e5e06e37ddd324897bd41ba5cd47937f.ppt
- Количество слайдов: 45

CS 5263 Bioinformatics Lecture 22 Introduction to Microarray
Outline • • What is microarray Basic categories of microarray How can microarray be used Computational and statistical methods involved in microarray – – – Probe design Image processing Pre-processing Differentially expressed gene identification Clustering / classification Network / pathway modeling
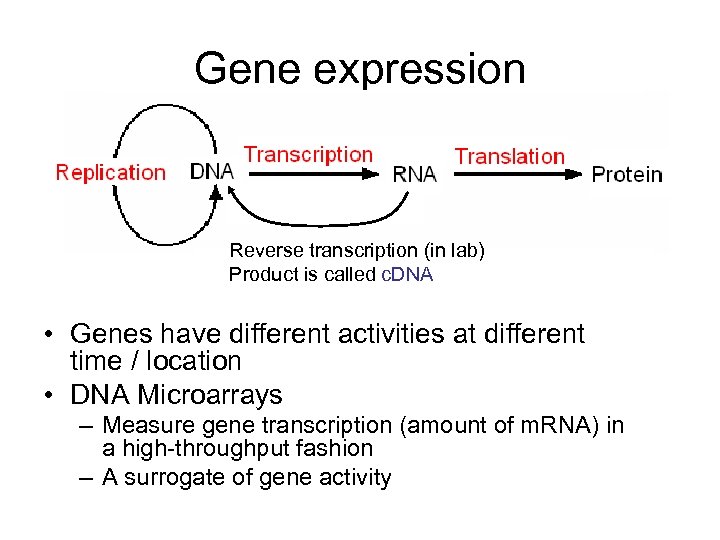
Gene expression Reverse transcription (in lab) Product is called c. DNA • Genes have different activities at different time / location • DNA Microarrays – Measure gene transcription (amount of m. RNA) in a high-throughput fashion – A surrogate of gene activity
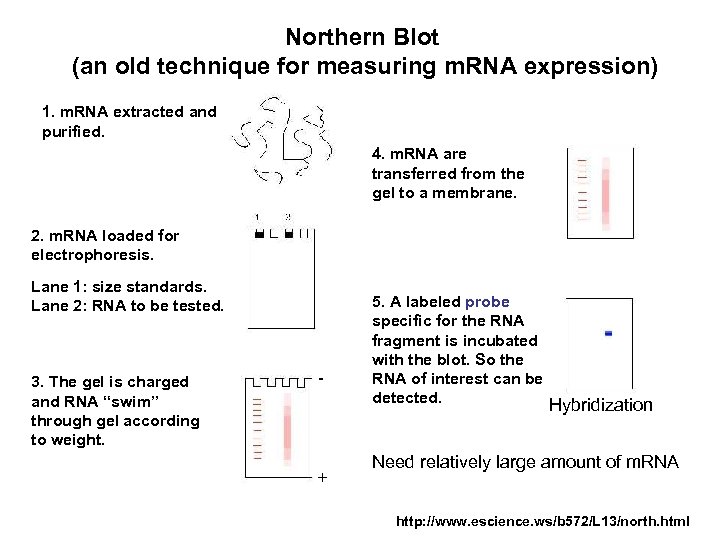
Northern Blot (an old technique for measuring m. RNA expression) 1. m. RNA extracted and purified. 4. m. RNA are transferred from the gel to a membrane. 2. m. RNA loaded for electrophoresis. Lane 1: size standards. Lane 2: RNA to be tested. 3. The gel is charged and RNA “swim” through gel according to weight. - + 5. A labeled probe specific for the RNA fragment is incubated with the blot. So the RNA of interest can be detected. Hybridization Need relatively large amount of m. RNA http: //www. escience. ws/b 572/L 13/north. html
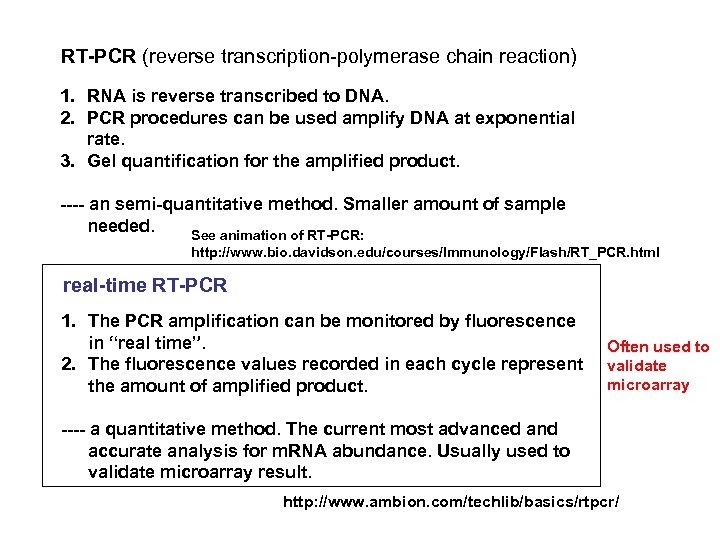
