dcdc3ece63ba334de5ab482e1abf6986.ppt
- Количество слайдов: 63

Copy Number Variations DTC Bio. Informatics Course Hillary Term 2010 WTHCG, Thursday 12 th of February Jean-Baptiste Cazier http: //www. well. ox. ac. uk/dr-jean-baptiste-cazier Jean-Baptiste. Cazier@well. ox. ac. uk
Outline • Lecture – Definitions • More important than it may seem – Identification • Technology, Algorithmic, Design – Recent studies • • Mc. Carrol & Korn, GSV, WTCCC Break – The special case of Cancer • More problems – Conclusions • • Break Practical – Applications with CGH data in R – Application with SNP data in Illumina’s Bead. Studio (PC only) 2

Definitions • Acronyms: – CNP: • Copy Number Polymorphisms – CNV: • Copy Number Variations – CNA: • • Copy Number Aberrations Copy Number Alterations Finding the missing heritability of complex diseases TA Manolio et al. Nature 461, 747 -753 (2009) • Creation: Germline vs Somatic – Is the CNV coming from the original cell or did it evolve only in a few ? • • There are very many CNVs shared among population like SNPs or STRs Somatic propagation of CNVs is a mark of Cancer 3
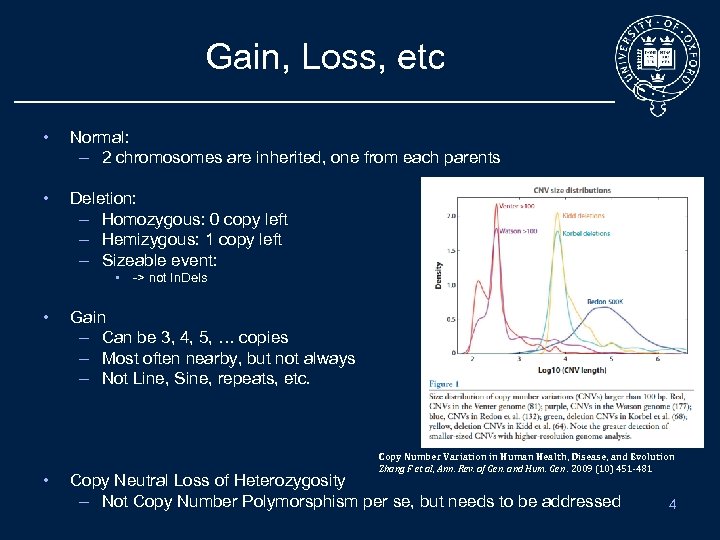
Gain, Loss, etc • Normal: – 2 chromosomes are inherited, one from each parents • Deletion: – Homozygous: 0 copy left – Hemizygous: 1 copy left – Sizeable event: • -> not In. Dels • • Gain – Can be 3, 4, 5, … copies – Most often nearby, but not always – Not Line, Sine, repeats, etc. Copy Number Variation in Human Health, Disease, and Evolution Zhang F et al, Ann. Rev. of Gen. and Hum. Gen. 2009 (10) 451 -481 Copy Neutral Loss of Heterozygosity – Not Copy Number Polymorsphism per se, but needs to be addressed 4
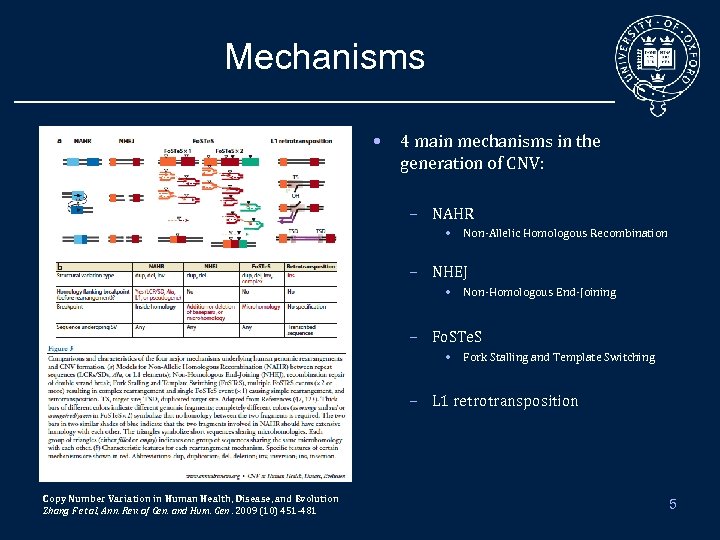
Mechanisms • 4 main mechanisms in the generation of CNV: – NAHR • Non-Allelic Homologous Recombination – NHEJ • Non-Homologous End-Joining – Fo. STe. S • Fork Stalling and Template Switching – L 1 retrotransposition Copy Number Variation in Human Health, Disease, and Evolution Zhang F et al, Ann. Rev. of Gen. and Hum. Gen. 2009 (10) 451 -481 5
Characterization • Identification: a Genome-Wide test – Karyotyping – Multi color chromosome painting – Comparative Genetic Hybridization (CGH) – Array CGH (a. CGH) – “SNP”- array • Validation: a local test – – q. PCR: quantitative Polymerase Chain Reaction MLPA: Multiplex Ligation-dependent Probe Amplification Fluorescent In-Situ Hybridization (FISH) Sequencing 6
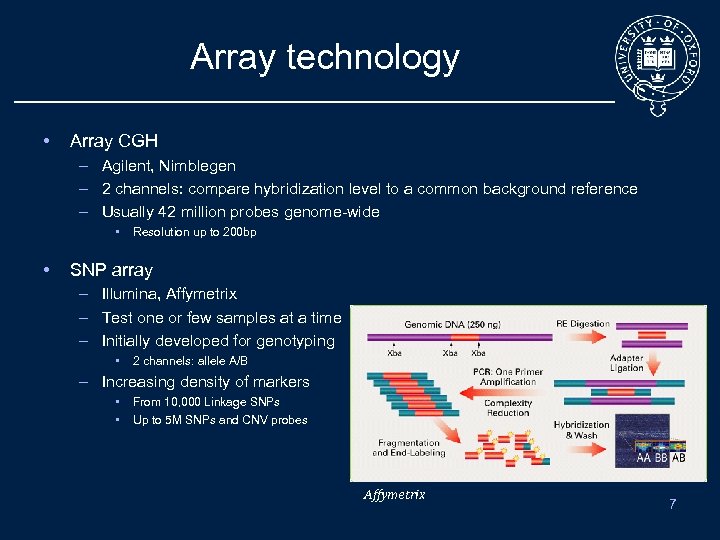
Array technology • Array CGH – Agilent, Nimblegen – 2 channels: compare hybridization level to a common background reference – Usually 42 million probes genome-wide • • Resolution up to 200 bp SNP array – Illumina, Affymetrix – Test one or few samples at a time – Initially developed for genotyping • 2 channels: allele A/B – Increasing density of markers • • From 10, 000 Linkage SNPs Up to 5 M SNPs and CNV probes Affymetrix 7
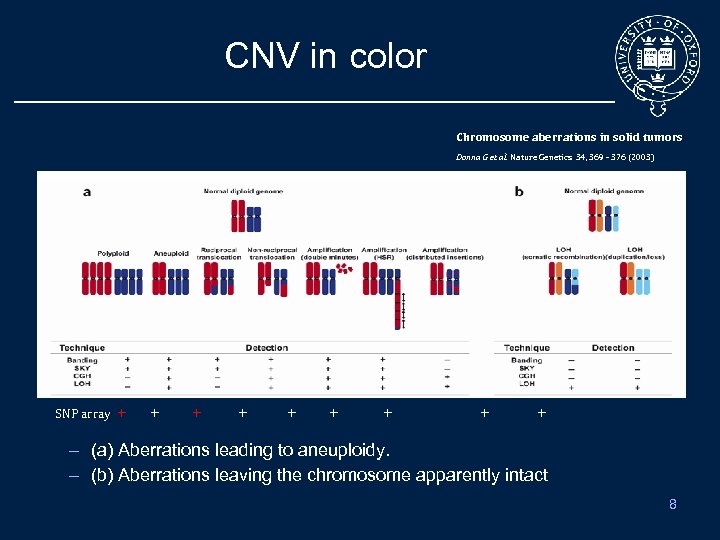
CNV in color Chromosome aberrations in solid tumors Donna G et al. Nature Genetics 34, 369 - 376 (2003) SNP array + + + + + + – (a) Aberrations leading to aneuploidy. – (b) Aberrations leaving the chromosome apparently intact 8

