8c3fb4b1da91cf5c4e9a70b21c118c56.ppt
- Количество слайдов: 51
 Courtroom Knowledge of Forensic Technology Workshop Forensic DNA Analysis and New Technologies John Picciano Automation/Validation/Research Supervisor Pennsylvania State Police Forensic DNA Division
Courtroom Knowledge of Forensic Technology Workshop Forensic DNA Analysis and New Technologies John Picciano Automation/Validation/Research Supervisor Pennsylvania State Police Forensic DNA Division
 Agenda • • • The Basics of Genetics DNA Structure and Function Types of evidence The Process of Creating a DNA Profile Example DNA Profiles What DNA Can/Can’t Tell You Report Wording DNA Statistics CODIS Q&A
Agenda • • • The Basics of Genetics DNA Structure and Function Types of evidence The Process of Creating a DNA Profile Example DNA Profiles What DNA Can/Can’t Tell You Report Wording DNA Statistics CODIS Q&A
 My Attempt at Legalese • Disclaimer: – Any discussion of manufacturers, vendors, or companies in the following presentation should not be considered as endorsed by the Pennsylvania State Police – Furthermore opinions offered may be considered my own and not necessarily reflect the official stance of the Pennsylvania State Police or the Forensic DNA Division.
My Attempt at Legalese • Disclaimer: – Any discussion of manufacturers, vendors, or companies in the following presentation should not be considered as endorsed by the Pennsylvania State Police – Furthermore opinions offered may be considered my own and not necessarily reflect the official stance of the Pennsylvania State Police or the Forensic DNA Division.
 Basics of Genetics The genome of each individual is unique (with the exception of identical twins) and is inherited from parents Unit of heredity is DNA is located in the nucleus of cells packaged into 23 pairs of chromosomes (46 total) Half of your DNA comes from your mother and half from your father DNA is the molecule that governs all of your physical and chemical traits (hair/eye/skin color, gender, height, body chemistry)
Basics of Genetics The genome of each individual is unique (with the exception of identical twins) and is inherited from parents Unit of heredity is DNA is located in the nucleus of cells packaged into 23 pairs of chromosomes (46 total) Half of your DNA comes from your mother and half from your father DNA is the molecule that governs all of your physical and chemical traits (hair/eye/skin color, gender, height, body chemistry)
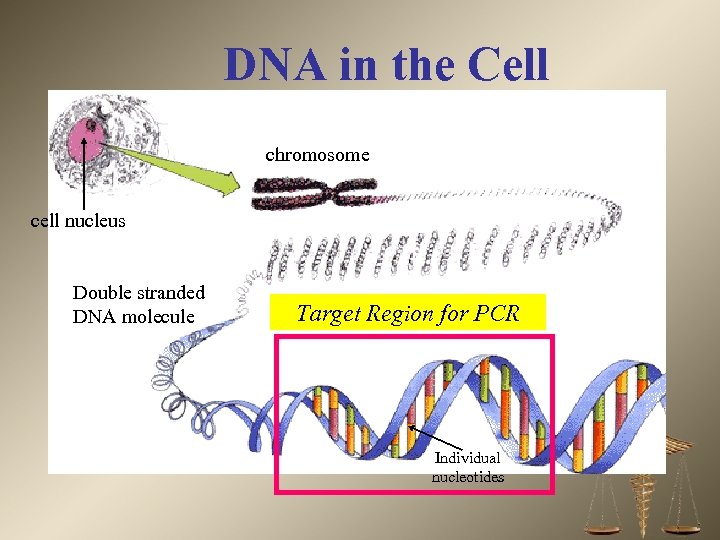 DNA in the Cell chromosome cell nucleus Double stranded DNA molecule Target Region for PCR Individual nucleotides
DNA in the Cell chromosome cell nucleus Double stranded DNA molecule Target Region for PCR Individual nucleotides
 Genetic Terms • Allele: For forensic applications, an allele refers to the number of short tandem repeats (STR) at a specific location. • You get one from each parent. • If you get the same allele from each parent, you are a “homozygote” at that location. • If you get two different alleles from each parent, you are a “heterozygote” at that location.
Genetic Terms • Allele: For forensic applications, an allele refers to the number of short tandem repeats (STR) at a specific location. • You get one from each parent. • If you get the same allele from each parent, you are a “homozygote” at that location. • If you get two different alleles from each parent, you are a “heterozygote” at that location.
