ab21f2abe09c081183bf07734c1e097b.ppt
- Количество слайдов: 18

• Criblage virtuel Alexandre Varnek Faculté de Chimie, ULP, Strasbourg, FRANCE
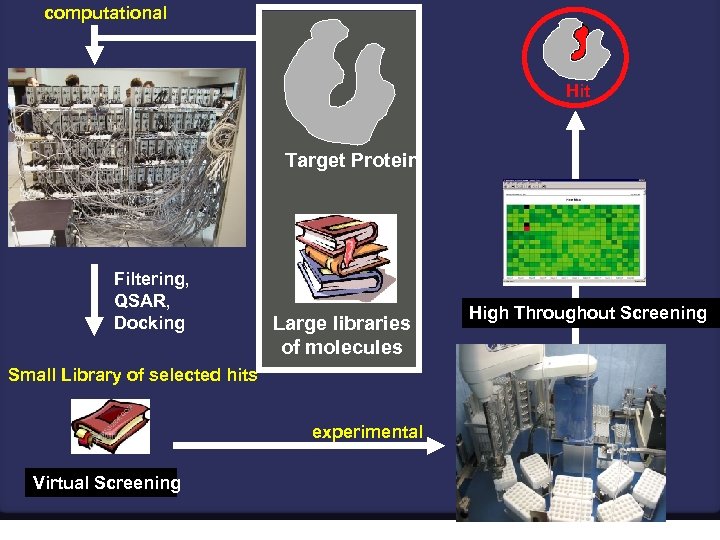
computational Hit Target Protein Filtering, QSAR, Docking Large libraries of molecules Small Library of selected hits experimental Virtual Screening High Throughout Screening
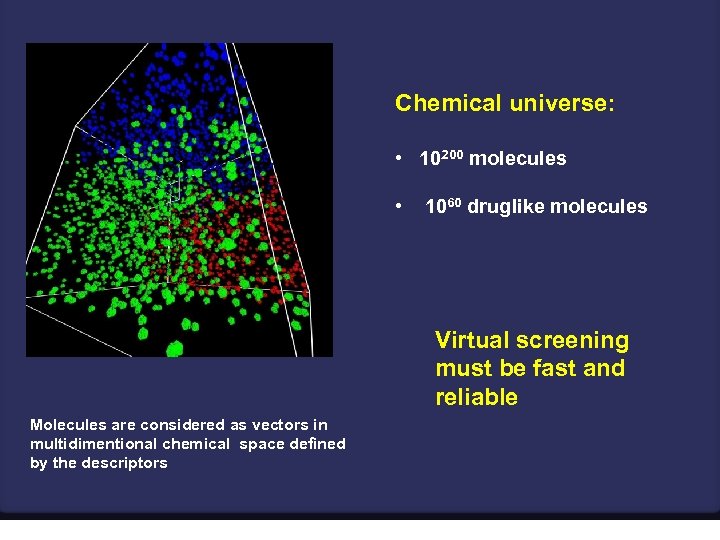
Chemical universe: • 10200 molecules • 1060 druglike molecules Virtual screening must be fast and reliable Molecules are considered as vectors in multidimentional chemical space defined by the descriptors
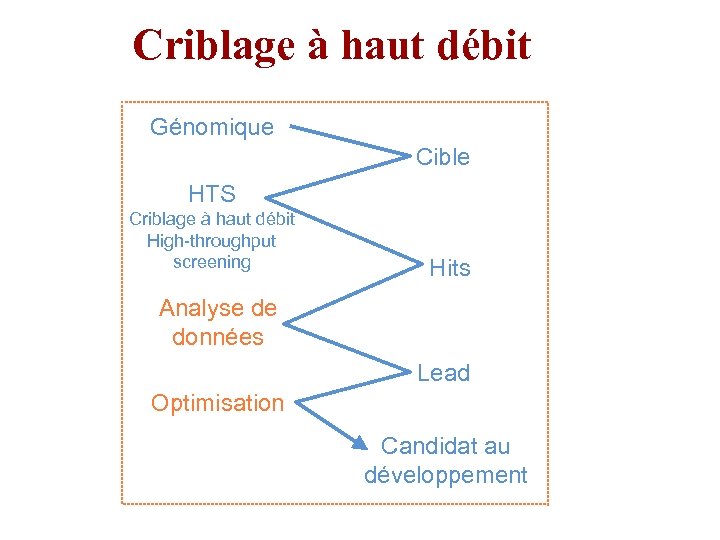
Criblage à haut débit Génomique Cible HTS Criblage à haut débit High-throughput screening Hits Analyse de données Lead Optimisation Candidat au développement