Revival • Genome-Wide Association provided some success in the identification of variants for many diseases: – AMD, Coeliac disease, Type 2 Diabetes, Prostate Cancer, Colorectal Cancer, etc. • However most variants are ‘only’ statistically significant: – 80% fall outside of coding regions • The case of Missing Heritability: – Whatever the number of variants identified, they usually account for only a small proportion of the heritability Finding the missing heritability of complex diseases TA Manolio et al. Nature 461, 747 -753 (2009) 9
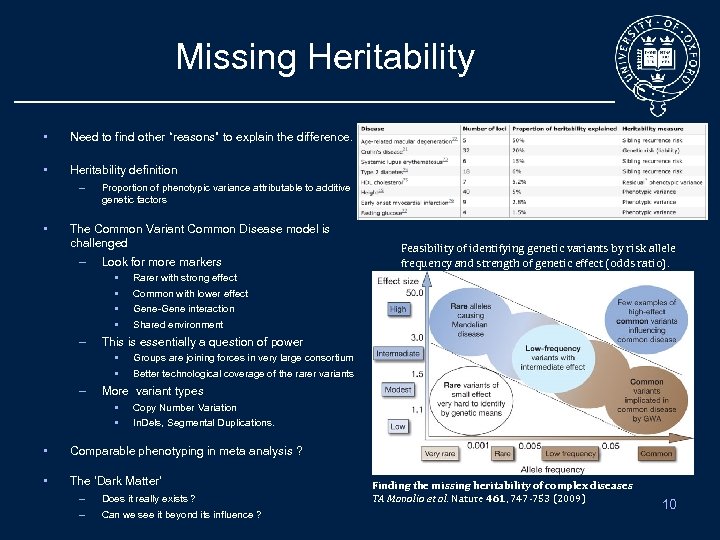
Missing Heritability • Need to find other “reasons” to explain the difference. • Heritability definition – • Proportion of phenotypic variance attributable to additive genetic factors The Common Variant Common Disease model is challenged – Look for more markers • • – Rarer with strong effect Common with lower effect Gene-Gene interaction Shared environment This is essentially a question of power • • – Feasibility of identifying genetic variants by risk allele frequency and strength of genetic effect (odds ratio). Groups are joining forces in very large consortium Better technological coverage of the rarer variants More variant types • • Copy Number Variation In. Dels, Segmental Duplications. • Comparable phenotyping in meta analysis ? • The ‘Dark Matter’ – Does it really exists ? – Can we see it beyond its influence ? Finding the missing heritability of complex diseases TA Manolio et al. Nature 461, 747 -753 (2009) 10
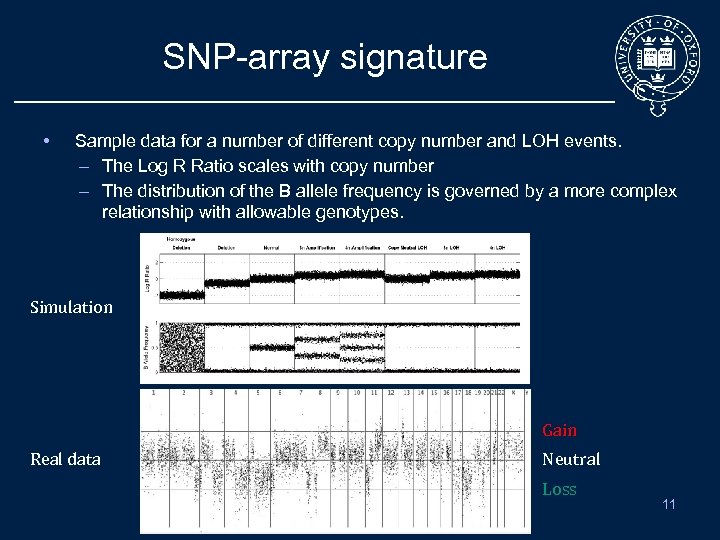
SNP-array signature • Sample data for a number of different copy number and LOH events. – The Log R Ratio scales with copy number – The distribution of the B allele frequency is governed by a more complex relationship with allowable genotypes. Simulation Gain Real data Neutral Loss 11
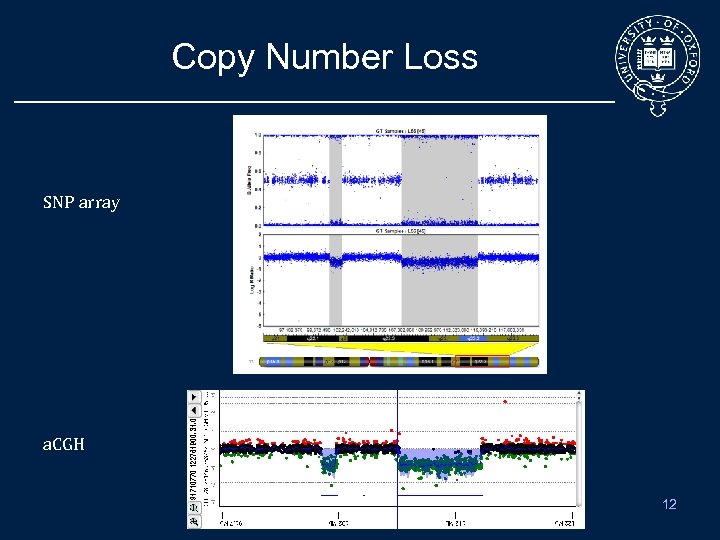
Copy Number Loss SNP array a. CGH 12
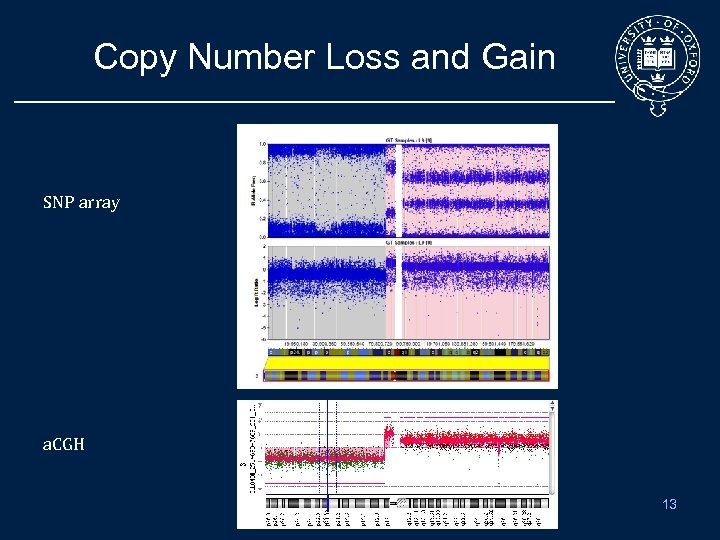
Copy Number Loss and Gain SNP array a. CGH 13
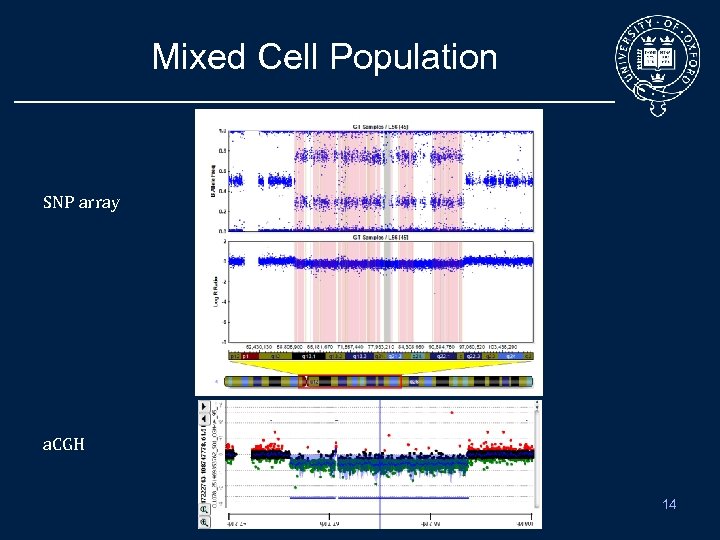
Mixed Cell Population SNP array a. CGH 14
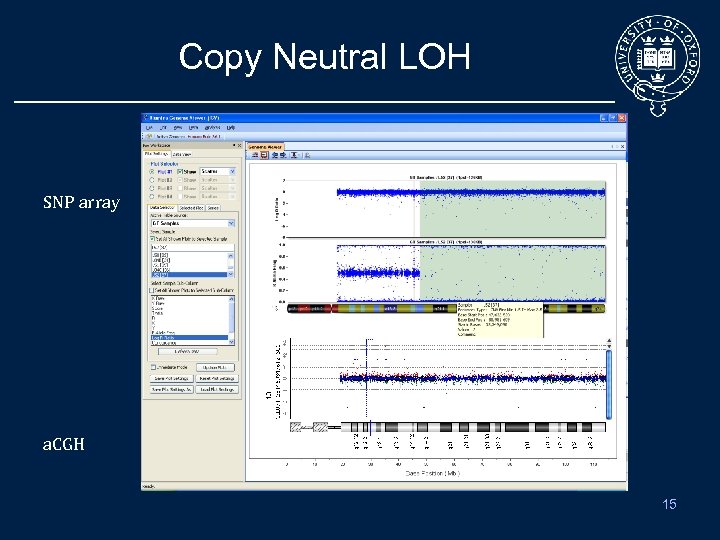
Copy Neutral LOH SNP array a. CGH 15

Automatic recognition of CNVs • Originally done by visual inspection – Problem of reproducibility – Problem of accuracy – With increasing density, problem of possibility to see • Automation and test – Moving average – Probe selection / compilation – Segmentation, Hidden Markov Model – Significance testing • Need to compile data with uncertainty 16
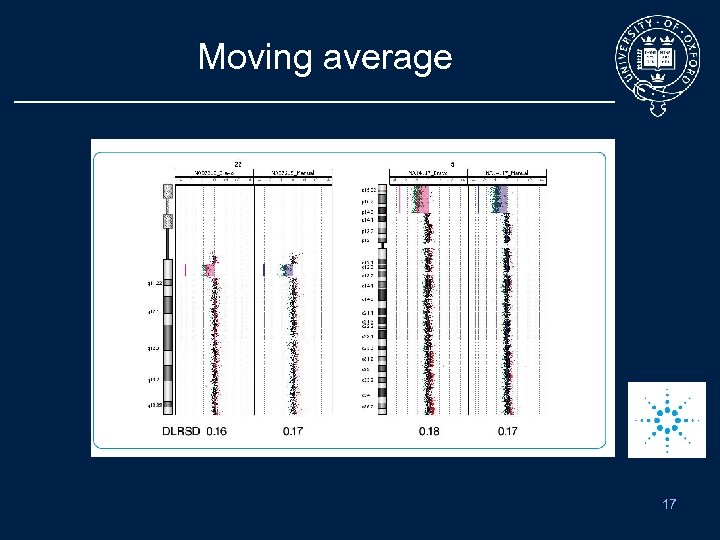
Moving average 17
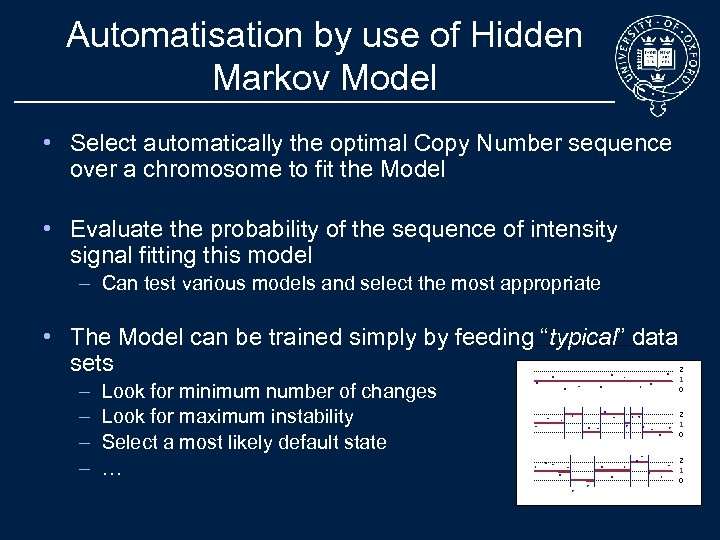
Automatisation by use of Hidden Markov Model • Select automatically the optimal Copy Number sequence over a chromosome to fit the Model • Evaluate the probability of the sequence of intensity signal fitting this model – Can test various models and select the most appropriate • The Model can be trained simply by feeding “typical” data sets – – Look for minimum number of changes Look for maximum instability Select a most likely default state … 2 1 0
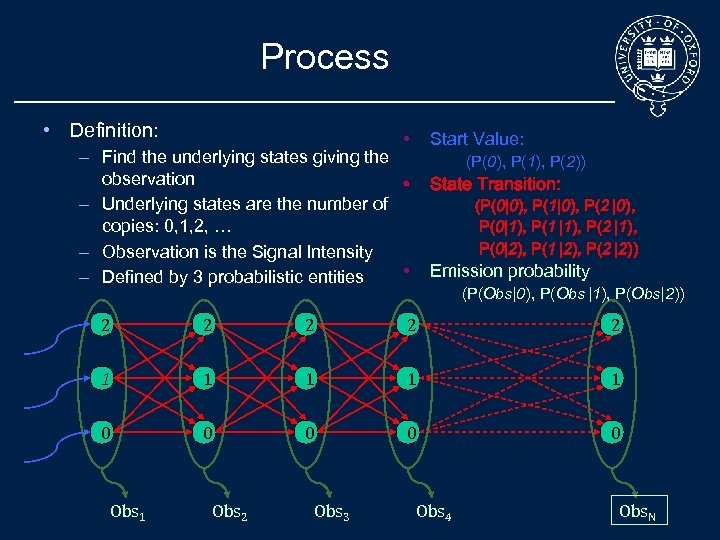