 Genetic Terms • Locus: a unique physical location of a gene on a chromosome • Loci: plural of locus • For forensic purposes a locus is the specific location of the STR alleles on a chromosome
Genetic Terms • Locus: a unique physical location of a gene on a chromosome • Loci: plural of locus • For forensic purposes a locus is the specific location of the STR alleles on a chromosome
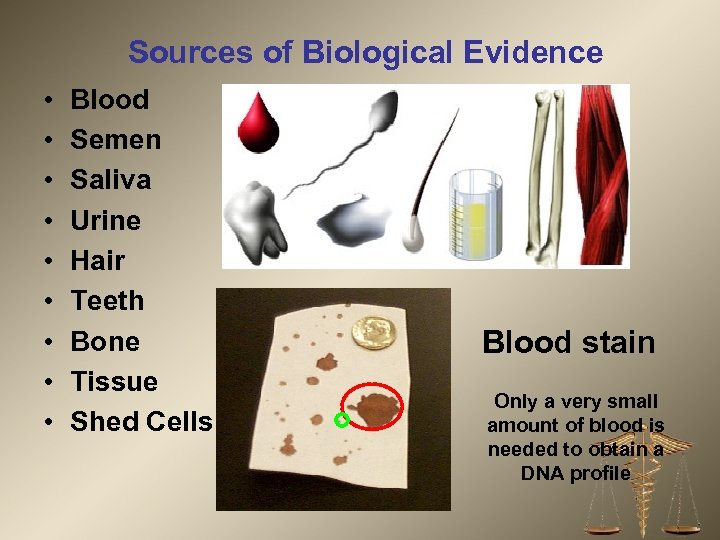 Sources of Biological Evidence • • • Blood Semen Saliva Urine Hair Teeth Bone Tissue Shed Cells Blood stain Only a very small amount of blood is needed to obtain a DNA profile
Sources of Biological Evidence • • • Blood Semen Saliva Urine Hair Teeth Bone Tissue Shed Cells Blood stain Only a very small amount of blood is needed to obtain a DNA profile
 The Cornerstones of DNA Analysis • Extraction – Isolation and purification of DNA from evidence • Quantitation – Determination of how much human/human male DNA present in a sample • Amplification – Making copies of STR’s • Analysis – Determine matches, inclusions, exclusions and statistics
The Cornerstones of DNA Analysis • Extraction – Isolation and purification of DNA from evidence • Quantitation – Determination of how much human/human male DNA present in a sample • Amplification – Making copies of STR’s • Analysis – Determine matches, inclusions, exclusions and statistics
 DNA Extraction • • Remove stain or cells from material Break open cells releasing DNA Purify DNA from remaining cellular debris Concentrate DNA
DNA Extraction • • Remove stain or cells from material Break open cells releasing DNA Purify DNA from remaining cellular debris Concentrate DNA
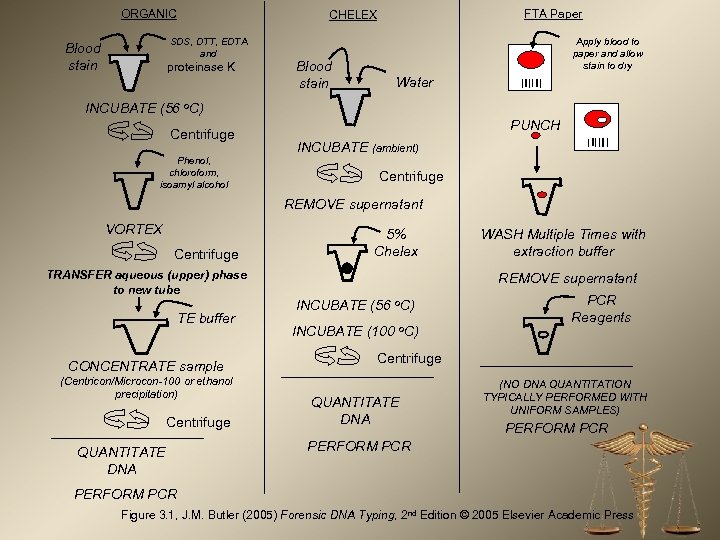 ORGANIC SDS, DTT, EDTA and Blood stain FTA Paper CHELEX proteinase K Blood stain Apply blood to paper and allow stain to dry Water INCUBATE (56 o. C) Centrifuge Phenol, chloroform, isoamyl alcohol PUNCH INCUBATE (ambient) Centrifuge REMOVE supernatant VORTEX Centrifuge 5% Chelex TRANSFER aqueous (upper) phase to new tube TE buffer CONCENTRATE sample (Centricon/Microcon-100 or ethanol precipitation) Centrifuge QUANTITATE DNA WASH Multiple Times with extraction buffer REMOVE supernatant INCUBATE (56 o. C) INCUBATE (100 o. C) PCR Reagents Centrifuge QUANTITATE DNA (NO DNA QUANTITATION TYPICALLY PERFORMED WITH UNIFORM SAMPLES) PERFORM PCR Figure 3. 1, J. M. Butler (2005) Forensic DNA Typing, 2 nd Edition © 2005 Elsevier Academic Press
ORGANIC SDS, DTT, EDTA and Blood stain FTA Paper CHELEX proteinase K Blood stain Apply blood to paper and allow stain to dry Water INCUBATE (56 o. C) Centrifuge Phenol, chloroform, isoamyl alcohol PUNCH INCUBATE (ambient) Centrifuge REMOVE supernatant VORTEX Centrifuge 5% Chelex TRANSFER aqueous (upper) phase to new tube TE buffer CONCENTRATE sample (Centricon/Microcon-100 or ethanol precipitation) Centrifuge QUANTITATE DNA WASH Multiple Times with extraction buffer REMOVE supernatant INCUBATE (56 o. C) INCUBATE (100 o. C) PCR Reagents Centrifuge QUANTITATE DNA (NO DNA QUANTITATION TYPICALLY PERFORMED WITH UNIFORM SAMPLES) PERFORM PCR Figure 3. 1, J. M. Butler (2005) Forensic DNA Typing, 2 nd Edition © 2005 Elsevier Academic Press
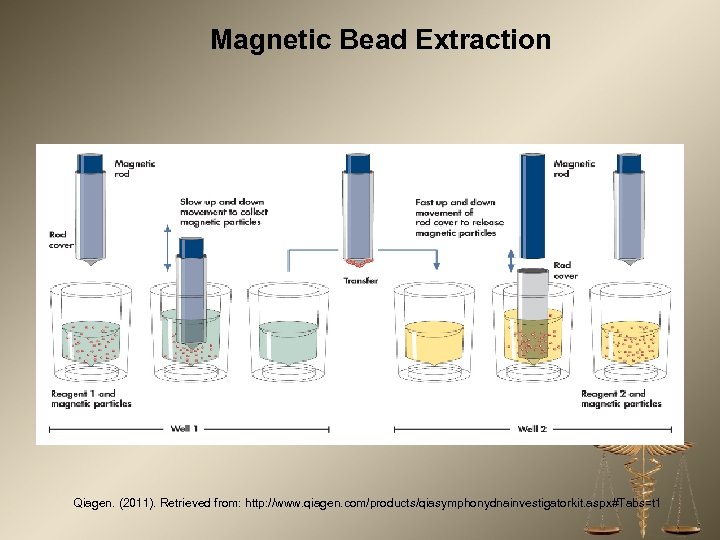 Magnetic Bead Extraction Qiagen. (2011). Retrieved from: http: //www. qiagen. com/products/qiasymphonydnainvestigatorkit. aspx#Tabs=t 1
Magnetic Bead Extraction Qiagen. (2011). Retrieved from: http: //www. qiagen. com/products/qiasymphonydnainvestigatorkit. aspx#Tabs=t 1
 Automated Extraction Platforms Utilizing Magnetic Beads Benefits to automated extraction include reduced time, limited analyst manipulation, multiple samples processed at once, and higher throughput. Downsides to automated extraction include increased cost per sample and significant time necessary for validation of new technology in the laboratory. QIAgen QIAsymphony Promega Maxwell 16 ABI Automate
Automated Extraction Platforms Utilizing Magnetic Beads Benefits to automated extraction include reduced time, limited analyst manipulation, multiple samples processed at once, and higher throughput. Downsides to automated extraction include increased cost per sample and significant time necessary for validation of new technology in the laboratory. QIAgen QIAsymphony Promega Maxwell 16 ABI Automate