RT-PCR (reverse transcription-polymerase chain reaction) 1. RNA is reverse transcribed to DNA. 2. PCR procedures can be used amplify DNA at exponential rate. 3. Gel quantification for the amplified product. ---- an semi-quantitative method. Smaller amount of sample needed. See animation of RT-PCR: http: //www. bio. davidson. edu/courses/Immunology/Flash/RT_PCR. html real-time RT-PCR 1. The PCR amplification can be monitored by fluorescence in “real time”. 2. The fluorescence values recorded in each cycle represent the amount of amplified product. Often used to validate microarray ---- a quantitative method. The current most advanced and accurate analysis for m. RNA abundance. Usually used to validate microarray result. http: //www. ambion. com/techlib/basics/rtpcr/
Limitation of the old techniques 1. Labor intensive 2. Can only detect up to dozens of genes. (gene-by-gene analysis)
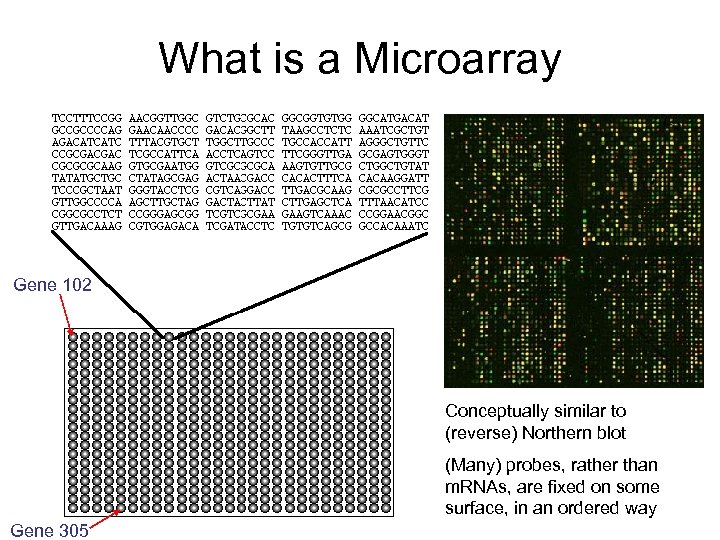
What is a Microarray Gene 102 Conceptually similar to (reverse) Northern blot (Many) probes, rather than m. RNAs, are fixed on some surface, in an ordered way Gene 305
What is a microarray (2) • A 2 D array of DNA sequences from thousands of genes • Each spot has many copies of same gene (probe) • Allow m. RNAs from a sample to hybridize • Measure number of hybridizations per spot