Process • Definition: • Start Value: – Find the underlying states giving the observation • – Underlying states are the number of copies: 0, 1, 2, … – Observation is the Signal Intensity • – Defined by 3 probabilistic entities (P(0), P(1), P(2)) State Transition: (P(0|0), P(1|0), P(2 |0), P(0|1), P(1 |1), P(2 |1), P(0|2), P(1 |2), P(2 |2)) Emission probability (P(Obs|0), P(Obs |1), P(Obs|2)) 2 2 2 1 1 1 0 0 0 Obs 1 Obs 2 Obs 3 Obs 4 Obs. N
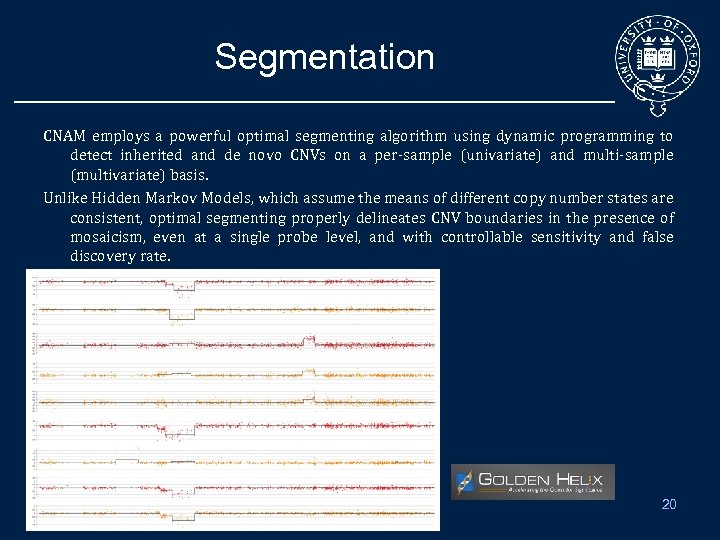
Segmentation CNAM employs a powerful optimal segmenting algorithm using dynamic programming to detect inherited and de novo CNVs on a per-sample (univariate) and multi-sample (multivariate) basis. Unlike Hidden Markov Models, which assume the means of different copy number states are consistent, optimal segmenting properly delineates CNV boundaries in the presence of mosaicism, even at a single probe level, and with controllable sensitivity and false discovery rate. 20
Available software • Graphical Interface: – – – – – Agilent Golden Helix Partek Bead. Studio/Genome. Studio Golf CNAT CNAG d. Chip Penn. CNV … • Uneven field of quality and specificity • Command line – – Quanti. SNP Bird. Suite Onco. SNP * … • R packages – – Somatics * DNACopy Aroma … * Cancer Specific tools 21
Development of recent array • In 2008 Mc. Carroll and Korn published the identification of CNPs and CNVs using/designing Affymetrix SNP 6. 0 high resolution array 22
SNP 6. 0 by Mc. Carroll • “ We designed a hybrid genotyping array (Affymetrix SNP 6. 0) to simultaneously measure 906, 600 SNPs and copy number at 1. 8 million genomic locations. By characterizing 270 Hap. Map samples, we developed a map of human CNV (at 2 -kb breakpoint resolution) informed by integer genotypes for 1, 320 copy number polymorphisms (CNPs)” Mc. Carroll • Published both analysis with chip design and algorithm suite: Bird. Suite – Perform both genotyping and CNV identification – First call for known CNP – Look for new CNV • • • 80% of observed copy number differences due to common CNPs (MAF>5%), > 99% derived from inheritance rather than new mutation. Found a common deletion polymorphism in perfect LD with Crohn’s disease SNPs – 2 kb upstream IRGM – Affect level of expression 23
High density of probes • Can identify smaller events – E. g. Important to spot residual event in translocation/fusion genes • Gain confidence in SNP-regions by increasing the number of probes • Can get better resolutions, i. e. more accurate breakpoints: – Can split existing large regions into smaller ones • Better coverage of CNP – These regions were mainly not be covered by SNP-only arrays – Beware of overrepresentation of these regions • Tiling across the genome – More exhaustive picture 24
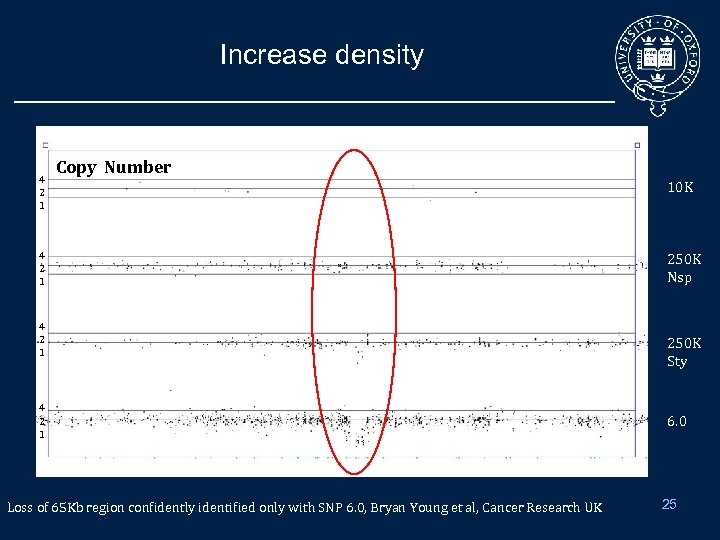
Increase density 4 2 1 Copy Number 4 2 1 Loss of 65 Kb region confidently identified only with SNP 6. 0, Bryan Young et al, Cancer Research UK 10 K 250 K Nsp 250 K Sty 6. 0 25
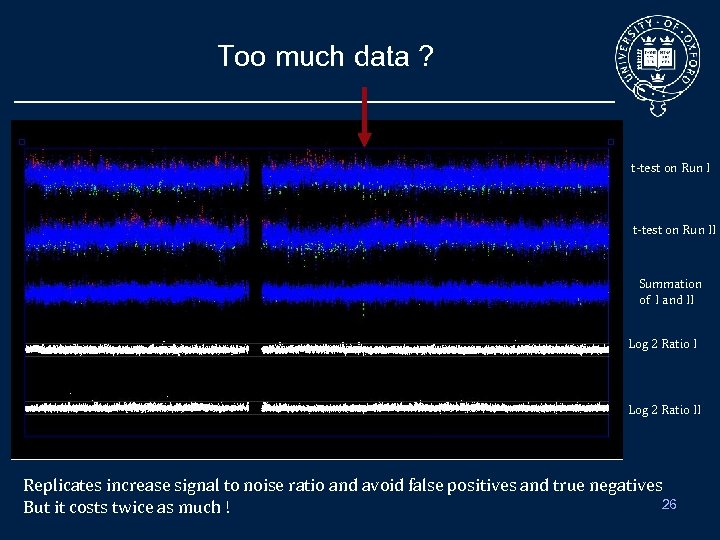
Too much data ? t-test on Run II Summation of I and II 4 2 1 Copy Number Log 2 Ratio II Replicates increase signal to noise ratio and avoid false positives and true negatives 26 But it costs twice as much !