 Differential DNA Extractions • Used when sperm is present in a sample • Designed to separate sperm cells from non-sperm cells • One sample becomes split into two different fractions (sperm and non-sperm) • Involves additional washing steps • After washing, fractions receive the same purification and concentration steps
Differential DNA Extractions • Used when sperm is present in a sample • Designed to separate sperm cells from non-sperm cells • One sample becomes split into two different fractions (sperm and non-sperm) • Involves additional washing steps • After washing, fractions receive the same purification and concentration steps
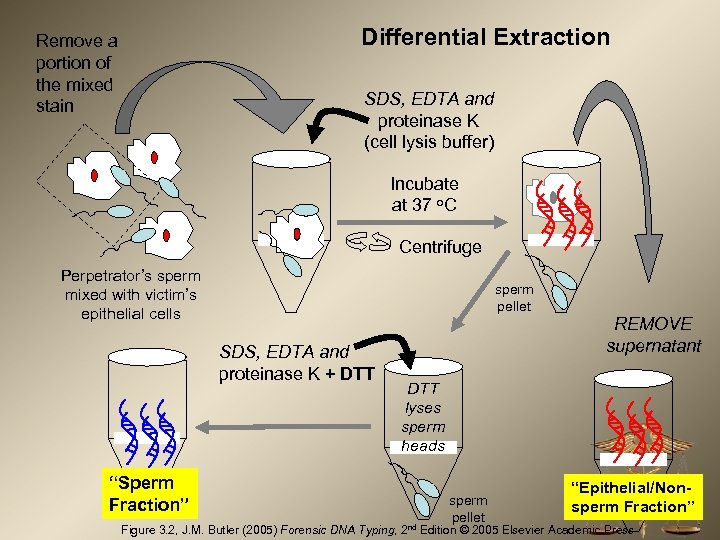 Differential Extraction Remove a portion of the mixed stain SDS, EDTA and proteinase K (cell lysis buffer) Incubate at 37 o. C Centrifuge Perpetrator’s sperm mixed with victim’s epithelial cells sperm pellet SDS, EDTA and proteinase K + DTT “Sperm Fraction” REMOVE supernatant DTT lyses sperm heads sperm pellet “Epithelial/Nonsperm Fraction” Figure 3. 2, J. M. Butler (2005) Forensic DNA Typing, 2 nd Edition © 2005 Elsevier Academic Press
Differential Extraction Remove a portion of the mixed stain SDS, EDTA and proteinase K (cell lysis buffer) Incubate at 37 o. C Centrifuge Perpetrator’s sperm mixed with victim’s epithelial cells sperm pellet SDS, EDTA and proteinase K + DTT “Sperm Fraction” REMOVE supernatant DTT lyses sperm heads sperm pellet “Epithelial/Nonsperm Fraction” Figure 3. 2, J. M. Butler (2005) Forensic DNA Typing, 2 nd Edition © 2005 Elsevier Academic Press
 DNA Quantitation • All sources of DNA are extracted when biological evidence from a crime scene is processed to isolate the DNA present. • Thus, non-human DNA such as bacterial, fungal, plant, or animal material may also be present in the total DNA recovered from the sample along with the relevant human DNA of interest. • Current forensic DNA quantitation kits can detect both human and human male DNA in one step. New kits go a step further and can give indications of DNA degradation. • Multiplex STR typing works best with a fairly narrow range of human DNA – typically 0. 5 to 2. 0 ng of input DNA works best with commercial STR kits. http: //www. cstl. nist. gov/biotech/strbase/training. htm
DNA Quantitation • All sources of DNA are extracted when biological evidence from a crime scene is processed to isolate the DNA present. • Thus, non-human DNA such as bacterial, fungal, plant, or animal material may also be present in the total DNA recovered from the sample along with the relevant human DNA of interest. • Current forensic DNA quantitation kits can detect both human and human male DNA in one step. New kits go a step further and can give indications of DNA degradation. • Multiplex STR typing works best with a fairly narrow range of human DNA – typically 0. 5 to 2. 0 ng of input DNA works best with commercial STR kits. http: //www. cstl. nist. gov/biotech/strbase/training. htm
 DNA Amplification - Polymerase Chain Reaction • • Developed in 1984 by Kary Mullis Mimics the cells natural ability to make copies of its DNA Targets specific loci (locations) on the DNA molecule Makes millions of copies of targets in short amount of time • Fluorescent probe attached to primers allow visualization of STR’s when run on genetic analyzers • Can target autosomal or Y-chromosome (male) DNA
DNA Amplification - Polymerase Chain Reaction • • Developed in 1984 by Kary Mullis Mimics the cells natural ability to make copies of its DNA Targets specific loci (locations) on the DNA molecule Makes millions of copies of targets in short amount of time • Fluorescent probe attached to primers allow visualization of STR’s when run on genetic analyzers • Can target autosomal or Y-chromosome (male) DNA
 Autosomal DNA Amplification Kits • Looking for DNA from chromosomes not involved in sex determination. • Labs have been using kits that included at a minimum the Core CODIS Loci (13) plus a few extra • New “Next Generation” kits include the core loci, but now include the loci tested by the European Union, plus a few extra (24 total loci now) • FBI and CODIS require all participating labs to implement a “Next Generation” kit by January 2017