Goals of a Microarray Experiment 1. Find the genes that change expression between experimental and control samples 2. Classify samples based on a gene expression profile 3. Find patterns: Groups of biologically related genes that change expression together across samples/treatments
Microarray categories • c. DNAs microarray – Each probe is the c. DNA of a gene (hundreds to thousands bp) – Stanford, Brown Lab • Oligonucleotide microarray – Each probe is a synthesized short DNA (uniquely corresponding to a substring of a gene) – Affymetrix: ~ 25 mers – Aglient: ~ 60 mers • Others

Spotted c. DNA microarray
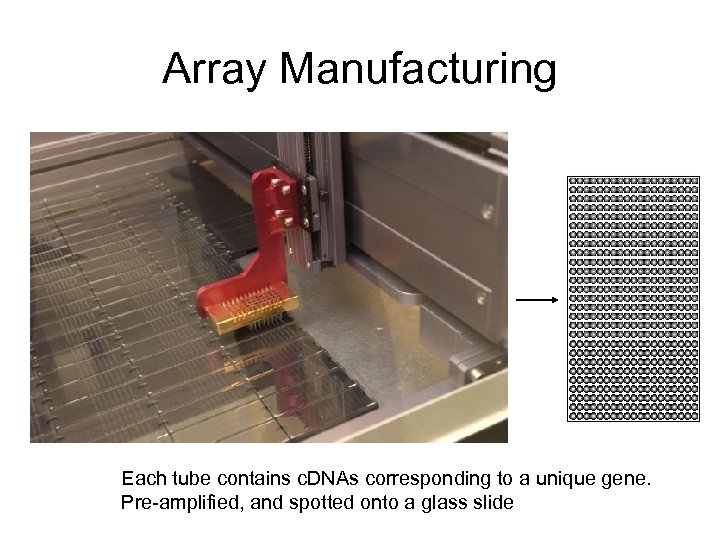
Array Manufacturing Each tube contains c. DNAs corresponding to a unique gene. Pre-amplified, and spotted onto a glass slide
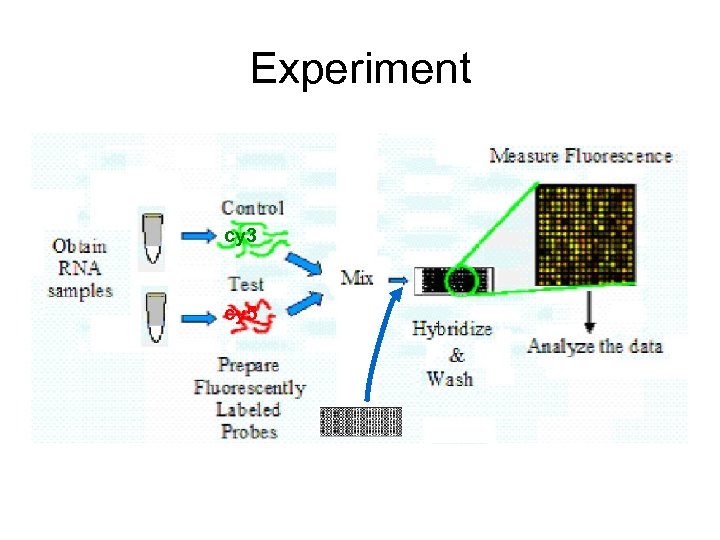
Experiment cy 3 cy 5
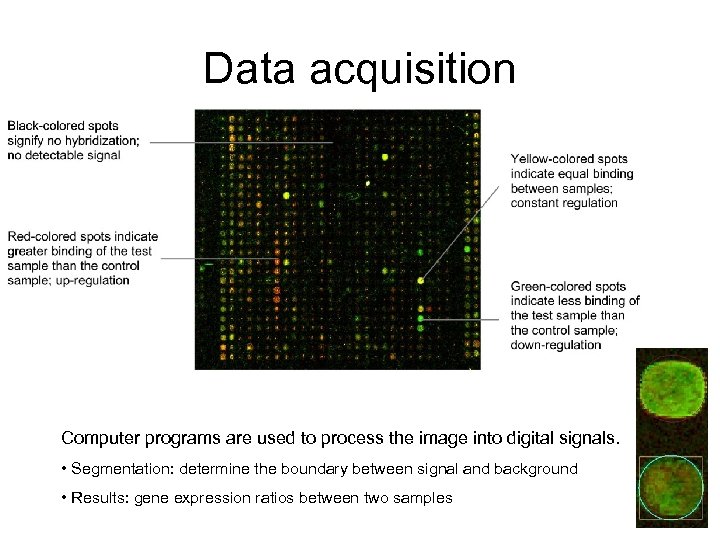
Data acquisition Computer programs are used to process the image into digital signals. • Segmentation: determine the boundary between signal and background • Results: gene expression ratios between two samples

c. DNA Microarray Methodology Animation

Affymetrix Gene. Chip®
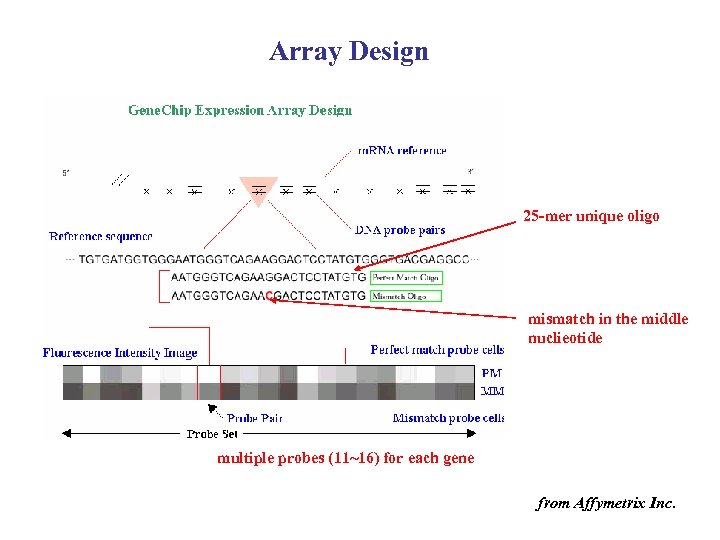
Array Design 25 -mer unique oligo mismatch in the middle nuclieotide multiple probes (11~16) for each gene from Affymetrix Inc.
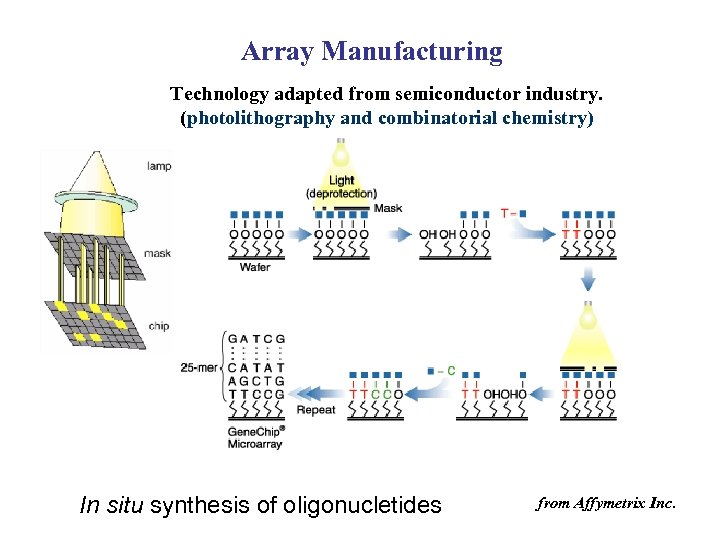
Array Manufacturing Technology adapted from semiconductor industry. (photolithography and combinatorial chemistry) In situ synthesis of oligonucletides from Affymetrix Inc.