Potential Issues • Interpretation – What to use as a baseline ? i. e. define the Ratio • Variations in probe coverage: – Gaps – Overlapping probes • Inaccurate reference – Reference build is inaccurate – Probes cannot match the locus accurately • Systematic error – Autocorrelation with GC content – Preparation, e. g. genome amplification 27
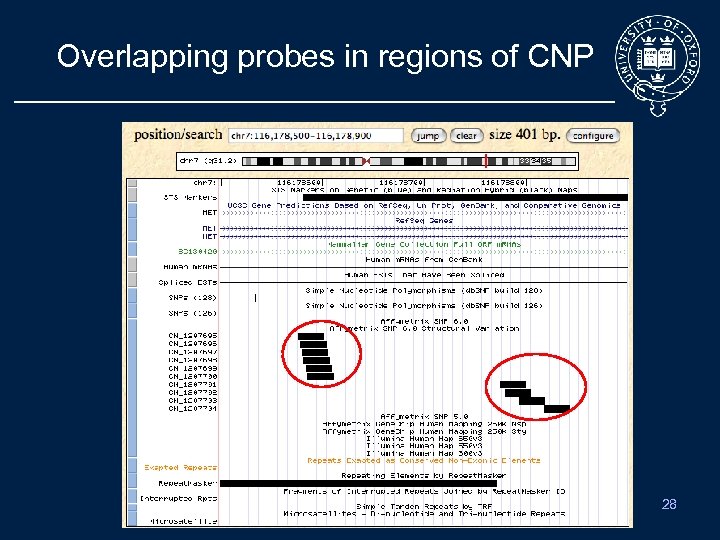
Overlapping probes in regions of CNP 28
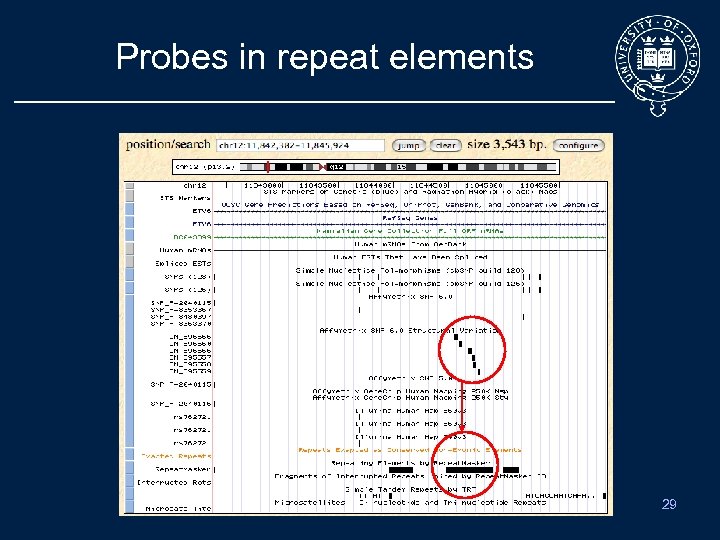
Probes in repeat elements 29
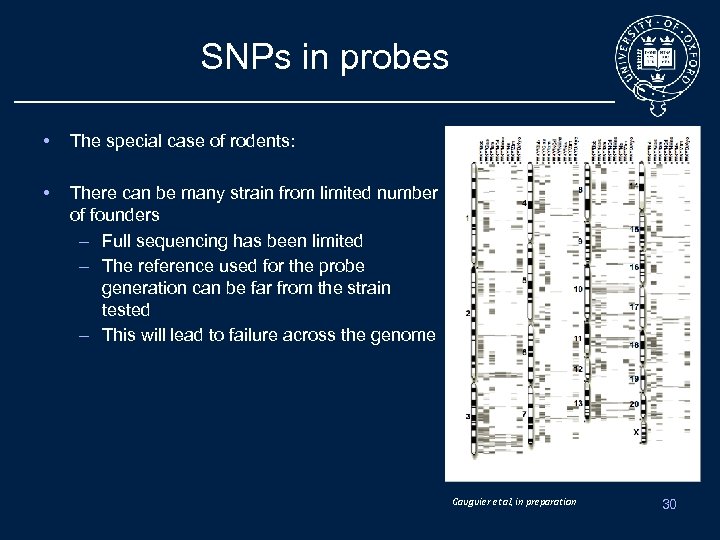
SNPs in probes • The special case of rodents: • There can be many strain from limited number of founders – Full sequencing has been limited – The reference used for the probe generation can be far from the strain tested – This will lead to failure across the genome Gauguier et al, in preparation 30
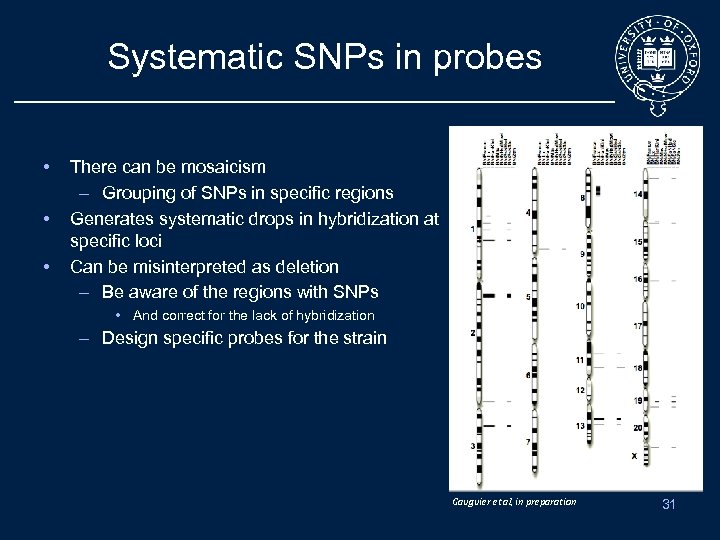
Systematic SNPs in probes • • • There can be mosaicism – Grouping of SNPs in specific regions Generates systematic drops in hybridization at specific loci Can be misinterpreted as deletion – Be aware of the regions with SNPs • And correct for the lack of hybridization – Design specific probes for the strain Gauguier et al, in preparation 31

Recent CNV Survey • Recently 2 projects started in parallel to identify and characterize CNVs in Human: – The Genome Structural Variation Consortium (GSV) • CNV discovery project to identify common CNVs using a. CGH by Nimblegen, • Detection in 20 CEU, 20 YRI, 1 reference • Assayed in 450 Hap. Map samples – The Wellcome Trust Case Control Consortium (WTCCC) • Test for association to diseases of CNVs in the WTCCC – 16, 000 cases, WTCCC plus Breast cancer – 3, 000 common ontrols 32
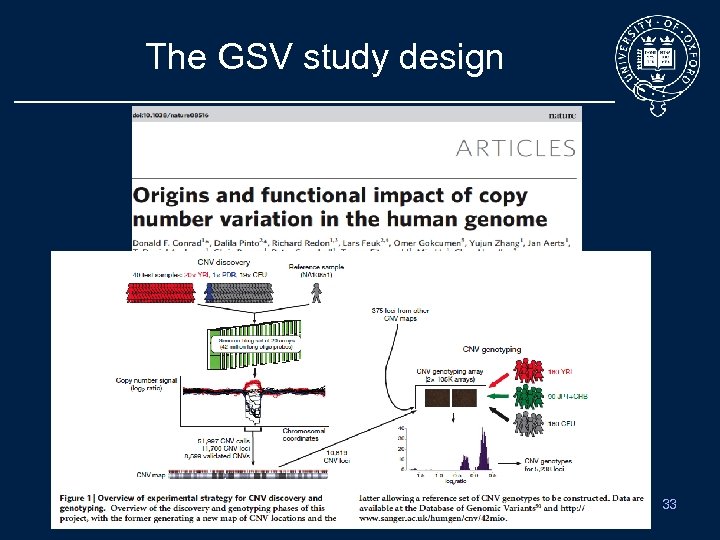
The GSV study design 33
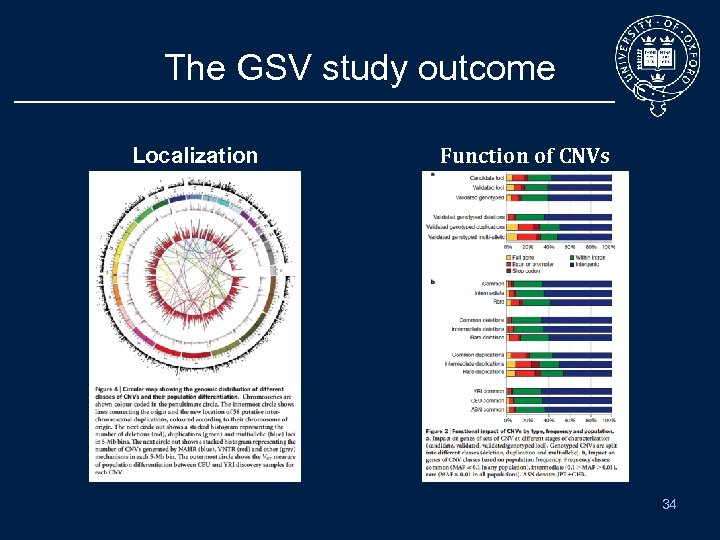
The GSV study outcome Localization Function of CNVs 34

The GSV study outcome (II) • Designed an array with 42 million probes – – cover 11, 700 CNV larger than 443 bp 8, 599 validated independently • Generate reference genotype for 4, 978 on 450 samples • • Identified 30 loci with CNV candidate for influencing phenotype Striking effect of purifying selection – Act on exonic and intronic deletions – So functional variants should be rare • But most of common CNVs are already well tagged by the existing SNParray – May need to look elsewhere to solve the missing heritability 35

The WTCCC study • Use the WTCCC cohort of 16, 000 samples and 3, 000 common controls. – Bipolar, type 1 diabetes, type 2 diabetes, coronary artery disease, hypertension, rheumatoid arthritis, Crohn’s disease + Breast Cancer – 1, 500 1958 Birth Cohort and 1, 500 National Blood Donor • Designed a specific array using GSV set, Mc. Carroll, 1 M and WTCCC 1 – 104, 000 probes targeting 12, 000 putatitve loci • Perform assay using the Agilent platform by Oxford Gene Technology (OGT) against a common pooled reference sample • Attempt to design a robust pipeline to call CNV across the different studies – Use CNVtools by Plagnol and local by Cardin (“Chiamesque”) http: //www. wtccc. org. uk/ccc 1/plus_typing_array. shtml 36

The WTCCC results • 3, 900 CNV identified • 3, 100 validated after QC • Concordance of 99. 8% on known 420 duplicates • Remaining 8, 000 CNVs from original selection: – False positive in discovery – Too noisy, but genuine – Genuine but very rare • 19 CNVs taken forward to replication with Bayes Factor: ~10 -4 p-value – 14 failed to replicate either using tagged SNPs or direct typing – 5 associations 37