Autosomal DNA Amplification Kits • Looking for DNA from chromosomes not involved in sex determination. • Labs have been using kits that included at a minimum the Core CODIS Loci (13) plus a few extra • New “Next Generation” kits include the core loci, but now include the loci tested by the European Union, plus a few extra (24 total loci now) • FBI and CODIS require all participating labs to implement a “Next Generation” kit by January 2017
 Next Generation Kits - Why • Compatibility with the European Union Kits – Allows comparison of profiles generated in U. S. to other countries • Increased Power of Discrimination – Can better differentiate close relatives – More discriminating statistics • More information for determining possible number of contributors/mixture deconvolution
Next Generation Kits - Why • Compatibility with the European Union Kits – Allows comparison of profiles generated in U. S. to other countries • Increased Power of Discrimination – Can better differentiate close relatives – More discriminating statistics • More information for determining possible number of contributors/mixture deconvolution
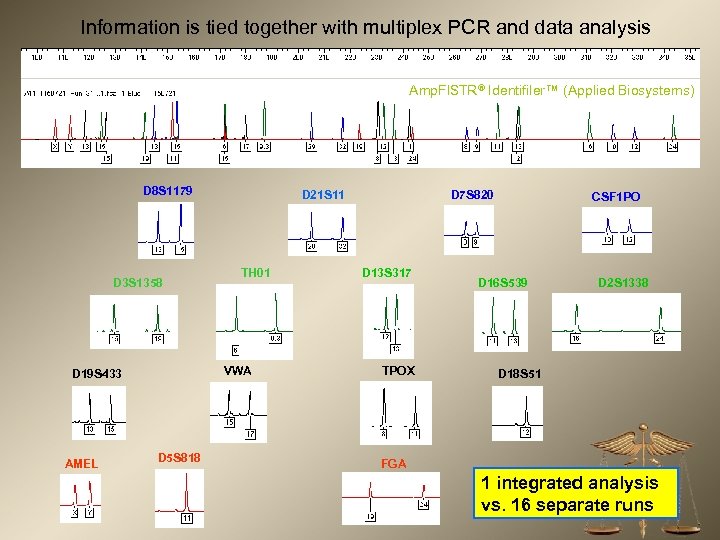 Information is tied together with multiplex PCR and data analysis Amp. Fl. STR® Identifiler™ (Applied Biosystems) D 8 S 1179 D 3 S 1358 TH 01 VWA D 19 S 433 AMEL D 21 S 11 D 5 S 818 D 7 S 820 D 13 S 317 TPOX CSF 1 PO D 16 S 539 D 2 S 1338 D 18 S 51 FGA 1 integrated analysis vs. 16 separate runs
Information is tied together with multiplex PCR and data analysis Amp. Fl. STR® Identifiler™ (Applied Biosystems) D 8 S 1179 D 3 S 1358 TH 01 VWA D 19 S 433 AMEL D 21 S 11 D 5 S 818 D 7 S 820 D 13 S 317 TPOX CSF 1 PO D 16 S 539 D 2 S 1338 D 18 S 51 FGA 1 integrated analysis vs. 16 separate runs
 YSTR Testing • Why do it? - To identify males based on their Y chromosome alleles, or establish/trace paternal male lineages • When is it helpful? -In situations where there is a large amount of female DNA present in comparison to male DNA (sexual assaults) • Limitations? -Alleles not independent (linked), are the same as all paternal relatives, less discriminating statistical calculation method. -Mixtures are problematic
YSTR Testing • Why do it? - To identify males based on their Y chromosome alleles, or establish/trace paternal male lineages • When is it helpful? -In situations where there is a large amount of female DNA present in comparison to male DNA (sexual assaults) • Limitations? -Alleles not independent (linked), are the same as all paternal relatives, less discriminating statistical calculation method. -Mixtures are problematic
 YSTR Testing – New Technologies • Newer kits available test more loci – More discrimination power – Include rapidly mutating loci • May help to differentiate paternally related individuals based on mutations between generations • New statistical methods being developed to help with YSTR mixtures
YSTR Testing – New Technologies • Newer kits available test more loci – More discrimination power – Include rapidly mutating loci • May help to differentiate paternally related individuals based on mutations between generations • New statistical methods being developed to help with YSTR mixtures
 DNA Analysis – Capillary Electrophoresis • Visualization of the DNA profile – Accomplished by passing copies of the STR loci with their attached fluorescent probes through a polymer matrix – Separates the STR’s by size and color – Produces an electropherogram used by the DNA analyst to interpret results
DNA Analysis – Capillary Electrophoresis • Visualization of the DNA profile – Accomplished by passing copies of the STR loci with their attached fluorescent probes through a polymer matrix – Separates the STR’s by size and color – Produces an electropherogram used by the DNA analyst to interpret results
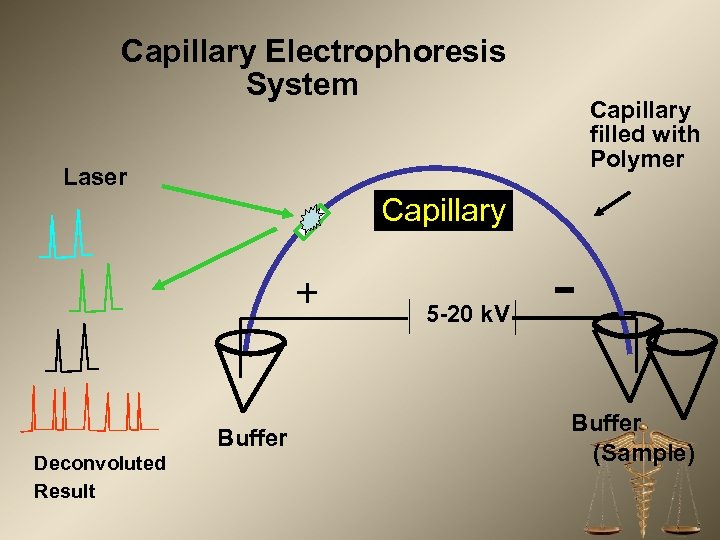 Capillary Electrophoresis System Capillary filled with Polymer Laser Capillary + Buffer Deconvoluted Result 5 -20 k. V Buffer (Sample)
Capillary Electrophoresis System Capillary filled with Polymer Laser Capillary + Buffer Deconvoluted Result 5 -20 k. V Buffer (Sample)
 Now Lets Have Some Fun! • Time for you guys and gals to be DNA Analysts!
