4a61304204d942328aa8814e08ad8b11.ppt
- Количество слайдов: 13
 CSC Conference 2. 6. 2010 Next generation sequencing data analysis Assembling the Glanville fritillary genome Panu Somervuo University of Helsinki MRG group & DNA sequencing and genomics lab
CSC Conference 2. 6. 2010 Next generation sequencing data analysis Assembling the Glanville fritillary genome Panu Somervuo University of Helsinki MRG group & DNA sequencing and genomics lab
 Next generation sequencing • Roche 454 • Illumina Solexa • ABI SOLi. D
Next generation sequencing • Roche 454 • Illumina Solexa • ABI SOLi. D
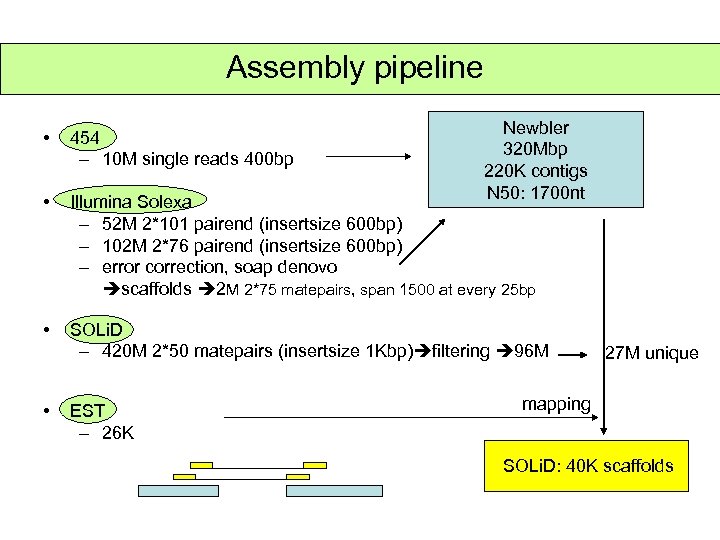 Assembly pipeline Newbler 320 Mbp 220 K contigs N 50: 1700 nt • 454 – 10 M single reads 400 bp • Illumina Solexa – 52 M 2*101 pairend (insertsize 600 bp) – 102 M 2*76 pairend (insertsize 600 bp) – error correction, soap denovo scaffolds 2 M 2*75 matepairs, span 1500 at every 25 bp • SOLi. D – 420 M 2*50 matepairs (insertsize 1 Kbp) filtering 96 M • EST – 26 K 27 M unique mapping SOLi. D: 40 K scaffolds
Assembly pipeline Newbler 320 Mbp 220 K contigs N 50: 1700 nt • 454 – 10 M single reads 400 bp • Illumina Solexa – 52 M 2*101 pairend (insertsize 600 bp) – 102 M 2*76 pairend (insertsize 600 bp) – error correction, soap denovo scaffolds 2 M 2*75 matepairs, span 1500 at every 25 bp • SOLi. D – 420 M 2*50 matepairs (insertsize 1 Kbp) filtering 96 M • EST – 26 K 27 M unique mapping SOLi. D: 40 K scaffolds
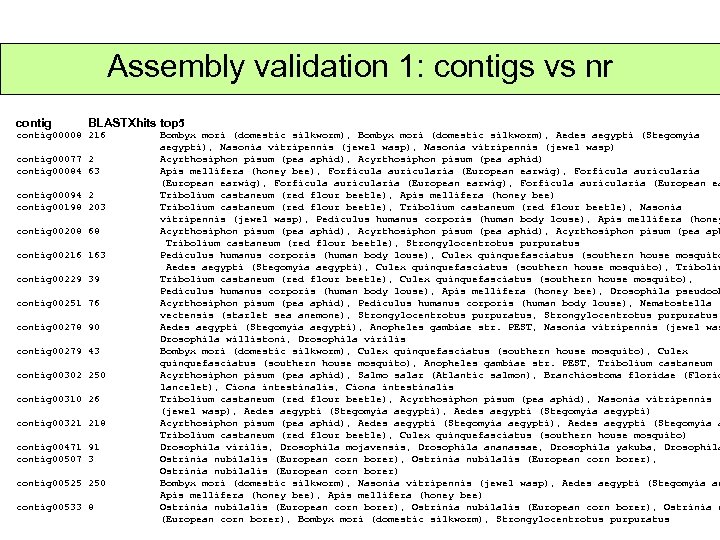 Assembly validation 1: contigs vs nr contig BLASTXhits top 5 contig 00008 216 contig 00077 2 contig 00084 63 contig 00094 2 contig 00198 203 contig 00208 68 contig 00216 163 contig 00229 39 contig 00251 76 contig 00278 90 contig 00279 43 contig 00302 250 contig 00310 26 contig 00321 218 contig 00471 91 contig 00507 3 contig 00525 250 contig 00533 8 Bombyx mori (domestic silkworm), Aedes