The WTCCC conclusions • Each CNV behaves uniquely • Size, genomic location, biological sample type, sample preparation – Designed 16 different pipelines • Key paramaters: – – Normalization Integration of the 10 probes • Impossible to define one-pipe-fits all – Show importance to have duplicates and large amount of diverse data • Confirmed the overrepresentation of CNVs in intronic regions • Confirm the high level of tag with SNP 6. 0 or Hap. Map 2 – MAF > 10% : 75% tagged at r 2>0. 8 – MAF <5% : 40% tagged at r 2>0. 8 • Found few new CNV associated with phenotype 38

Conclusions of these studies • Both identified many CNV in the human genome • Characterization of CNV is very difficult, and not easily stream lined – Careful interpretation of association results – Some artifacts will survive confirmation • Many CNVs co-localize with variants identified by GWAS – Good functional candidate • But, most of the common CNVs are already well tagged with SNPs – This will not bring new common variant in common disease • i. e. these will not solve the mystery of missing heritability. • Still rare CNVs can be associated to diseases, but just as much as SNPs 39
What with CNV then ? • Copy Number Variations are key in Cancer • Cancers are typical of somatic variations – They are therefore mostly unique – Cannot be tagged – Relatively common event – Although still difficult to identify it is essential 40
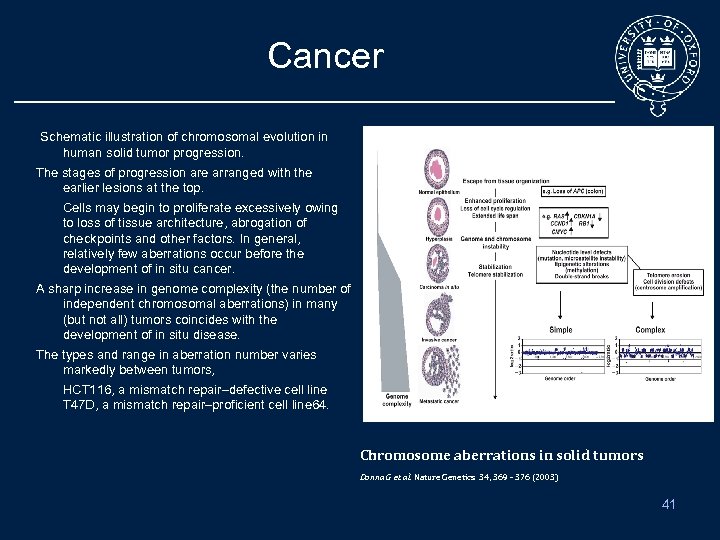
Cancer Schematic illustration of chromosomal evolution in human solid tumor progression. The stages of progression are arranged with the earlier lesions at the top. Cells may begin to proliferate excessively owing to loss of tissue architecture, abrogation of checkpoints and other factors. In general, relatively few aberrations occur before the development of in situ cancer. A sharp increase in genome complexity (the number of independent chromosomal aberrations) in many (but not all) tumors coincides with the development of in situ disease. The types and range in aberration number varies markedly between tumors, HCT 116, a mismatch repair–defective cell line T 47 D, a mismatch repair–proficient cell line 64. Chromosome aberrations in solid tumors Donna G et al. Nature Genetics 34, 369 - 376 (2003) 41
Germline vs. Somatic • Germline variants – The aberration exists from the start, and is inherited – Such variants are more likely to be common Copy Number Polymorphisms, predisposing variants. – Approach similar to non-cancer studies • Somatic events – Aberrations happen during the life-time – Happen more than once – Heterogeneous events; => Each cancer is unique – In Tumours, recurrent aberrations are more likely to be linked to the cancer as a selective advantage We want to identify the regions with recurrent events 42

More issues • Interpretation – What to use as a baseline ? i. e. define the Ratio • Within sample baseline of 2 is not an easy assumption anymore • Heterogeneity of tissue – Biopsy can be “contaminated” by normal tissue – Cancer are usually made up of a set of co-existing clones • CNVs are unique – Each one has its own breakpoints • Systematic error – Preparation, e. g. genome amplification – Sample quality 43