Now Lets Have Some Fun! • Time for you guys and gals to be DNA Analysts!
 What can forensic DNA testing tell you? • Simple answer: WHO!!! • Forensic cases – matching/including/excluding suspects/victims/witnesses with evidence • • • Paternity testing -- identifying father Mass disasters -- putting pieces back together Historical investigations Missing persons investigations Military DNA “dog tag” Convicted felon DNA databases
What can forensic DNA testing tell you? • Simple answer: WHO!!! • Forensic cases – matching/including/excluding suspects/victims/witnesses with evidence • • • Paternity testing -- identifying father Mass disasters -- putting pieces back together Historical investigations Missing persons investigations Military DNA “dog tag” Convicted felon DNA databases
 What forensic DNA testing can’t tell you…yet? • When it was deposited (there are environmental factors that can have influences on results) • How or the manner in which it was deposited (secondary transfer/consent) • Whether or not sexual acts were performed with consent • Presence of a DNA profile doesn’t mean it was deposited during a crime • Lack of a DNA profile doesn’t mean an individual was not at the scene (there are environmental factors that can have influences on results) • The difference between identical twins (fingerprints can) • Physical appearance (hair color, eye color, height, etc…) • Ethnicity • Predisposition for disease
What forensic DNA testing can’t tell you…yet? • When it was deposited (there are environmental factors that can have influences on results) • How or the manner in which it was deposited (secondary transfer/consent) • Whether or not sexual acts were performed with consent • Presence of a DNA profile doesn’t mean it was deposited during a crime • Lack of a DNA profile doesn’t mean an individual was not at the scene (there are environmental factors that can have influences on results) • The difference between identical twins (fingerprints can) • Physical appearance (hair color, eye color, height, etc…) • Ethnicity • Predisposition for disease
 DNA Report Terminology • Partial Profile (Autosomal or Y Chromosome): refers to a profile where complete results are not returned at all locations tested. • Match: when genetic profiles exhibit the same alleles at loci tested and no unexplainable differences exist. In a criminal case a match doesn’t necessarily equate guilt or innocence. • Exclusion: when a genetic profile from a known standard does not match or is absent from the genetic profile from an evidentiary sample. In a criminal case an exclusion doesn’t necessarily equate guilt or innocence.
DNA Report Terminology • Partial Profile (Autosomal or Y Chromosome): refers to a profile where complete results are not returned at all locations tested. • Match: when genetic profiles exhibit the same alleles at loci tested and no unexplainable differences exist. In a criminal case a match doesn’t necessarily equate guilt or innocence. • Exclusion: when a genetic profile from a known standard does not match or is absent from the genetic profile from an evidentiary sample. In a criminal case an exclusion doesn’t necessarily equate guilt or innocence.
 DNA Report Terminology • Inclusion: when a genetic profile from a known standard cannot be excluded from the genetic profile from an evidentiary sample (usually reserved for DNA mixtures). In a criminal case an inclusion doesn’t necessarily equate guilt or innocence. • Inconclusive/Uninterpretable: when no conclusion can be reached regarding testing done due to one of many possible reasons (e. g. , no DNA profiles obtained, uninterpretable mixtures, partial profiles, no known standards to compare to, etc…)
DNA Report Terminology • Inclusion: when a genetic profile from a known standard cannot be excluded from the genetic profile from an evidentiary sample (usually reserved for DNA mixtures). In a criminal case an inclusion doesn’t necessarily equate guilt or innocence. • Inconclusive/Uninterpretable: when no conclusion can be reached regarding testing done due to one of many possible reasons (e. g. , no DNA profiles obtained, uninterpretable mixtures, partial profiles, no known standards to compare to, etc…)
 DNA Report Terminology • Mixture: a DNA profile originating from a combination of more than one contributor. • Major Component/Contributor: the individual(s) that contributed the majority of the DNA in the mixture • Minor Component/Contributor: the individual(s) that contributed the lesser portion of the DNA in the mixture. • Indistinguishable Mixture: a mixed DNA profile where no major or minor contributors can be identified. Does not necessarily mean the mixture is uninterpretable.
DNA Report Terminology • Mixture: a DNA profile originating from a combination of more than one contributor. • Major Component/Contributor: the individual(s) that contributed the majority of the DNA in the mixture • Minor Component/Contributor: the individual(s) that contributed the lesser portion of the DNA in the mixture. • Indistinguishable Mixture: a mixed DNA profile where no major or minor contributors can be identified. Does not necessarily mean the mixture is uninterpretable.