aegypti (Stegomyia aegypti), Nasonia vitripennis (jewel wasp) Acyrthosiphon pisum (pea aphid), Acyrthosiphon pisum (pea aphid) Apis mellifera (honey bee), Forficula auricularia (European earwig), Forficula auricularia (European ea Tribolium castaneum (red flour beetle), Apis mellifera (honey bee) Tribolium castaneum (red flour beetle), Nasonia vitripennis (jewel wasp), Pediculus humanus corporis (human body louse), Apis mellifera (honey Acyrthosiphon pisum (pea aphid), Acyrthosiphon pisum (pea aph Tribolium castaneum (red flour beetle), Strongylocentrotus purpuratus Pediculus humanus corporis (human body louse), Culex quinquefasciatus (southern house mosquito Aedes aegypti (Stegomyia aegypti), Culex quinquefasciatus (southern house mosquito), Tribolium castaneum (red flour beetle), Culex quinquefasciatus (southern house mosquito), Pediculus humanus corporis (human body louse), Apis mellifera (honey bee), Drosophila pseudoob Acyrthosiphon pisum (pea aphid), Pediculus humanus corporis (human body louse), Nematostella vectensis (starlet sea anemone), Strongylocentrotus purpuratus Aedes aegypti (Stegomyia aegypti), Anopheles gambiae str. PEST, Nasonia vitripennis (jewel was Drosophila willistoni, Drosophila virilis Bombyx mori (domestic silkworm), Culex quinquefasciatus (southern house mosquito), Anopheles gambiae str. PEST, Tribolium castaneum ( Acyrthosiphon pisum (pea aphid), Salmo salar (Atlantic salmon), Branchiostoma floridae (Florid lancelet), Ciona intestinalis Tribolium castaneum (red flour beetle), Acyrthosiphon pisum (pea aphid), Nasonia vitripennis (jewel wasp), Aedes aegypti (Stegomyia aegypti) Acyrthosiphon pisum (pea aphid), Aedes aegypti (Stegomyia aegypti), Aedes aegypti (Stegomyia a Tribolium castaneum (red flour beetle), Culex quinquefasciatus (southern house mosquito) Drosophila virilis, Drosophila mojavensis, Drosophila