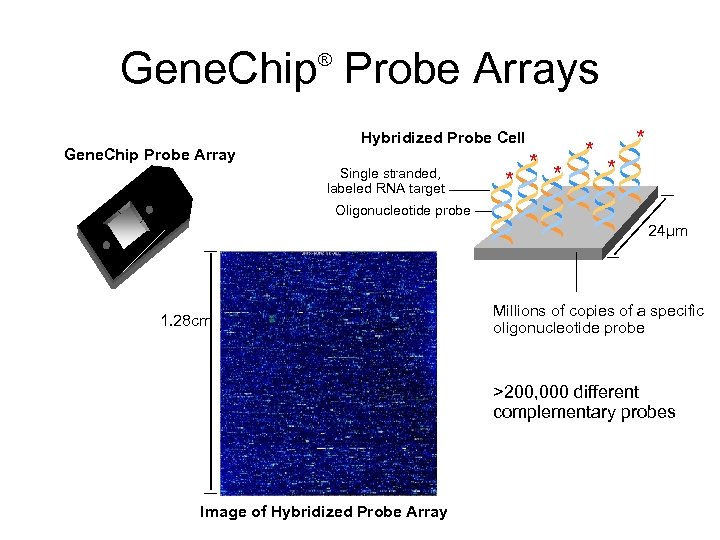
Gene. Chip Probe Arrays ® Gene. Chip Probe Array Hybridized Probe Cell Single stranded, labeled RNA target * * * Oligonucleotide probe 24µm 1. 28 cm Millions of copies of a specific oligonucleotide probe >200, 000 different complementary probes Image of Hybridized Probe Array
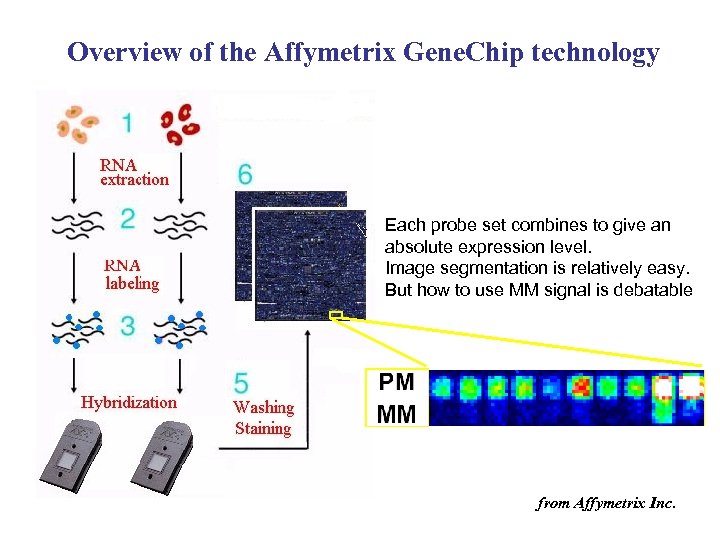
Overview of the Affymetrix Gene. Chip technology Each probe set combines to give an absolute expression level. Image segmentation is relatively easy. But how to use MM signal is debatable from Affymetrix Inc.