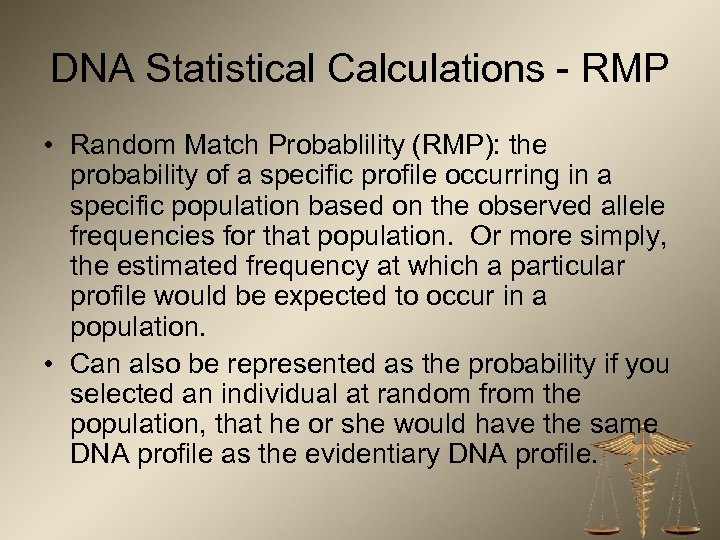 DNA Statistical Calculations - RMP • Random Match Probablility (RMP): the probability of a specific profile occurring in a specific population based on the observed allele frequencies for that population. Or more simply, the estimated frequency at which a particular profile would be expected to occur in a population. • Can also be represented as the probability if you selected an individual at random from the population, that he or she would have the same DNA profile as the evidentiary DNA profile.
DNA Statistical Calculations - RMP • Random Match Probablility (RMP): the probability of a specific profile occurring in a specific population based on the observed allele frequencies for that population. Or more simply, the estimated frequency at which a particular profile would be expected to occur in a population. • Can also be represented as the probability if you selected an individual at random from the population, that he or she would have the same DNA profile as the evidentiary DNA profile.
 DNA Statistical Calculations - RMP • RMP is NOT: - The chance that someone else is guilty or that someone else left the biological material on the evidence. Or, in a given population, anyone with the same DNA profile as the evidence sample is as likely to have left the sample as the suspect. Or in a mixture, every possible genotype has an equal chance of having committed the crime. (Defense Attorney’s Fallacy) - The chance of the defendant not being guilty or that someone else in reality would have the same DNA profile. If the DNA samples match then they are 1 quadrillion times more likely to have come from the same person than different persons. (Prosecutor’s Fallacy)
DNA Statistical Calculations - RMP • RMP is NOT: - The chance that someone else is guilty or that someone else left the biological material on the evidence. Or, in a given population, anyone with the same DNA profile as the evidence sample is as likely to have left the sample as the suspect. Or in a mixture, every possible genotype has an equal chance of having committed the crime. (Defense Attorney’s Fallacy) - The chance of the defendant not being guilty or that someone else in reality would have the same DNA profile. If the DNA samples match then they are 1 quadrillion times more likely to have come from the same person than different persons. (Prosecutor’s Fallacy)
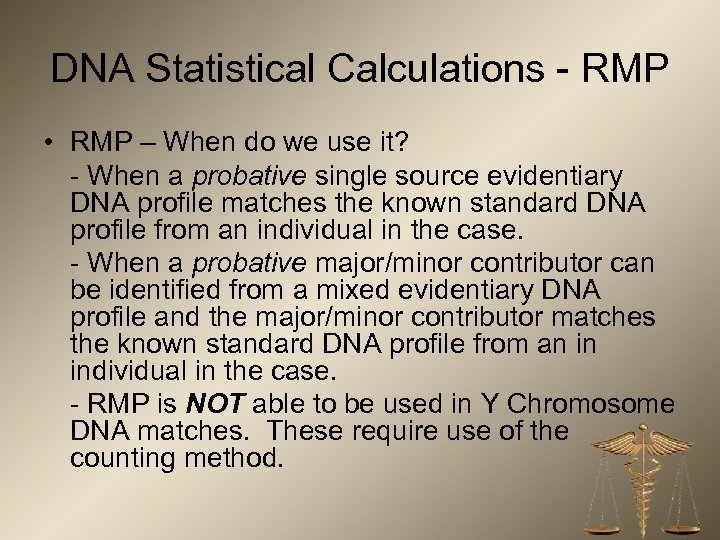 DNA Statistical Calculations - RMP • RMP – When do we use it? - When a probative single source evidentiary DNA profile matches the known standard DNA profile from an individual in the case. - When a probative major/minor contributor can be identified from a mixed evidentiary DNA profile and the major/minor contributor matches the known standard DNA profile from an in individual in the case. - RMP is NOT able to be used in Y Chromosome DNA matches. These require use of the counting method.
DNA Statistical Calculations - RMP • RMP – When do we use it? - When a probative single source evidentiary DNA profile matches the known standard DNA profile from an individual in the case. - When a probative major/minor contributor can be identified from a mixed evidentiary DNA profile and the major/minor contributor matches the known standard DNA profile from an in individual in the case. - RMP is NOT able to be used in Y Chromosome DNA matches. These require use of the counting method.
