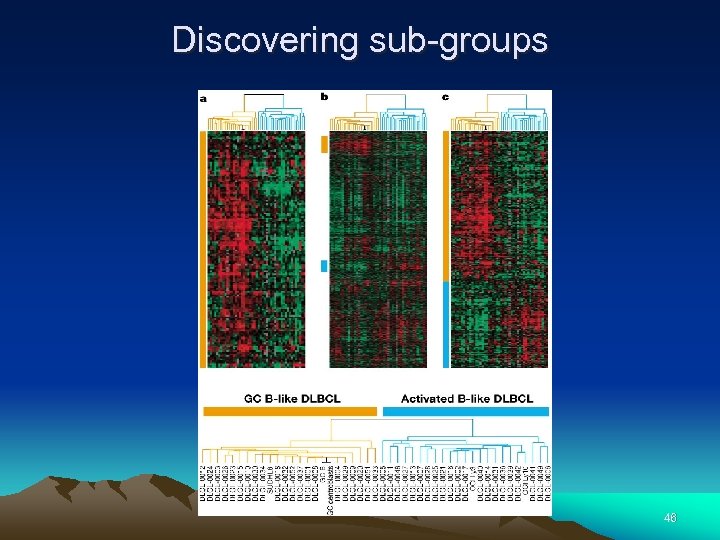
Discovering sub-groups 46
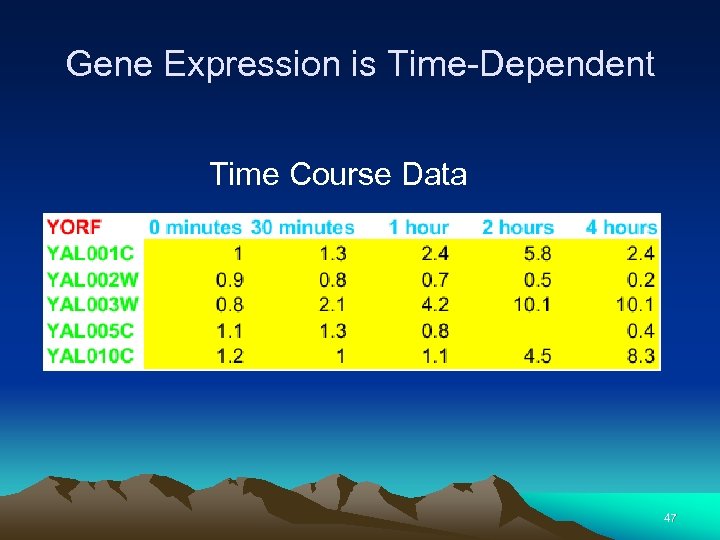
Gene Expression is Time-Dependent Time Course Data 47
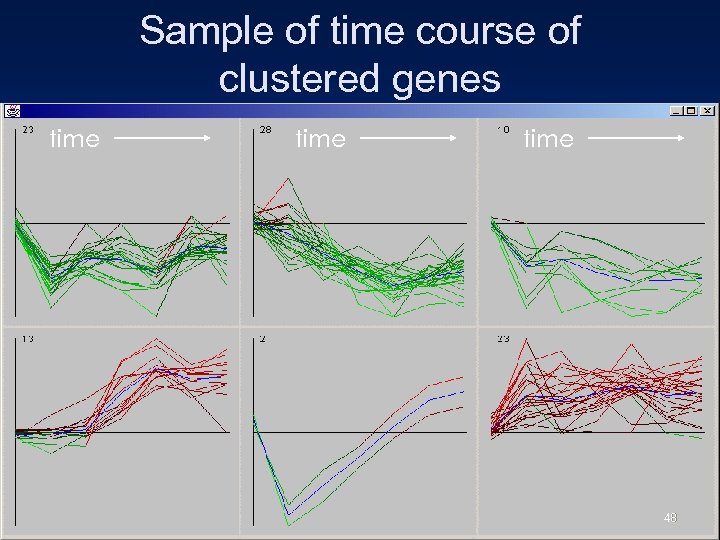
Sample of time course of clustered genes time 48
Limitations • Cluster analyses: – Usually outside the normal framework of statistical inference – Less appropriate when only a few genes are likely to change – Needs lots of experiments • Single gene tests: – May be too noisy in general to show much – May not reveal coordinated effects of positively correlated genes. – Hard to relate to pathways 49

Useful Links • Affymetrix www. affymetrix. com • Michael Eisen Lab at LBL (hierarchical clustering software “Cluster” and “Tree View” (Windows)) rana. lbl. gov/ • Review of Currently Available Microarray Software www. the-scientist. com/yr 2001/apr/profile 1_010430. html • Array. Express at the EBI http: //www. ebi. ac. uk/arrayexpress/ • Stanford Micro. Array Database http: //genome-www 5. stanford. edu/ • Yale Microarray Database http: //info. med. yale. edu/microarray/ • Microarray DB www. biologie. ens. fr/en/genetiqu/puces/bddeng. html 50
0aff262ca18492b09b95ccb93b8c43b0.ppt