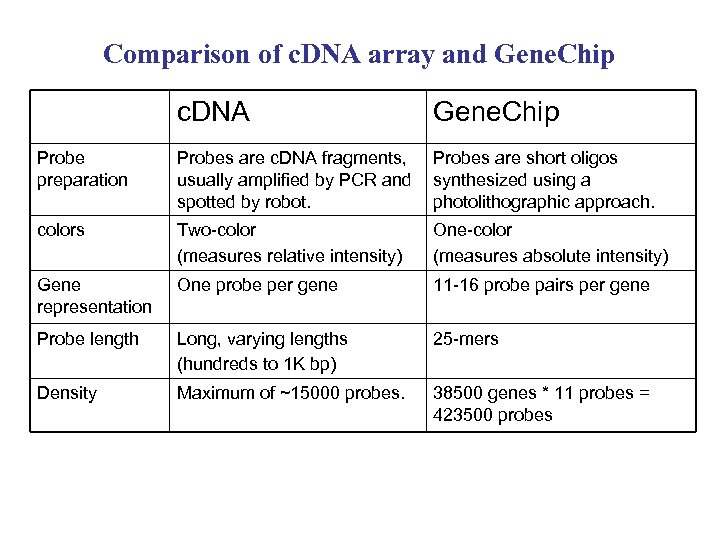
Comparison of c. DNA array and Gene. Chip c. DNA Gene. Chip Probe preparation Probes are c. DNA fragments, usually amplified by PCR and spotted by robot. Probes are short oligos synthesized using a photolithographic approach. colors Two-color (measures relative intensity) One-color (measures absolute intensity) Gene representation One probe per gene 11 -16 probe pairs per gene Probe length Long, varying lengths (hundreds to 1 K bp) 25 -mers Density Maximum of ~15000 probes. 38500 genes * 11 probes = 423500 probes
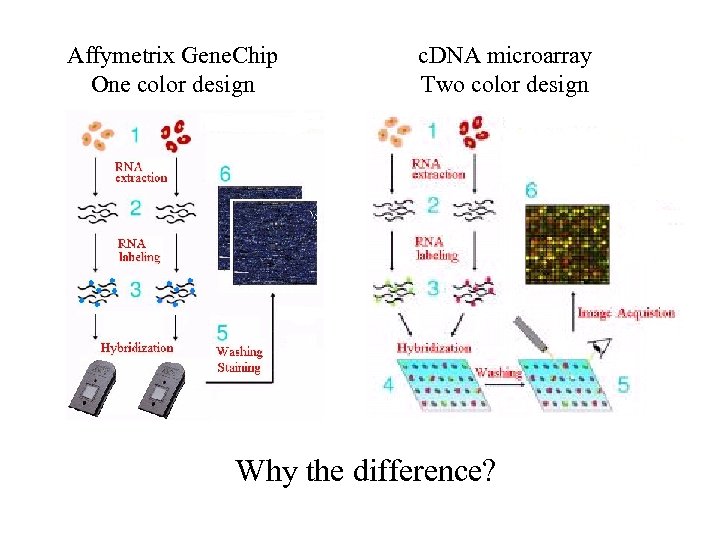
Affymetrix Gene. Chip One color design c. DNA microarray Two color design Why the difference?
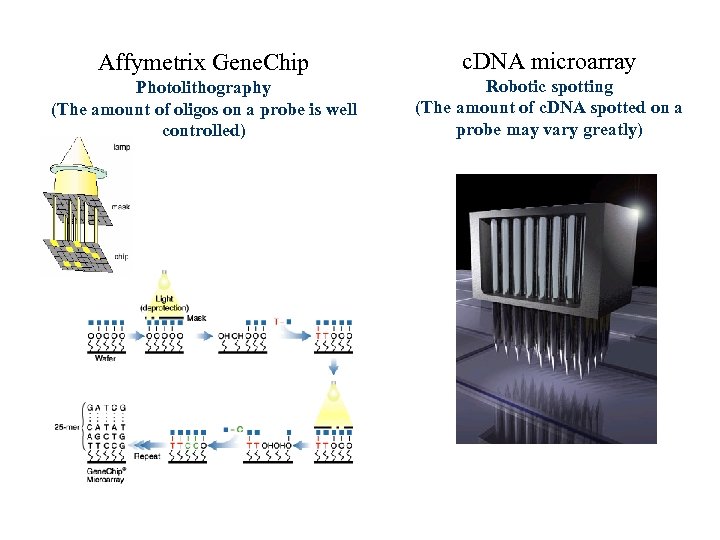
Affymetrix Gene. Chip c. DNA microarray Photolithography (The amount of oligos on a probe is well controlled) Robotic spotting (The amount of c. DNA spotted on a probe may vary greatly)
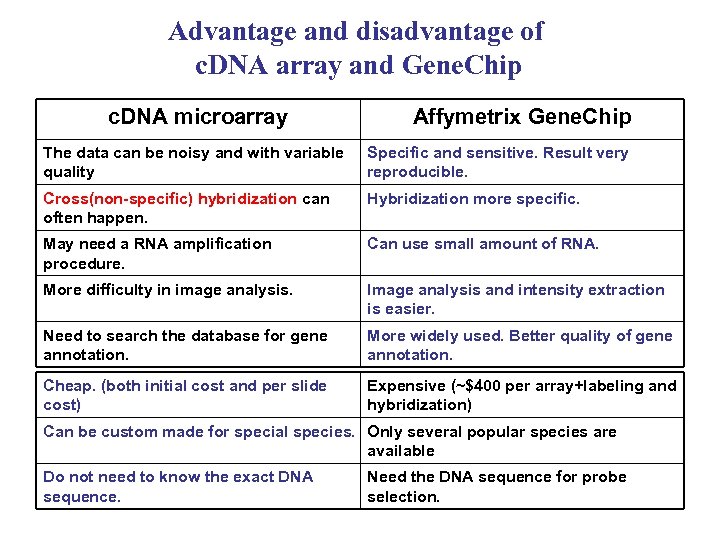
Advantage and disadvantage of c. DNA array and Gene. Chip c. DNA microarray Affymetrix Gene. Chip The data can be noisy and with variable quality Specific and sensitive. Result very reproducible. Cross(non-specific) hybridization can often happen. Hybridization more specific. May need a RNA amplification procedure. Can use small amount of RNA. More difficulty in image analysis. Image analysis and intensity extraction is easier. Need to search the database for gene annotation. More widely used. Better quality of gene annotation. Cheap. (both initial cost and per slide cost) Expensive (~$400 per array+labeling and hybridization) Can be custom made for special species. Only several popular species are available Do not need to know the exact DNA sequence. Need the DNA sequence for probe selection.