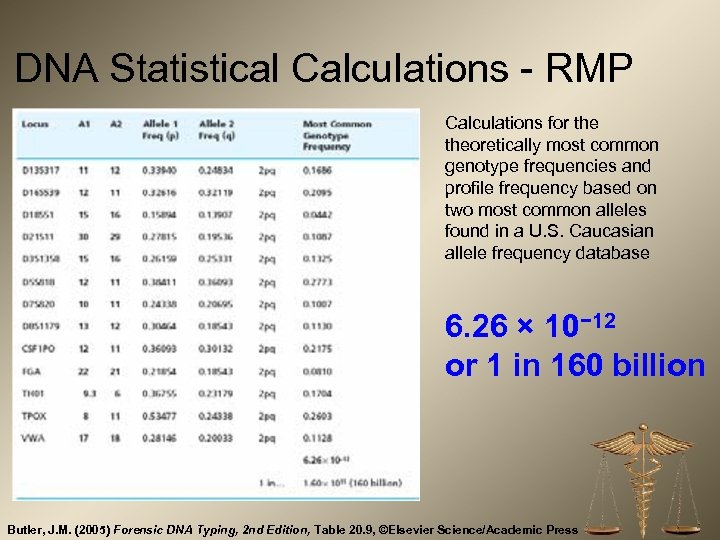 DNA Statistical Calculations - RMP Calculations for theoretically most common genotype frequencies and profile frequency based on two most common alleles found in a U. S. Caucasian allele frequency database 6. 26 × 10− 12 or 1 in 160 billion Butler, J. M. (2005) Forensic DNA Typing, 2 nd Edition, Table 20. 9, ©Elsevier Science/Academic Press
DNA Statistical Calculations - RMP Calculations for theoretically most common genotype frequencies and profile frequency based on two most common alleles found in a U. S. Caucasian allele frequency database 6. 26 × 10− 12 or 1 in 160 billion Butler, J. M. (2005) Forensic DNA Typing, 2 nd Edition, Table 20. 9, ©Elsevier Science/Academic Press
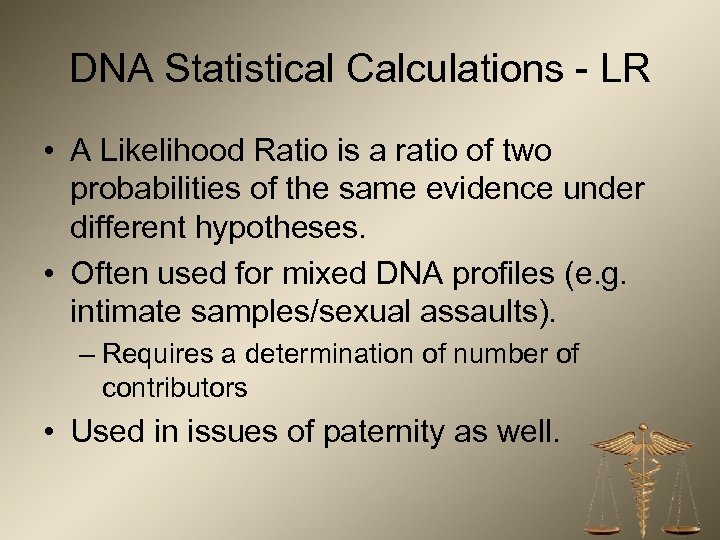 DNA Statistical Calculations - LR • A Likelihood Ratio is a ratio of two probabilities of the same evidence under different hypotheses. • Often used for mixed DNA profiles (e. g. intimate samples/sexual assaults). – Requires a determination of number of contributors • Used in issues of paternity as well.
DNA Statistical Calculations - LR • A Likelihood Ratio is a ratio of two probabilities of the same evidence under different hypotheses. • Often used for mixed DNA profiles (e. g. intimate samples/sexual assaults). – Requires a determination of number of contributors • Used in issues of paternity as well.
 DNA Statistical Calculations - LR • For example, in a sexual assault the non-sperm fraction of the vaginal swabs is a mixture of the victim and the suspect. • The hypotheses of the likelihood ratio would be: -Prosecution’s Hypothesis = the mixture is a result of the victim’s and suspect’s DNA profiles. -Defense’s Hypothesis = the mixture is a result of the victim’s DNA profile and an unidentified random individual in the population.
DNA Statistical Calculations - LR • For example, in a sexual assault the non-sperm fraction of the vaginal swabs is a mixture of the victim and the suspect. • The hypotheses of the likelihood ratio would be: -Prosecution’s Hypothesis = the mixture is a result of the victim’s and suspect’s DNA profiles. -Defense’s Hypothesis = the mixture is a result of the victim’s DNA profile and an unidentified random individual in the population.
 DNA Statistical Calculations - LR • If the likelihood ratio is < 1, then the defense’s hypothesis is supported. • If the likelihood ratio is >1, then the prosecution’s hypothesis is supported. • The greater the likelihood ratio, the greater the support for the prosecution’s hypothesis.
DNA Statistical Calculations - LR • If the likelihood ratio is < 1, then the defense’s hypothesis is supported. • If the likelihood ratio is >1, then the prosecution’s hypothesis is supported. • The greater the likelihood ratio, the greater the support for the prosecution’s hypothesis.
 DNA Statistical Calculations – CPI/CPE • Represents an estimate of the portion of the population that has at least one allele not observed in the mixed DNA profile. • Do not require an assumption as to the number of contributors to mixture.
DNA Statistical Calculations – CPI/CPE • Represents an estimate of the portion of the population that has at least one allele not observed in the mixed DNA profile. • Do not require an assumption as to the number of contributors to mixture.
 DNA Statistical Calculations – CPI/CPE • For example, a mixed DNA profile was found on a gun. The suspect cannot be excluded as a possible contributor to the mixed DNA profile on the gun. • The Combined Probability of Exclusion would be the % of the population that you would expect to be excluded from the mixed DNA profile on the gun based on the frequencies of the alleles present in the mixture (e. g. , 99. 99999%) • The Combined Probability of Inclusion would be the probability of selecting and individual at random and having their DNA profile be included in the mixed DNA profile on the gun based on the frequencies of the alleles present in the mixture (e. g. , 1 in 1, 000)
DNA Statistical Calculations – CPI/CPE • For example, a mixed DNA profile was found on a gun. The suspect cannot be excluded as a possible contributor to the mixed DNA profile on the gun. • The Combined Probability of Exclusion would be the % of the population that you would expect to be excluded from the mixed DNA profile on the gun based on the frequencies of the alleles present in the mixture (e. g. , 99. 99999%) • The Combined Probability of Inclusion would be the probability of selecting and individual at random and having their DNA profile be included in the mixed DNA profile on the gun based on the frequencies of the alleles present in the mixture (e. g. , 1 in 1, 000)
 DNA Statistical Calculations – Counting Method • Reserved for YSTR calculations. • Cannot use RMP because loci are linked and inherited paternally, causing statistics to be much lower than RMP. • Calculated by dividing the number of times a specific YSTR profile is seen in a database by the total number of profiles in the database and applying a confidence interval (95%). • The confidence interval reflects the probability that in 95% of the samples tested, the interval should contain the actual value measured.