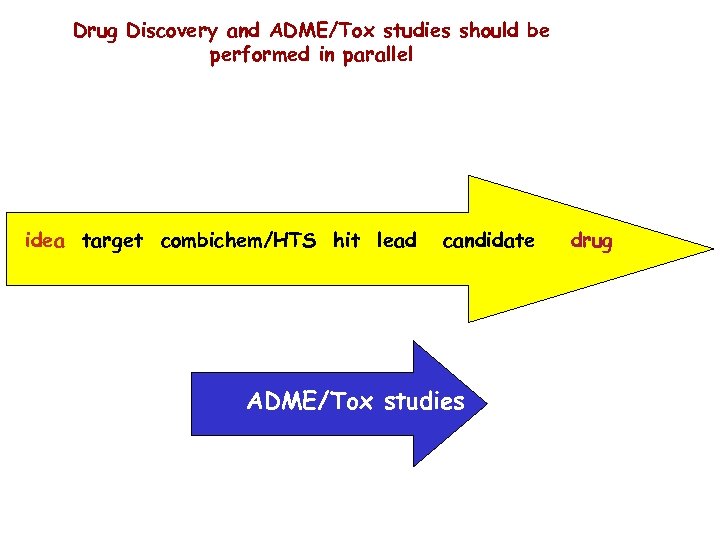
Drug Discovery and ADME/Tox studies should be performed in parallel idea target combichem/HTS hit lead candidate ADME/Tox studies drug
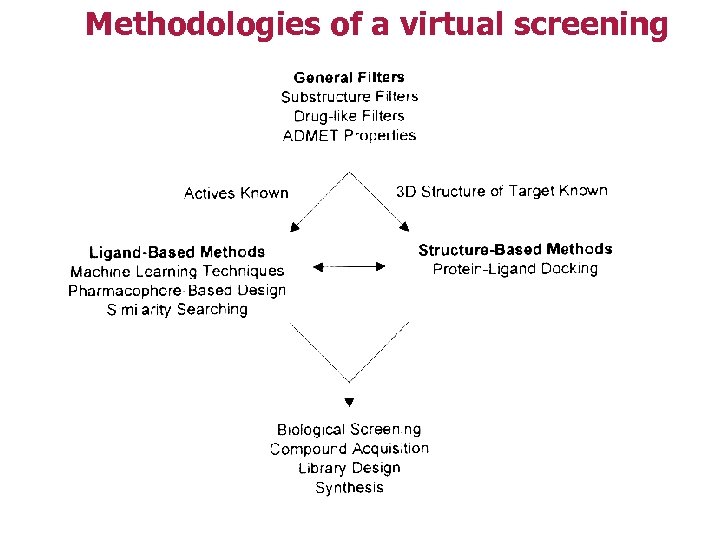
Methodologies of a virtual screening
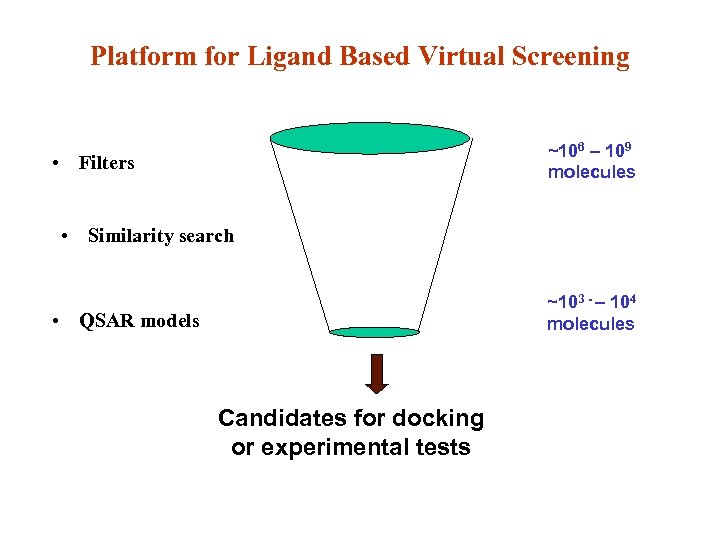
Platform for Ligand Based Virtual Screening ~106 – 109 molecules • Filters • Similarity search ~103 - – 104 molecules • QSAR models Candidates for docking or experimental tests
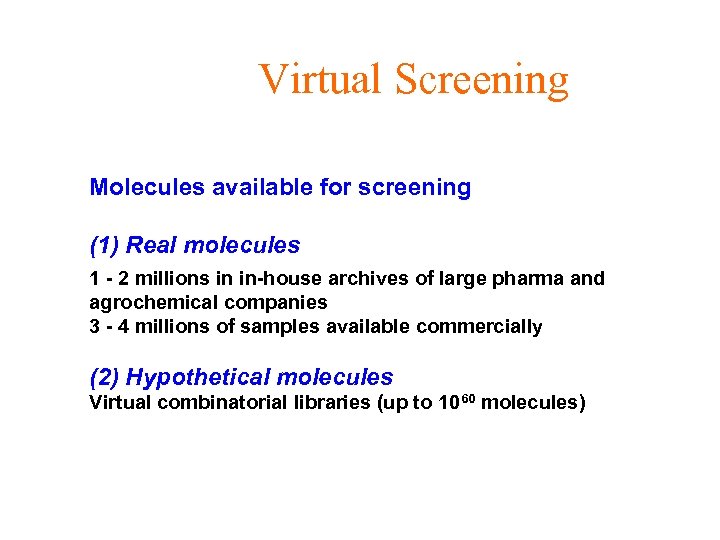
Virtual Screening Molecules available for screening (1) Real molecules 1 - 2 millions in in-house archives of large pharma and agrochemical companies 3 - 4 millions of samples available commercially (2) Hypothetical molecules Virtual combinatorial libraries (up to 1060 molecules)