ananassae, Drosophila yakuba, Drosophila Ostrinia nubilalis (European corn borer), Ostrinia nubilalis (European corn borer) Bombyx mori (domestic silkworm), Nasonia vitripennis (jewel wasp), Aedes aegypti (Stegomyia ae Apis mellifera (honey bee), Apis mellifera (honey bee) Ostrinia nubilalis (European corn borer), Ostrinia n (European corn borer), Bombyx mori (domestic silkworm), Strongylocentrotus purpuratus
Assembly validation 1: contigs vs nr contig BLASTXhits top 5 contig 00008 216 contig 00077 2 contig 00084 63 contig 00094 2 contig 00198 203 contig 00208 68 contig 00216 163 contig 00229 39 contig 00251 76 contig 00278 90 contig 00279 43 contig 00302 250 contig 00310 26 contig 00321 218 contig 00471 91 contig 00507 3 contig 00525 250 contig 00533 8 Bombyx mori (domestic silkworm), Aedes aegypti (Stegomyia aegypti), Nasonia vitripennis (jewel wasp) Acyrthosiphon pisum (pea aphid), Acyrthosiphon pisum (pea aphid) Apis mellifera (honey bee), Forficula auricularia (European earwig), Forficula auricularia (European ea Tribolium castaneum (red flour beetle), Apis mellifera (honey bee) Tribolium castaneum (red flour beetle), Nasonia vitripennis (jewel wasp), Pediculus humanus corporis (human body louse), Apis mellifera (honey Acyrthosiphon pisum (pea aphid), Acyrthosiphon pisum (pea aph Tribolium castaneum (red flour beetle), Strongylocentrotus purpuratus Pediculus humanus corporis (human body louse), Culex quinquefasciatus (southern house mosquito Aedes aegypti (Stegomyia aegypti), Culex quinquefasciatus (southern house mosquito), Tribolium castaneum (red flour beetle), Culex quinquefasciatus (southern house mosquito), Pediculus humanus corporis (human body louse), Apis mellifera (honey bee), Drosophila pseudoob Acyrthosiphon pisum (pea aphid), Pediculus humanus corporis (human body louse), Nematostella vectensis (starlet sea