Computational aspects • • • Probe design Image processing Pre-processing Differentially expressed gene identification Clustering / classification Network / pathway modeling

First step: pre-processing • Transformation – Transforms intensities or ratios to a different scale – Why? • For convenience • Convert data into a certain distribution (e. g. normal) assumed by many other statistical procedures • Normalization – Correct for systematic errors – Make data from different samples comparable Garbage in => Garbage out

Where errors could come from? • Random errors – Repeat the same experiment twice, get diff results – Using multiple replicates reduces the problem • Systematic errors – Arrays manufactured at different time – On the same array, probes printed with different printer tips may have different biases – Dye effect: difference between Cy 5 and Cy 3 labeling – Experimental factors • Array A being applied more m. RNAs than array B • Sample preparation procedure • Experiments carried out at different time, by different users, etc.

c. DNA microarray data preprocessing
Typical experiments Wide-type cells vs mutated cells Diseased cells with normal cells Cells under normal growth condition vs cells treated with chemicals Typically repeated for several times Ratios Probes (genes) • •
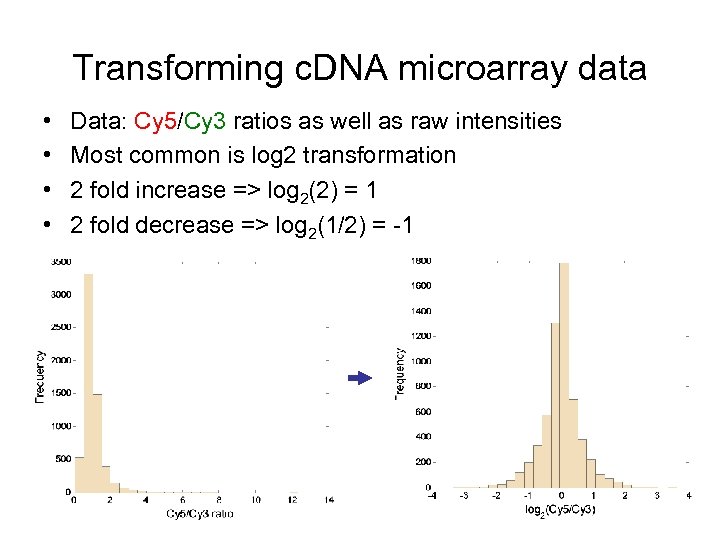
Transforming c. DNA microarray data • • Data: Cy 5/Cy 3 ratios as well as raw intensities Most common is log 2 transformation 2 fold increase => log 2(2) = 1 2 fold decrease => log 2(1/2) = -1
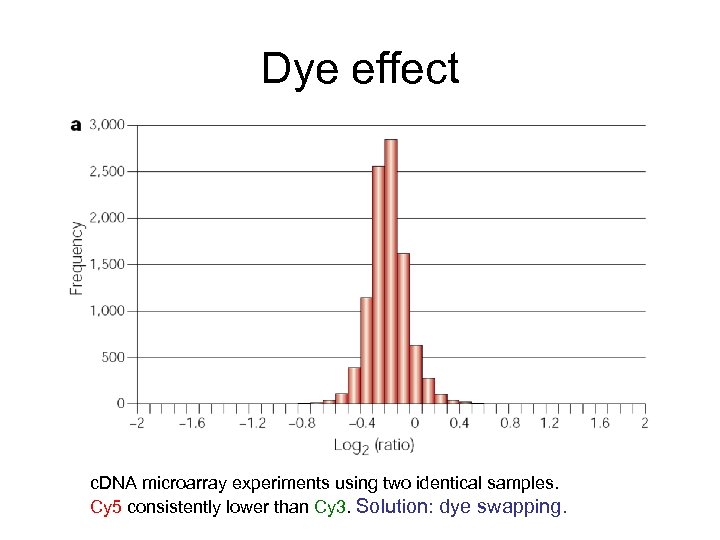
Dye effect c. DNA microarray experiments using two identical samples. Cy 5 consistently lower than Cy 3. Solution: dye swapping.