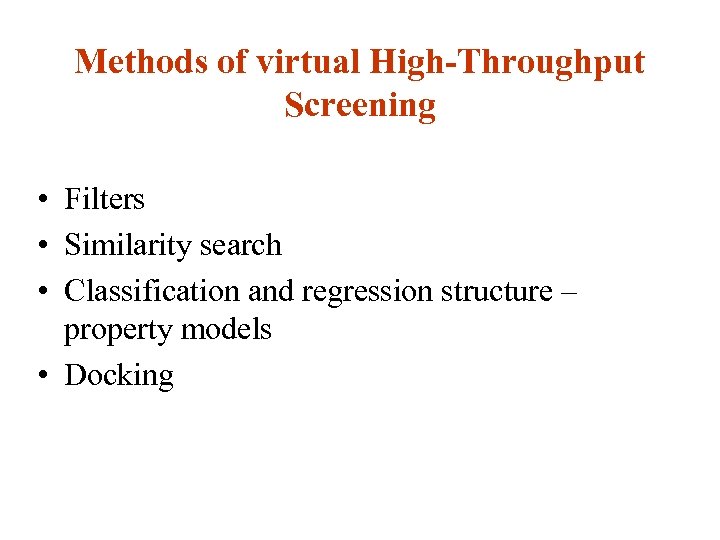
Methods of virtual High-Throughput Screening • Filters • Similarity search • Classification and regression structure – property models • Docking

Filters: Lipinski rules for drug-like molecules ( « Rules of 5 » ) • H-bond donors < 5 • (the sum of OH and NH groups); • MWT < 500; • Log. P < 5 • H-bond acceptors < 10 (the sum of N and O atoms without H attached).
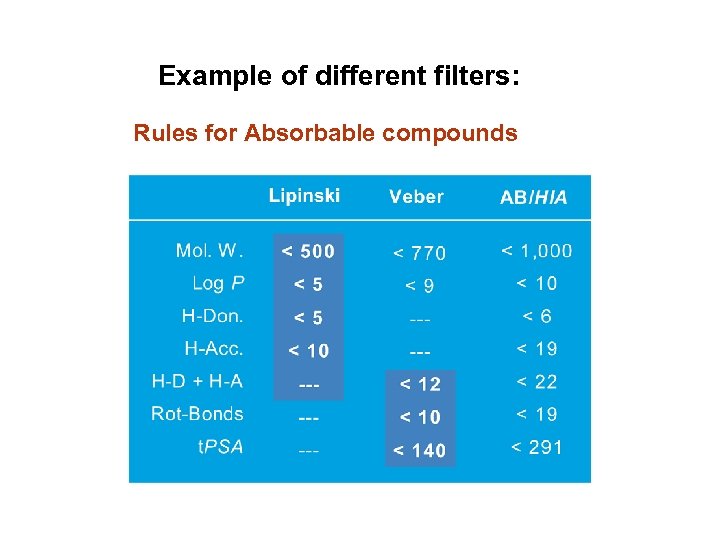
Example of different filters: Rules for Absorbable compounds

Similarity Search: unsupervised and supervised approaches
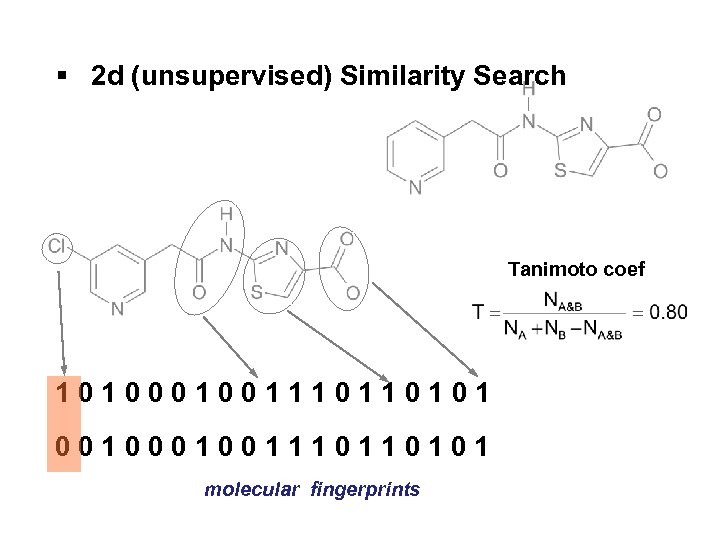
§ 2 d (unsupervised) Similarity Search Tanimoto coef 101001110110101 001001110110101 molecular fingerprints