DNA Statistical Calculations – Counting Method • Reserved for YSTR calculations. • Cannot use RMP because loci are linked and inherited paternally, causing statistics to be much lower than RMP. • Calculated by dividing the number of times a specific YSTR profile is seen in a database by the total number of profiles in the database and applying a confidence interval (95%). • The confidence interval reflects the probability that in 95% of the samples tested, the interval should contain the actual value measured.
 DNA Statistical Calculations – Counting Method • The statistics are determined by the size of the database. The larger the database, the better the statistics. • US YSTR Haplotype Database (25, 683 samples)
DNA Statistical Calculations – Counting Method • The statistics are determined by the size of the database. The larger the database, the better the statistics. • US YSTR Haplotype Database (25, 683 samples)
 Statistics – The Future • It has been recommended that the Likelihood Ratio is the preferred statistical calculation for mixture profiles • There is a movement to discontinue/limit the use of the CPE/CPI calculation • New semi-continuous and continuous methods of analysis provide additional mixture deconvolution aid and allows greater “use” of difficult DNA profiles
Statistics – The Future • It has been recommended that the Likelihood Ratio is the preferred statistical calculation for mixture profiles • There is a movement to discontinue/limit the use of the CPE/CPI calculation • New semi-continuous and continuous methods of analysis provide additional mixture deconvolution aid and allows greater “use” of difficult DNA profiles
 About those Continuous Methods • Use a mathematic modeling approach in an attempt to generate genotype probabilities – Deconvolute a mixture down to its individual contributors – Determine ratio of contributors – Not limited by thresholds • Applies Likelihood Ratio Calculations to give weight to inclusions/exclustions
About those Continuous Methods • Use a mathematic modeling approach in an attempt to generate genotype probabilities – Deconvolute a mixture down to its individual contributors – Determine ratio of contributors – Not limited by thresholds • Applies Likelihood Ratio Calculations to give weight to inclusions/exclustions
 Speaking of Databases • The FBI and NIST population databases are commonly used for the calculation of RMP, LR, and CPI. Other population databases are available and some are specific for certain minority populations (Native American Tribes, Amish, Island Nations, etc…) • The US YSTR Database is commonly used for calculation of YSTR statistics. • The CODIS database consists mainly of DNA profiles from crime scenes, convicted offenders, missing persons, and relatives of missing persons. • These databases are COMPLETELY INDEPENDENT of one another. One has absolutely no impact on any of the others. They all serve completely different purposes.
Speaking of Databases • The FBI and NIST population databases are commonly used for the calculation of RMP, LR, and CPI. Other population databases are available and some are specific for certain minority populations (Native American Tribes, Amish, Island Nations, etc…) • The US YSTR Database is commonly used for calculation of YSTR statistics. • The CODIS database consists mainly of DNA profiles from crime scenes, convicted offenders, missing persons, and relatives of missing persons. • These databases are COMPLETELY INDEPENDENT of one another. One has absolutely no impact on any of the others. They all serve completely different purposes.
 CODIS Combined DNA Index System
CODIS Combined DNA Index System
 FBI’s CODIS DNA Database Combined DNA Index System • Used for linking serial crimes and unsolved cases with repeat offenders • Launched October 1998 • Links all 50 states, 2 Federal Laboratories & Puerto Rico • Profiles must meet a minimum match estimate for inclusion into NDIS/SDIS
FBI’s CODIS DNA Database Combined DNA Index System • Used for linking serial crimes and unsolved cases with repeat offenders • Launched October 1998 • Links all 50 states, 2 Federal Laboratories & Puerto Rico • Profiles must meet a minimum match estimate for inclusion into NDIS/SDIS
 Database • Utilizes computer software to automatically search files for matches. • Administrated by the FBI • Enables state & local crime laboratories to exchange and compare DNA profiles electronically
Database • Utilizes computer software to automatically search files for matches. • Administrated by the FBI • Enables state & local crime laboratories to exchange and compare DNA profiles electronically
 Forensic Unknown File • Contains DNA profiles developed from crime scene samples (also referred to as forensic unknown samples). • When samples from crime scenes match one another it is referred to as a “forensic hit”
Forensic Unknown File • Contains DNA profiles developed from crime scene samples (also referred to as forensic unknown samples). • When samples from crime scenes match one another it is referred to as a “forensic hit”
 Convicted Offender File • Contains profiles from individuals convicted of certain crimes • When evidence from a crime scene matches a profile in this file, it is referred to as an “offender hit”
Convicted Offender File • Contains profiles from individuals convicted of certain crimes • When evidence from a crime scene matches a profile in this file, it is referred to as an “offender hit”
 What is NOT entered into CODIS • Victim profile(s) • Profiles developed from known/reference samples (excluding convicted offender samples) • Profiles developed from evidence which match the victim, witness, etc • Non-probative profiles
What is NOT entered into CODIS • Victim profile(s) • Profiles developed from known/reference samples (excluding convicted offender samples) • Profiles developed from evidence which match the victim, witness, etc • Non-probative profiles
 National Statistics • As of June 2016 the profile composition of the National DNA Index System (NDIS) is as follows: • Total forensic profiles: 714, 660 • Total convicted offender profiles: 12, 306, 681 • Total profiles = 15, 416, 332 • As of June 2016, CODIS has produced over 336, 000 hits assisting in more than 322, 000 investigations.
National Statistics • As of June 2016 the profile composition of the National DNA Index System (NDIS) is as follows: • Total forensic profiles: 714, 660 • Total convicted offender profiles: 12, 306, 681 • Total profiles = 15, 416, 332 • As of June 2016, CODIS has produced over 336, 000 hits assisting in more than 322, 000 investigations.