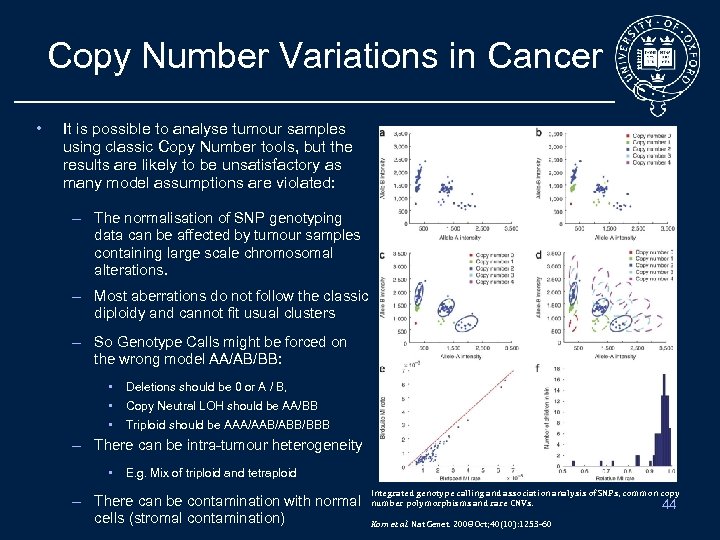
Copy Number Variations in Cancer • It is possible to analyse tumour samples using classic Copy Number tools, but the results are likely to be unsatisfactory as many model assumptions are violated: – The normalisation of SNP genotyping data can be affected by tumour samples containing large scale chromosomal alterations. – Most aberrations do not follow the classic diploidy and cannot fit usual clusters – So Genotype Calls might be forced on the wrong model AA/AB/BB: • Deletions should be 0 or A / B, • Copy Neutral LOH should be AA/BB • Triploid should be AAA/AAB/ABB/BBB – There can be intra-tumour heterogeneity • E. g. Mix of triploid and tetraploid – There can be contamination with normal cells (stromal contamination) Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. 44 Korn et al. Nat Genet. 2008 Oct; 40(10): 1253 -60
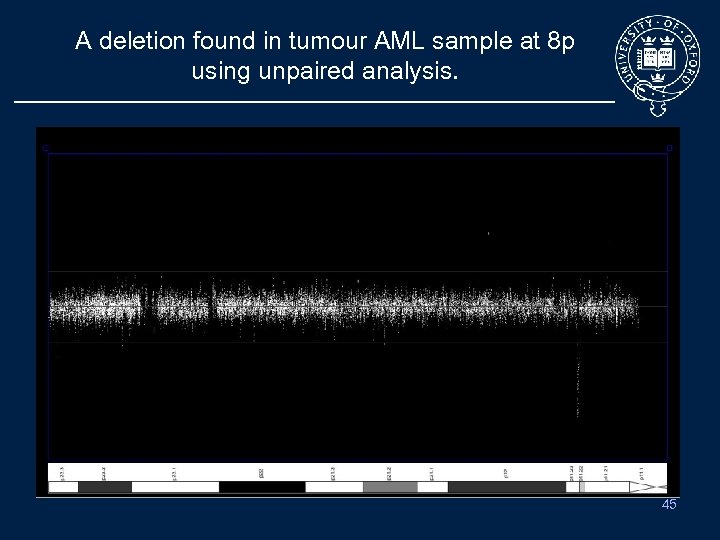
A deletion found in tumour AML sample at 8 p using unpaired analysis. Tumour sample vs Baseline 4 2 1 45
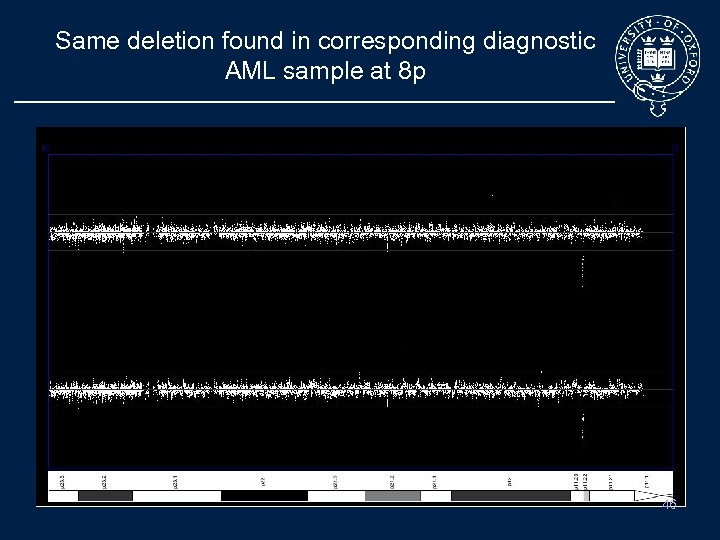
Same deletion found in corresponding diagnostic AML sample at 8 p Tumour sample vs Baseline 4 2 1 Normal sample vs Baseline 4 2 1 46
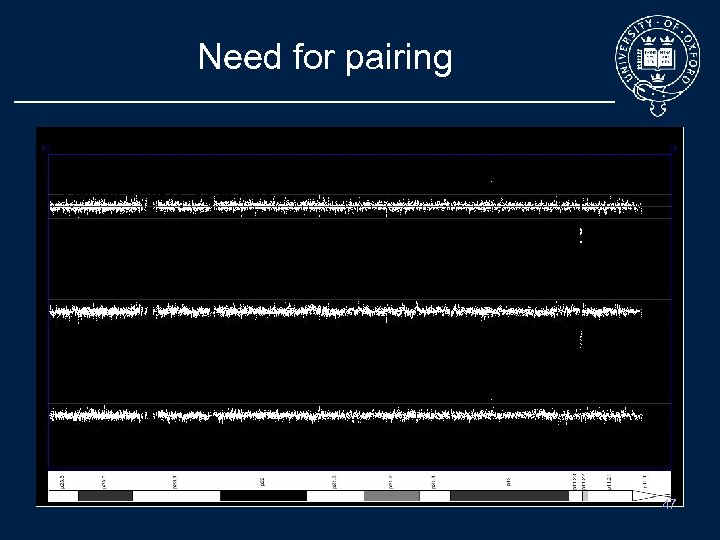
Need for pairing 4 2 1 Tumour sample vs Baseline Normal sample vs Baseline 4 2 1 Tumour sample vs Normal sample 47
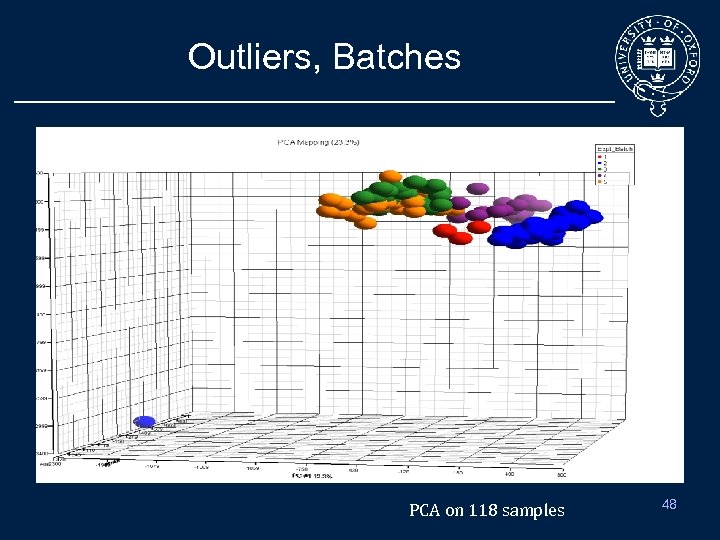
Outliers, Batches PCA on 118 samples 48
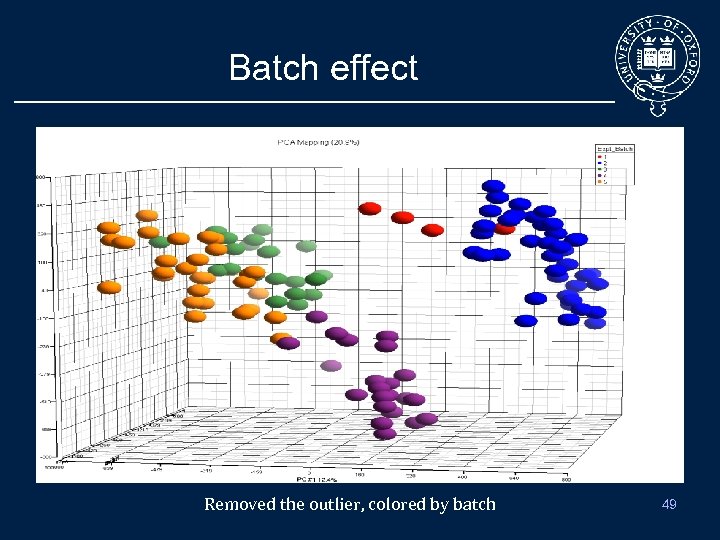
Batch effect Removed the outlier, colored by batch 49
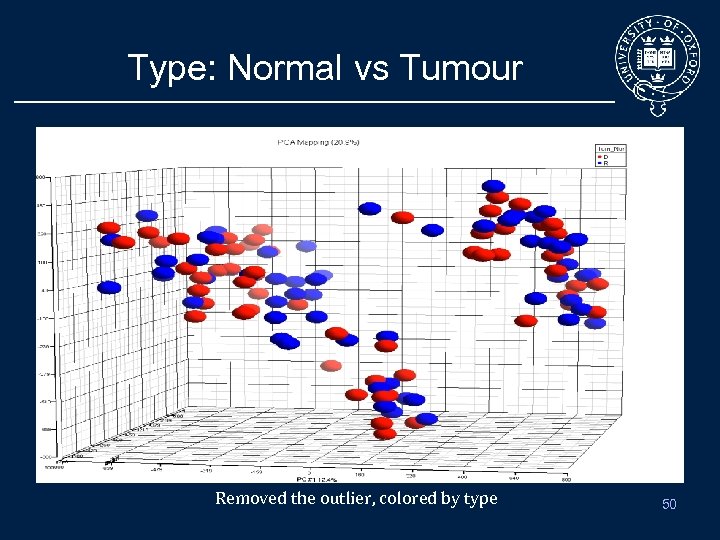
Type: Normal vs Tumour Removed the outlier, colored by type 50
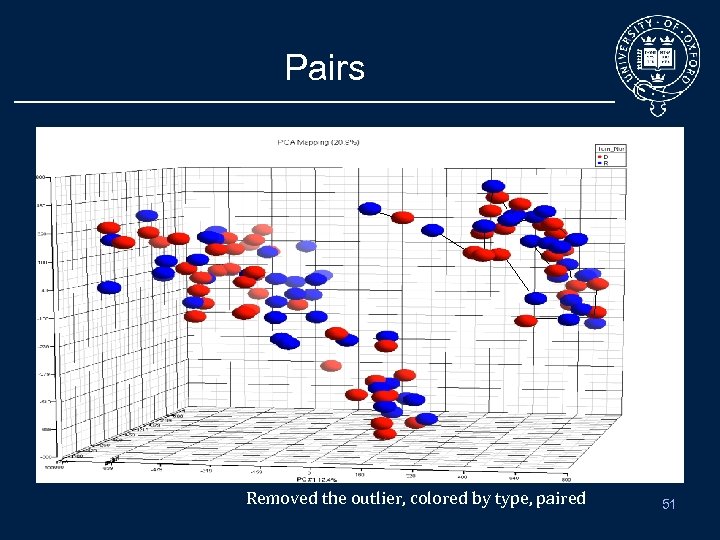
Pairs Removed the outlier, colored by type, paired 51
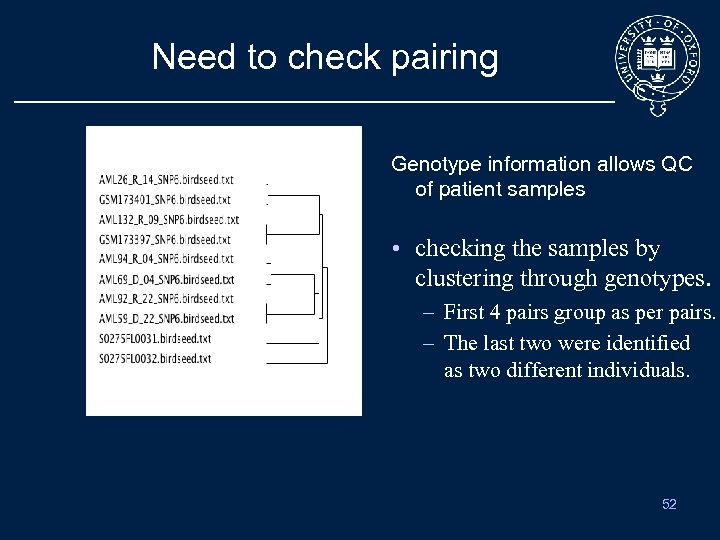
Need to check pairing Genotype information allows QC of patient samples • checking the samples by clustering through genotypes. – First 4 pairs group as per pairs. – The last two were identified as two different individuals. 52
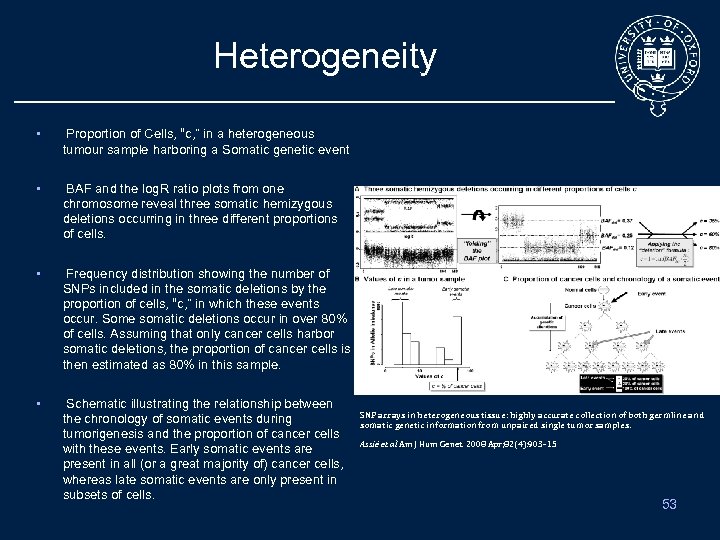
Heterogeneity • Proportion of Cells, “c, ” in a heterogeneous tumour sample harboring a Somatic genetic event • BAF and the log. R ratio plots from one chromosome reveal three somatic hemizygous deletions occurring in three different proportions of cells. • Frequency distribution showing the number of SNPs included in the somatic deletions by the proportion of cells, “c, ” in which these events occur. Some somatic deletions occur in over 80% of cells. Assuming that only cancer cells harbor somatic deletions, the proportion of cancer cells is then estimated as 80% in this sample. • Schematic illustrating the relationship between the chronology of somatic events during tumorigenesis and the proportion of cancer cells with these events. Early somatic events are present in all (or a great majority of) cancer cells, whereas late somatic events are only present in subsets of cells. SNP arrays in heterogeneous tissue: highly accurate collection of both germline and somatic genetic information from unpaired single tumor samples. Assié et al Am J Hum Genet. 2008 Apr; 82(4): 903 -15 53
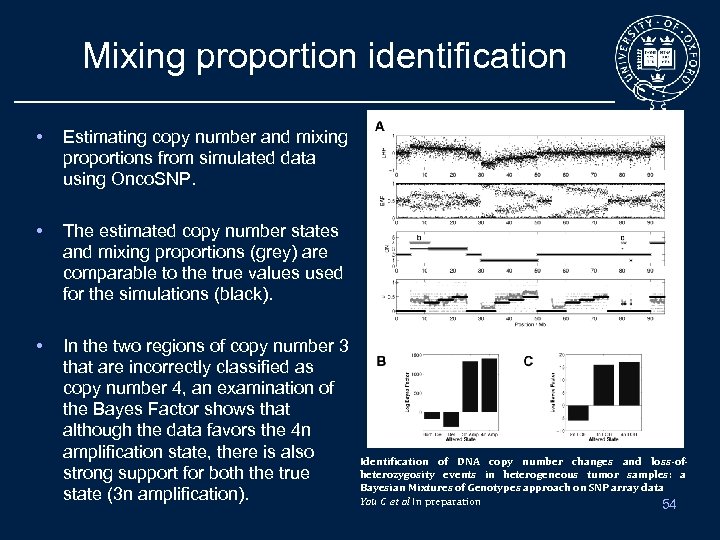