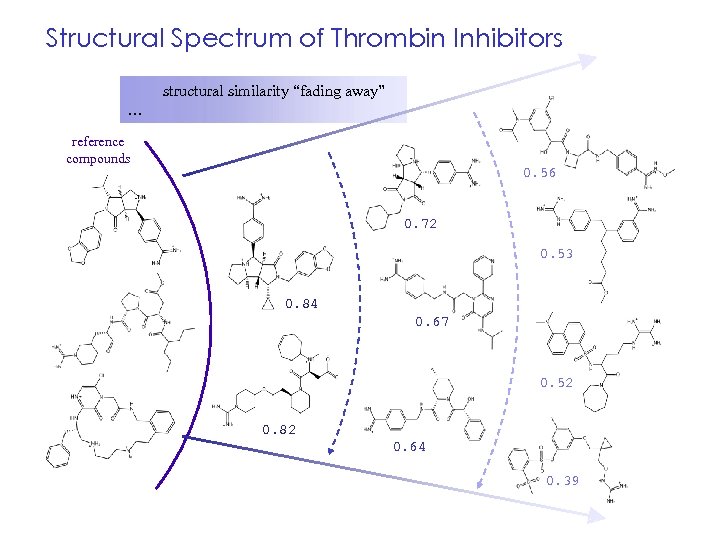
Structural Spectrum of Thrombin Inhibitors structural similarity “fading away” … reference compounds 0. 56 0. 72 0. 53 0. 84 0. 67 0. 52 0. 82 0. 64 0. 39
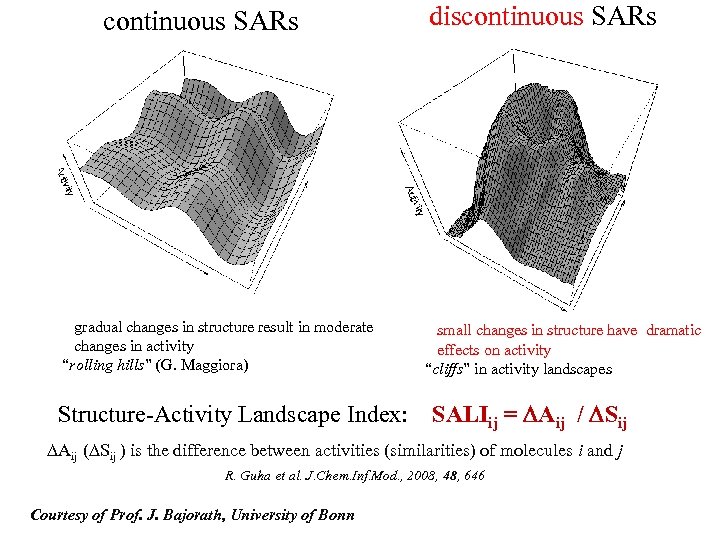
continuous SARs gradual changes in structure result in moderate changes in activity “rolling hills” (G. Maggiora) Structure-Activity Landscape Index: discontinuous SARs small changes in structure have dramatic effects on activity “cliffs” in activity landscapes SALIij = DAij / DSij DAij (DSij ) is the difference between activities (similarities) of molecules i and j R. Guha et al. J. Chem. Inf. Mod. , 2008, 48, 646 Courtesy of Prof. J. Bajorath, University of Bonn
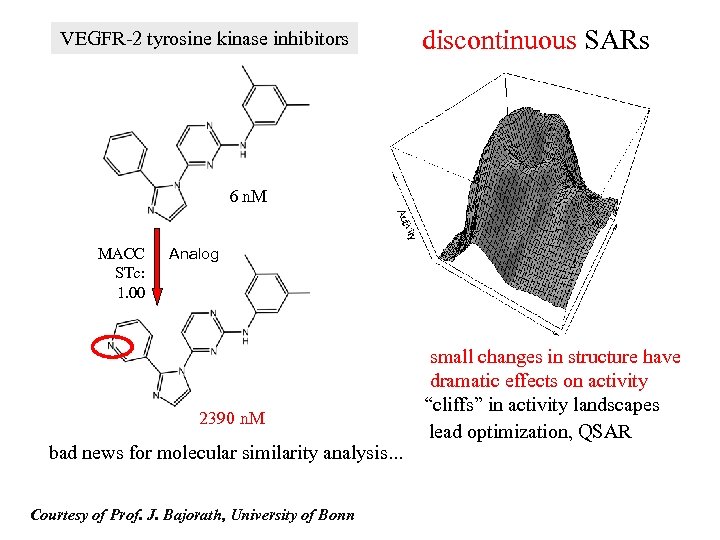