Dye swapping • • • Chip 1: label test by cy 5 and control by cy 3 Chip 2: label test by cy 3 and control by cy 5 Ideally cy 5/cy 3 = cy 3/cy 5 Not so due to dye effect Compute average ratio: ½ log 2 (cy 5/cy 3 on chip 1) + ½ log 2 (cy 3/cy 5 on chip 2)
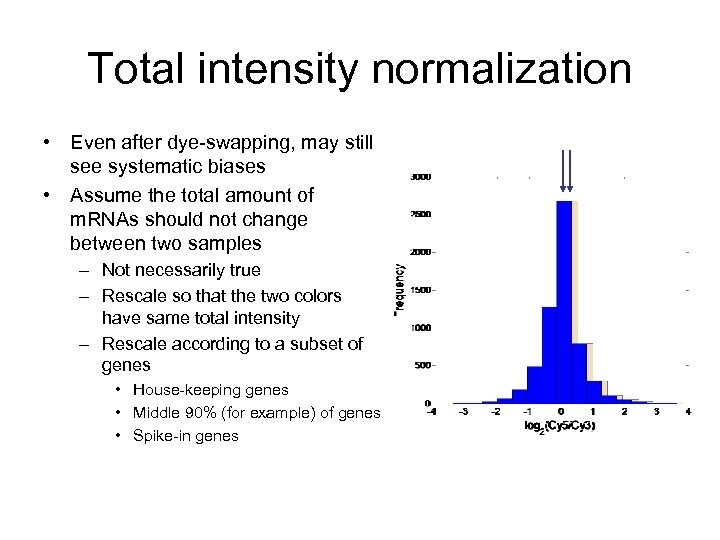
Total intensity normalization • Even after dye-swapping, may still see systematic biases • Assume the total amount of m. RNAs should not change between two samples – Not necessarily true – Rescale so that the two colors have same total intensity – Rescale according to a subset of genes • House-keeping genes • Middle 90% (for example) of genes • Spike-in genes
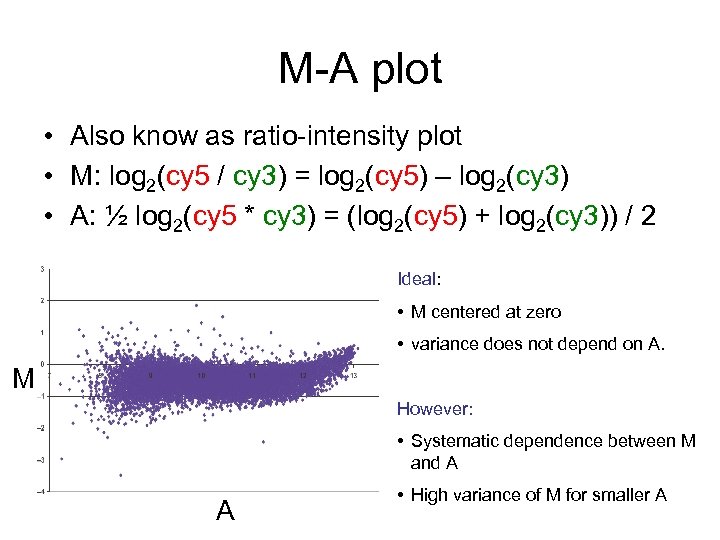
M-A plot • Also know as ratio-intensity plot • M: log 2(cy 5 / cy 3) = log 2(cy 5) – log 2(cy 3) • A: ½ log 2(cy 5 * cy 3) = (log 2(cy 5) + log 2(cy 3)) / 2 Ideal: • M centered at zero • variance does not depend on A. M However: • Systematic dependence between M and A A • High variance of M for smaller A
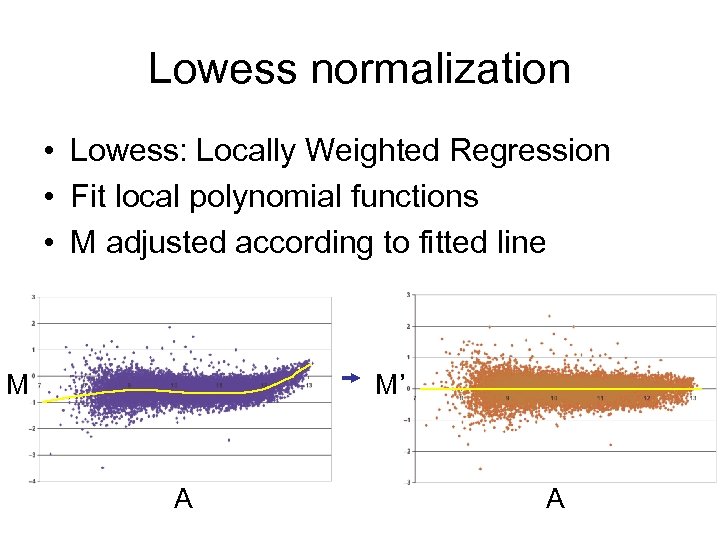
Lowess normalization • Lowess: Locally Weighted Regression • Fit local polynomial functions • M adjusted according to fitted line M M’ A A
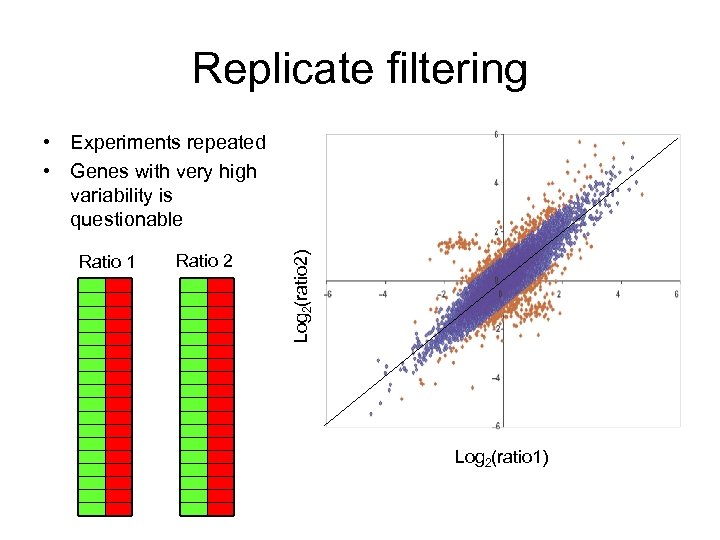
Replicate filtering Ratio 1 Ratio 2 Log 2(ratio 2) • Experiments repeated • Genes with very high variability is questionable Log 2(ratio 1)

oligo microarray data preprocessing (Affymetrix chip)

Typical experiments • Multiple microarrays – n samples (from different time, location, condition, treatment, etc. ) – k replicates for each samples • For example – Samples collected from 100 healthy people and 100 cancer patients – Cells treated with some drugs, take samples every 10 minutes • Repeat on 3 – 5 microarrays for each sample – Improve reliability of the results – Often averaged after some preprocessing
Main characteristics • For each gene, there are multiple PM and MM probes (11 -16 pairs) – how to obtain overall intensities from these probe-level intensities? • Array outputs are absolute values rather than ratios – Cross-array normalization is important for them to be comparable