Mixing proportion identification • Estimating copy number and mixing proportions from simulated data using Onco. SNP. • The estimated copy number states and mixing proportions (grey) are comparable to the true values used for the simulations (black). • In the two regions of copy number 3 that are incorrectly classified as copy number 4, an examination of the Bayes Factor shows that although the data favors the 4 n amplification state, there is also strong support for both the true state (3 n amplification). Identification of DNA copy number changes and loss-ofheterozygosity events in heterogeneous tumor samples: a Bayesian Mixtures of Genotypes approach on SNP array data Yau C et al In preparation 54
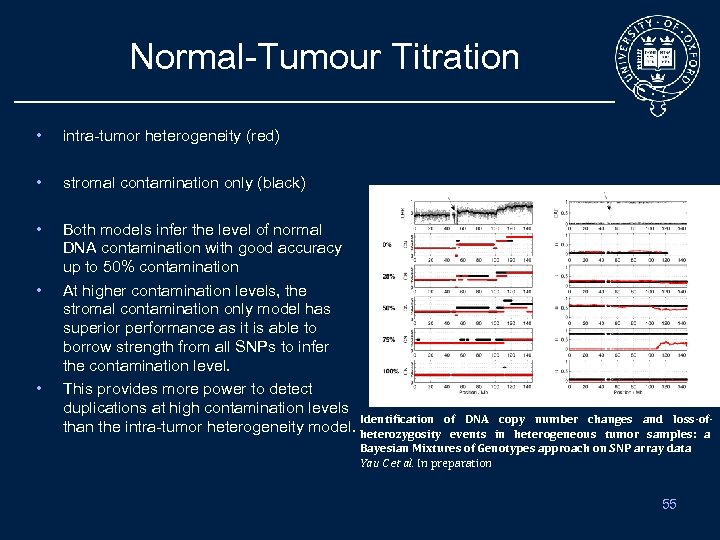
Normal-Tumour Titration • intra-tumor heterogeneity (red) • stromal contamination only (black) • Both models infer the level of normal DNA contamination with good accuracy up to 50% contamination At higher contamination levels, the stromal contamination only model has superior performance as it is able to borrow strength from all SNPs to infer the contamination level. This provides more power to detect duplications at high contamination levels Identification of DNA than the intra-tumor heterogeneity model. heterozygosity events • • copy number changes and loss-ofin heterogeneous tumor samples: a Bayesian Mixtures of Genotypes approach on SNP array data Yau C et al. In preparation 55
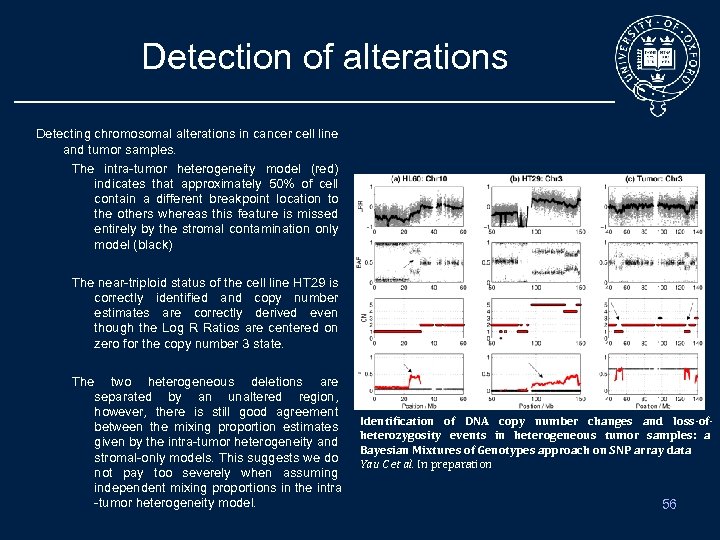
Detection of alterations Detecting chromosomal alterations in cancer cell line and tumor samples. The intra-tumor heterogeneity model (red) indicates that approximately 50% of cell contain a different breakpoint location to the others whereas this feature is missed entirely by the stromal contamination only model (black) The near-triploid status of the cell line HT 29 is correctly identified and copy number estimates are correctly derived even though the Log R Ratios are centered on zero for the copy number 3 state. The two heterogeneous deletions are separated by an unaltered region, however, there is still good agreement between the mixing proportion estimates given by the intra-tumor heterogeneity and stromal-only models. This suggests we do not pay too severely when assuming independent mixing proportions in the intra -tumor heterogeneity model. Identification of DNA copy number changes and loss-ofheterozygosity events in heterogeneous tumor samples: a Bayesian Mixtures of Genotypes approach on SNP array data Yau C et al. In preparation 56
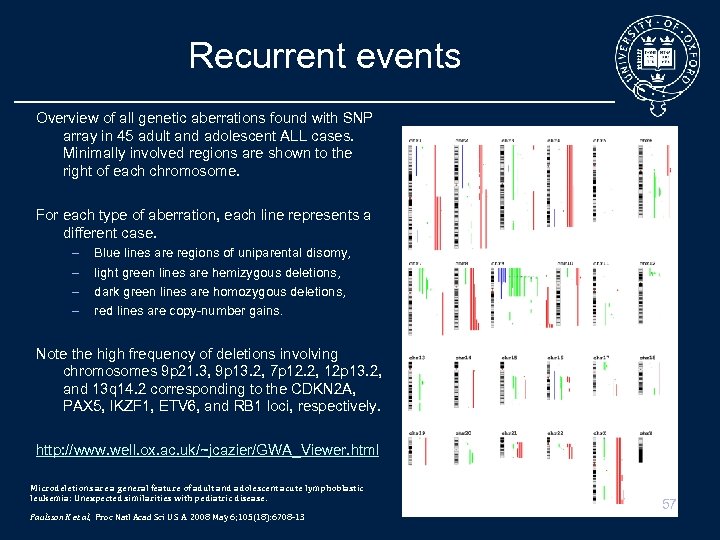
Recurrent events Overview of all genetic aberrations found with SNP array in 45 adult and adolescent ALL cases. Minimally involved regions are shown to the right of each chromosome. For each type of aberration, each line represents a different case. – – Blue lines are regions of uniparental disomy, light green lines are hemizygous deletions, dark green lines are homozygous deletions, red lines are copy-number gains. Note the high frequency of deletions involving chromosomes 9 p 21. 3, 9 p 13. 2, 7 p 12. 2, 12 p 13. 2, and 13 q 14. 2 corresponding to the CDKN 2 A, PAX 5, IKZF 1, ETV 6, and RB 1 loci, respectively. http: //www. well. ox. ac. uk/~jcazier/GWA_Viewer. html Microdeletions are a general feature of adult and adolescent acute lymphoblastic leukemia: Unexpected similarities with pediatric disease. Paulsson K et al, Proc Natl Acad Sci U S A. 2008 May 6; 105(18): 6708 -13 57
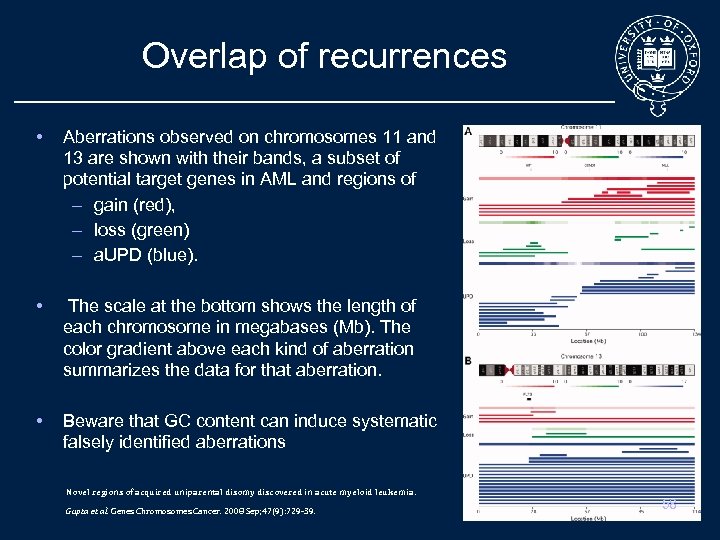