VEGFR-2 tyrosine kinase inhibitors discontinuous SARs 6 n. M MACC STc: 1. 00 Analog 2390 n. M bad news for molecular similarity analysis. . . Courtesy of Prof. J. Bajorath, University of Bonn small changes in structure have dramatic effects on activity “cliffs” in activity landscapes lead optimization, QSAR
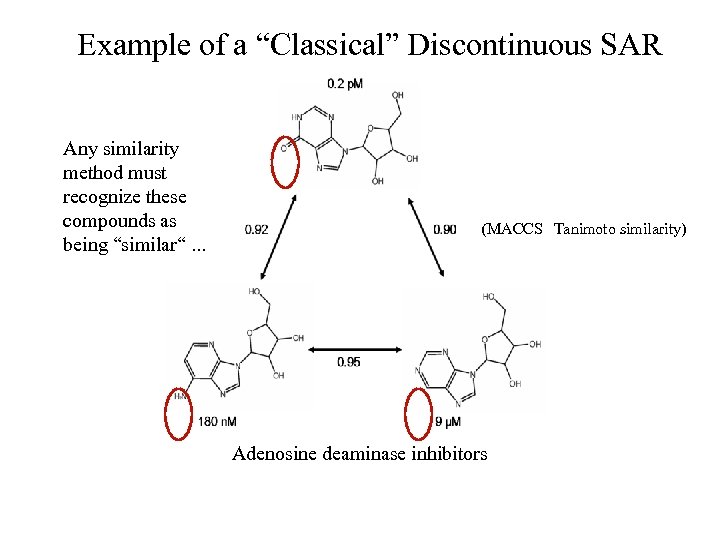
Example of a “Classical” Discontinuous SAR Any similarity method must recognize these compounds as being “similar“. . . (MACCS Tanimoto similarity) Adenosine deaminase inhibitors
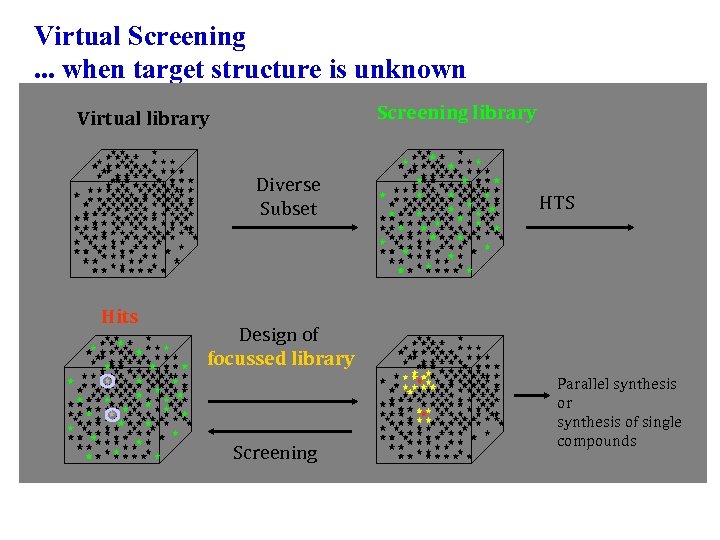
Virtual Screening. . . when target structure is unknown Screening library Virtual library Diverse Subset Hits HTS Design of focussed library Screening Parallel synthesis or synthesis of single compounds
ab21f2abe09c081183bf07734c1e097b.ppt