anemone), Strongylocentrotus purpuratus Aedes aegypti (Stegomyia aegypti), Anopheles gambiae str. PEST, Nasonia vitripennis (jewel was Drosophila willistoni, Drosophila virilis Bombyx mori (domestic silkworm), Culex quinquefasciatus (southern house mosquito), Anopheles gambiae str. PEST, Tribolium castaneum ( Acyrthosiphon pisum (pea aphid), Salmo salar (Atlantic salmon), Branchiostoma floridae (Florid lancelet), Ciona intestinalis Tribolium castaneum (red flour beetle), Acyrthosiphon pisum (pea aphid), Nasonia vitripennis (jewel wasp), Aedes aegypti (Stegomyia aegypti) Acyrthosiphon pisum (pea aphid), Aedes aegypti (Stegomyia aegypti), Aedes aegypti (Stegomyia a Tribolium castaneum (red flour beetle), Culex quinquefasciatus (southern house mosquito) Drosophila virilis, Drosophila mojavensis, Drosophila ananassae, Drosophila yakuba, Drosophila Ostrinia nubilalis (European corn borer), Ostrinia nubilalis (European corn borer) Bombyx mori (domestic silkworm), Nasonia vitripennis (jewel wasp), Aedes aegypti (Stegomyia ae Apis mellifera (honey bee), Apis mellifera (honey bee) Ostrinia nubilalis (European corn borer), Ostrinia n (European corn borer), Bombyx mori (domestic silkworm), Strongylocentrotus purpuratus
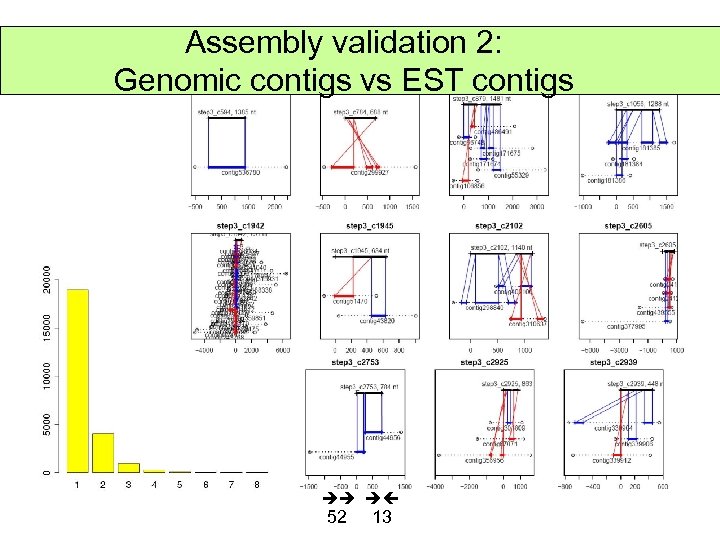 Assembly validation 2: Genomic contigs vs EST contigs 52 13
Assembly validation 2: Genomic contigs vs EST contigs 52 13
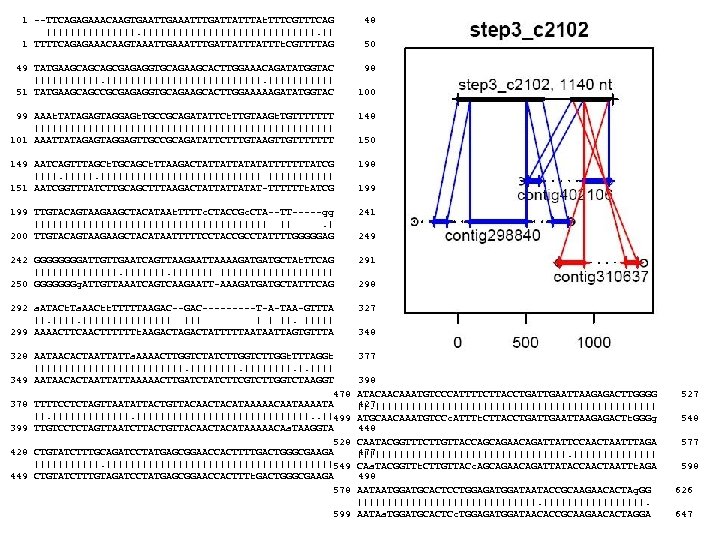 1 --TTCAGAGAAACAAGTGAATTGAAATTTGATTATTTAt. TTTCGTTTCAG |||||||||||||||. || 1 TTTTCAGAGAAACAAGTAAATTGAAATTTGATTATTTt. CGTTTTAG 48 49 TATGAAGCAGCAGCGAGAGGTGCAGAAGCACTTGGAAACAGATATGGTAC ||||||||||||| 51 TATGAAGCAGCCGCGAGAGGTGCAGAAGCACTTGGAAAAAGATATGGTAC 98 50 100 99 AAAt. TATAGAGTAGGAGt. TGCCGCAGATATTCt. TTGTAAGt. TGTTTTTTT ||||||||||||||||||||||||| 101 AAATTATAGAGTAGGAGTTGCCGCAGATATTCTTTGTAAGTTGTTTTTTT 148 149 