Overlap of recurrences • Aberrations observed on chromosomes 11 and 13 are shown with their bands, a subset of potential target genes in AML and regions of – gain (red), – loss (green) – a. UPD (blue). • The scale at the bottom shows the length of each chromosome in megabases (Mb). The color gradient above each kind of aberration summarizes the data for that aberration. • Beware that GC content can induce systematic falsely identified aberrations Novel regions of acquired uniparental disomy discovered in acute myeloid leukemia. Gupta et al. Genes Chromosomes Cancer. 2008 Sep; 47(9): 729 -39. 58

Typical workflow • • • Normalisation – GC Content Correction – Paired – Unpaired with appropriate baseline Determination of Aberrations – Correct Genotype – Copy Number Identification of recurrent locations Test against germline sample if possible – Could it be an at-risk variant ? Test against known variations Validation – Identify precisely breakpoints • Sequencing – Identify the frequency – Identify the Associated risk – Perform functional analysis 59

Summary • CNP are very common in the human genome – It is easier to have a functional role for them • Common ones are well tagged by existing markers – Does not bring much new loci, but function • Hard to characterize uniformly • Not yet much proven functional • Still very key in Cancer – More challenges to be identified – More essential for the understanding 60
Future • Catalogue of CNPs – GSV and WTCCC effort – Use of the 1000 genome project • Methods – Improvements of the algorithms – Improvements of the Computing power • Other technologies – Use of expression data – Use of Clonal Sequencing – Single molecule sequencing 61
Useful references • Collections of known aberrations: – Mitelman Database of Chromosome Aberration in Cancer • • http: //cgap. nci. nih. gov/Chromsomes/Mitelman cytogenetic confirmed – Database of Genomics Variants • Zhang, J et al. Development of bioinformatics resources for display and analysis of copy number and other structural variants in the human genome. Cytogenet. Genome Res. (2006). – Redon, R. et al. Global variation in copy number in the human genome. Nature, (2006). – Iafrate, A. J. et al. Detection of large-scale variation in the human genome. Nat. Genet. (2004). – Mc. Carrol & Korn (2008) • Based on SNP 6. 0 in 270 Hap. Map samples – Genome Structural Variation Consortium • Conrad D et al. Origins and functional impact of copy number variation in the human genome Nat. 62 Genet. (2009)

Practical • R- packages: – DNACopy: • A Package for Analyzing DNA Copy data • A faster circular binary segmentation algorithm for the analysis of array cgh data. Venkatraman, E. S. and Olshen, A. B. (2007). Bioinformatics, 23: 657 – 663 – snap. CGH: • Segmentation, Normalization and Processing of a. CGH Data • Bio. HMM: a heterogeneous hidden Markov model for segmenting array CGH data. Marioni, J. C. , Thorne, N. P. , and Tavaré, S. (2006). Bioinformatics 22: 1144 – 1146 – Beadarray. SNP: • • • package for the analysis of Illumina genotyping Bead. Array data High-resolution copy number analysis of paraffin-embedded archival tissue using SNP Bead. Arrays. Oosting J et al. Genome Res. 2007 Mar; 17(3): 368 -76 Web interface: – Integration of CNV results across multiple samples – http: //www. well. ox. ac. uk/~jcazier/GWA_Viewer. html 63
dcdc3ece63ba334de5ab482e1abf6986.ppt