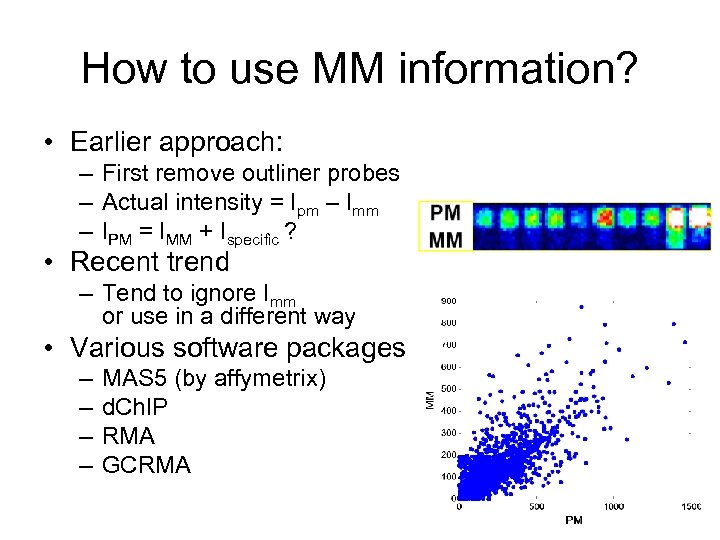
How to use MM information? • Earlier approach: – First remove outliner probes – Actual intensity = Ipm – Imm – IPM = IMM + Ispecific ? • Recent trend – Tend to ignore Imm or use in a different way • Various software packages – – MAS 5 (by affymetrix) d. Ch. IP RMA GCRMA
Normalization • Similar to c. DNA microarrays • Total intensity normalization – Each array has the same mean intensity – Can be based on all genes or a selected subset of genes • House-keeping genes • Middle 90% (for example) of genes • Spike-in genes • Lowess with a common reference • Many useful tools implemented in Bioconductor

Conclusions • Microarray provides a way to measure thousands of genes simultaneously and make the global monitoring of cellular activities possible. • The method produces noisy data and normalization is crucial. • Real Time RT-PCR for validation of small number of genes.
Limitation • Measures m. RNA instead of proteins. Actual protein abundance and post-translation modification can not be detected. • Suitable for global monitoring and should be used to generate further hypothesis or should combine with other carefully designed experiments.

Microarray preproc questions • What kind of array it is? – – Two-color? One-color? Oligo array? c. DNA array? • How is the experiment designed? – Time series? – Test vs control? • What kind of preprocessing has been done? – What value: raw intensity value or ratios? – Transformation? Log scale? Linear scale? – Normalization: within-array? Cross-array? • What are the next steps you want to proceed? – Identifying differentially expressed genes? – Clustering?

Some real data • Joseph L. De. Risi, Vishwanath R. Iyer, Patrick O. Brown, “Exploring the Metabolic and Genetic Control of Gene Expression on a Genomic Scale”, Science, 278: 680 – 686, 1997
e5e06e37ddd324897bd41ba5cd47937f.ppt