AATCAGTTTAGCt. TGCAGCt. TTAAGACTATTATTATATATTTTTTTATCG |||||||||||||| 151 AATCGGTTTATCTTGCAGCTTTAAGACTATTATTATAT-TTTTTTt. ATCG 198 199 TTGTACAGTAAGAAGCTACATAAt. TTTTc. CTACCGc. CTA--TT-----gg |||||||||||||||||||| ||. | 200 TTGTACAGTAAGAAGCTACATAATTTTTCCTACCGCCTATTTTGGGGGAG 241 242 GGGGATTGTTGAATCAGTTAAGAATTAAAAGATGATGCTAt. TTCAG |||||||||| 250 GGGGGGGg. ATTGTTAAATCAGTCAAGAATT-AAAGATGATGCTATTTCAG 291 292 a. ATACt. Ta. AACtt. TTTTTAAGAC---------T-A-TAA-GTTTA ||. ||||||||| ||| | | ||. ||||| 299 AAAACTTCAACTTTTTTt. AAGACTATTTTTAATAATTAGTGTTTA 327 328 AATAACACTAATTATTa. AAAACTTGGTCTATCTTGGt. TTTAGGt |||||||||||||. |. |||| 349 AATAACACTAATTATTAAAAACTTGATCTTCGTCTTGGTCTAAGGT 478 378 TTTTCCTCTAGTTAATATTACTGTTACAACTACATAAAAACAATAAAATA ||. ||||||||||||||||||||. . ||499 399 TTGTCCTCTAGTTAATCTTACTGTTACAACTACATAAAAACAa. TAAGGTA 528 428 CTGTATCTTTGCAGATCCTATGAGCGGAACCACTTTTGACTGGGCGAAGA |||||||||||||||||||549 449 CTGTATCTTTGTAGATCCTATGAGCGGAACCACTTTt. GACTGGGCGAAGA 578 377 150 199 249 298 348 398 ATACAACAAATGTCCCATTTTCTTACCTGATTGAATTAAGAGACTTGGGG 427 ||. |||||||||||||||||||||||| ATGCAACAAATGTCCc. ATTTt. CTTACCTGATTGAATTAAGAGACTt. GGGg 448 CAATACGGTTTCTTGTTACCAGCAGAACAGATTATTCCAACTAATTTAGA 477 |||||||||||||||||| CAa. TACGGTTt. CTTGTTACc. AGCAGAACAGATTATACCAACTAATTt. AGA 498 AATAATGGATGCACTCCTGGAGATGGATAATACCGCAAGAACACTAg. GG |||||||||||||||. 599 AATAa. TGGATGCACTCc. TGGAGATGGATAACACCGCAAGAACACTAGGA 527 548 577 598 626 647
1 --TTCAGAGAAACAAGTGAATTGAAATTTGATTATTTAt. TTTCGTTTCAG |||||||||||||||. || 1 TTTTCAGAGAAACAAGTAAATTGAAATTTGATTATTTt. CGTTTTAG 48 49 TATGAAGCAGCAGCGAGAGGTGCAGAAGCACTTGGAAACAGATATGGTAC ||||||||||||| 51 TATGAAGCAGCCGCGAGAGGTGCAGAAGCACTTGGAAAAAGATATGGTAC 98 50 100 99 AAAt. TATAGAGTAGGAGt. TGCCGCAGATATTCt. TTGTAAGt. TGTTTTTTT ||||||||||||||||||||||||| 101 AAATTATAGAGTAGGAGTTGCCGCAGATATTCTTTGTAAGTTGTTTTTTT 148 149 AATCAGTTTAGCt. TGCAGCt. TTAAGACTATTATTATATATTTTTTTATCG |||||||||||||| 151 AATCGGTTTATCTTGCAGCTTTAAGACTATTATTATAT-TTTTTTt. ATCG 198 199 TTGTACAGTAAGAAGCTACATAAt. TTTTc. CTACCGc. CTA--TT-----gg |||||||||||||||||||| ||. | 200 TTGTACAGTAAGAAGCTACATAATTTTTCCTACCGCCTATTTTGGGGGAG 241 242 GGGGATTGTTGAATCAGTTAAGAATTAAAAGATGATGCTAt. TTCAG |||||||||| 250 GGGGGGGg. ATTGTTAAATCAGTCAAGAATT-AAAGATGATGCTATTTCAG 291 292 a. ATACt. Ta. AACtt. TTTTTAAGAC---------T-A-TAA-GTTTA ||. ||||||||| ||| | | ||. ||||| 299 AAAACTTCAACTTTTTTt. AAGACTATTTTTAATAATTAGTGTTTA 327 328 AATAACACTAATTATTa. AAAACTTGGTCTATCTTGGt. TTTAGGt |||||||||||||. |. |||| 349 AATAACACTAATTATTAAAAACTTGATCTTCGTCTTGGTCTAAGGT 478 378 TTTTCCTCTAGTTAATATTACTGTTACAACTACATAAAAACAATAAAATA ||. ||||||||||||||||||||. . ||499 399 TTGTCCTCTAGTTAATCTTACTGTTACAACTACATAAAAACAa. TAAGGTA 528 428 CTGTATCTTTGCAGATCCTATGAGCGGAACCACTTTTGACTGGGCGAAGA |||||||||||||||||||549 449 CTGTATCTTTGTAGATCCTATGAGCGGAACCACTTTt. GACTGGGCGAAGA 578 377 150 199 249 298 348 398 ATACAACAAATGTCCCATTTTCTTACCTGATTGAATTAAGAGACTTGGGG 427 ||. |||||||||||||||||||||||| ATGCAACAAATGTCCc. ATTTt. CTTACCTGATTGAATTAAGAGACTt. GGGg 448 CAATACGGTTTCTTGTTACCAGCAGAACAGATTATTCCAACTAATTTAGA 477 |||||||||||||||||| CAa. TACGGTTt. CTTGTTACc. AGCAGAACAGATTATACCAACTAATTt. AGA 498 AATAATGGATGCACTCCTGGAGATGGATAATACCGCAAGAACACTAg. GG |||||||||||||||. 599 AATAa. TGGATGCACTCc. TGGAGATGGATAACACCGCAAGAACACTAGGA 527 548 577 598 626 647
 What now? Still more sequencing needed. . . • target enrichment: 55 K 120 nt probes ? ? • 5’ SAGE • longer matepairs longer contigs & scaffolds annotation
What now? Still more sequencing needed. . . • target enrichment: 55 K 120 nt probes ? ? • 5’ SAGE • longer matepairs longer contigs & scaffolds annotation
 Challenges • no elegant solution for combining SOLi. D colorspace reads with other platforms in denovo assembly • read quality: filtering vs error correction • difficulties generating long matepairs • how to finish the assembly project: validation Goal: to get contigs/scaffolds useful for gene prediction
Challenges • no elegant solution for combining SOLi. D colorspace reads with other platforms in denovo assembly • read quality: filtering vs error correction • difficulties generating long matepairs • how to finish the assembly project: validation Goal: to get contigs/scaffolds useful for gene prediction
 What is the best assembler? • soap, velvet, Newbler, CLC bio, Celera • #contigs, contig lengths, accuracy
What is the best assembler? • soap, velvet, Newbler, CLC bio, Celera • #contigs, contig lengths, accuracy
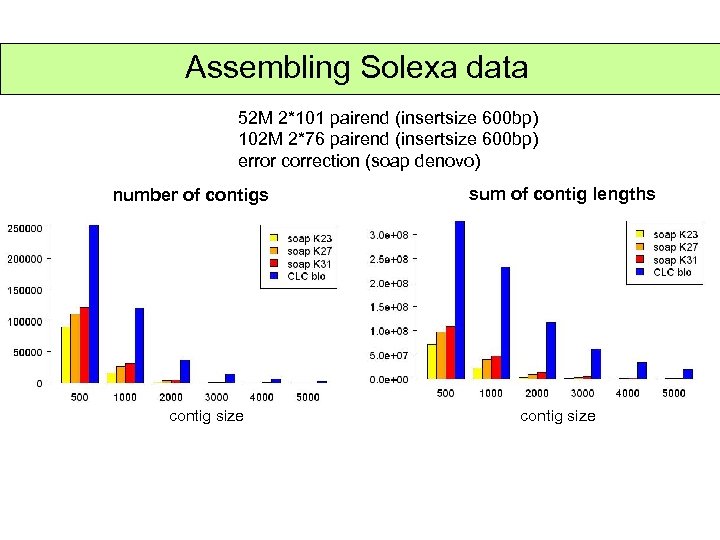 Assembling Solexa data 52 M 2*101 pairend (insertsize 600 bp) 102 M 2*76 pairend (insertsize 600 bp) error correction (soap denovo) number of contigs contig size sum of contig lengths contig size
Assembling Solexa data 52 M 2*101 pairend (insertsize 600 bp) 102 M 2*76 pairend (insertsize 600 bp) error correction (soap denovo) number of contigs contig size sum of contig lengths contig size
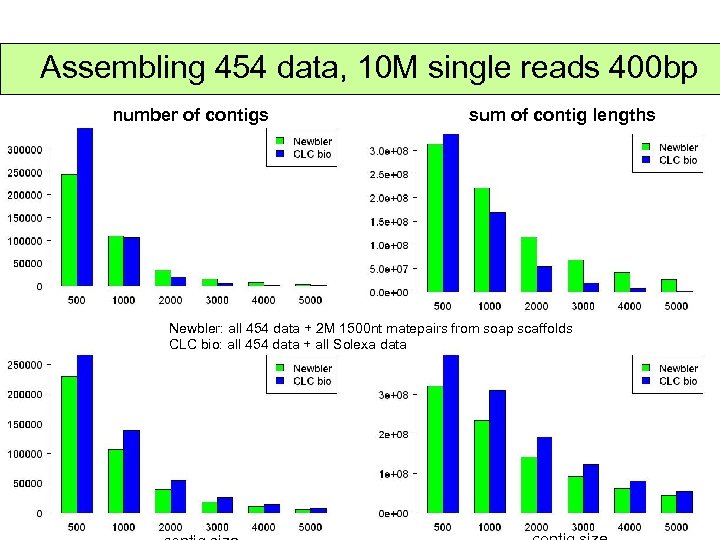 Assembling 454 data, 10 M single reads 400 bp number of contigs sum of contig lengths Newbler: all 454 data + 2 M 1500 nt matepairs from soap scaffolds CLC bio: all 454 data + all Solexa data
Assembling 454 data, 10 M single reads 400 bp number of contigs sum of contig lengths Newbler: all 454 data + 2 M 1500 nt matepairs from soap scaffolds CLC bio: all 454 data + all Solexa data
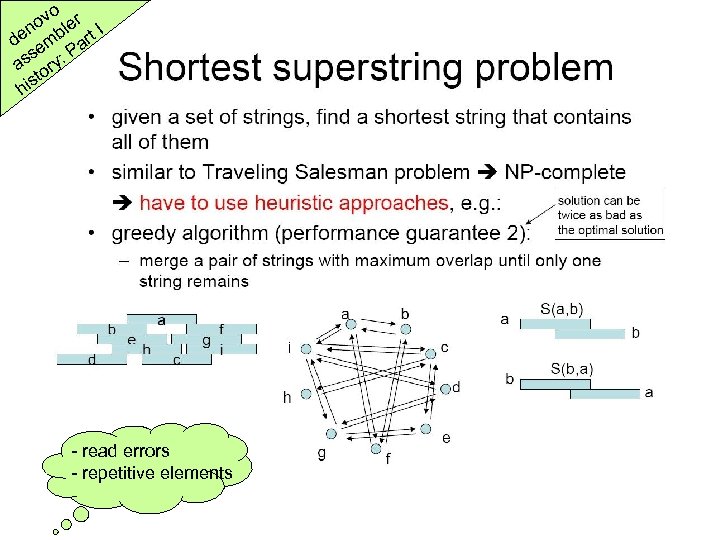 o ov ler I n de emb art P s as ory: t his - read errors - repetitive elements
o ov ler I n de emb art P s as ory: t his - read errors - repetitive elements
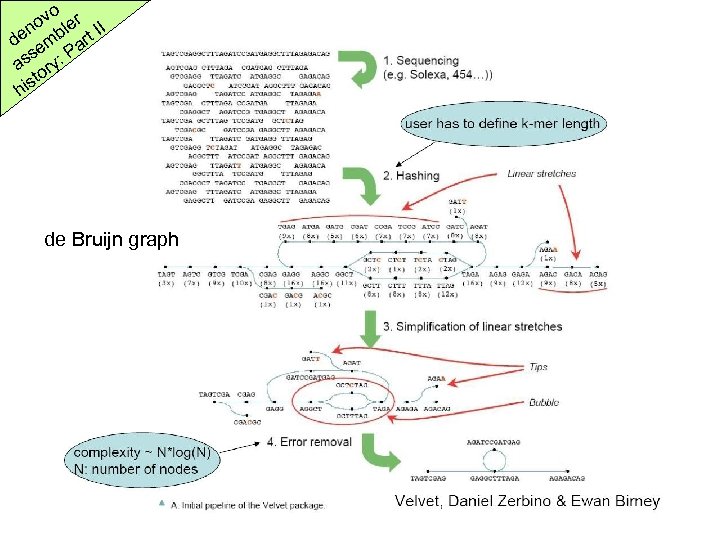 o ov ler II n de emb art P s as ory: ist h de Bruijn graph
o ov ler II n de emb art P s as ory: ist h de Bruijn graph


