79fe9df9c8b2db1af46504b628e7fe0b.ppt
- Количество слайдов: 47
 Control of Eukaryotic Genes (Ch. 19)
Control of Eukaryotic Genes (Ch. 19)
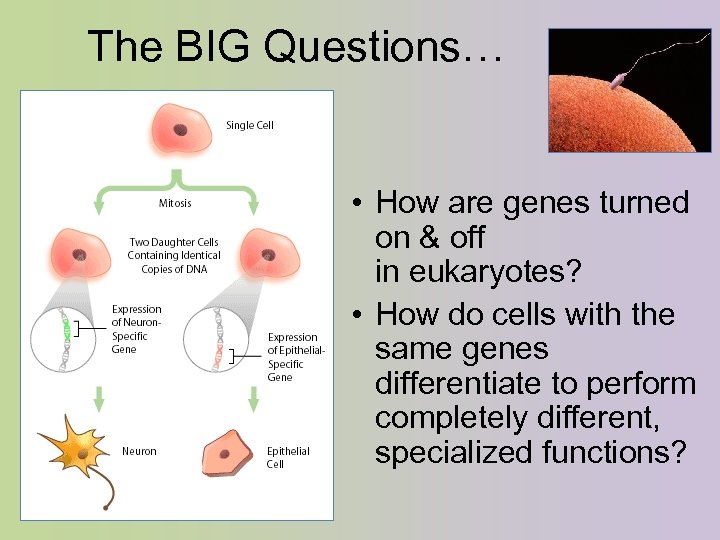 The BIG Questions… • How are genes turned on & off in eukaryotes? • How do cells with the same genes differentiate to perform completely different, specialized functions?
The BIG Questions… • How are genes turned on & off in eukaryotes? • How do cells with the same genes differentiate to perform completely different, specialized functions?
 Evolution of gene regulation • Prokaryotes – single-celled – evolved to grow & divide rapidly – must respond quickly to changes in external environment • exploit transient resources • Gene regulation- Operons – turn genes on & off rapidly • flexibility & reversibility – adjust levels of enzymes for synthesis & digestion
Evolution of gene regulation • Prokaryotes – single-celled – evolved to grow & divide rapidly – must respond quickly to changes in external environment • exploit transient resources • Gene regulation- Operons – turn genes on & off rapidly • flexibility & reversibility – adjust levels of enzymes for synthesis & digestion
 Evolution of gene regulation • Eukaryotes – multicellular – evolved to maintain homeostasis – regulate body as a whole • specialization – turn on & off large number of genes • must coordinate the body as a whole rather than serve the needs of individual cells
Evolution of gene regulation • Eukaryotes – multicellular – evolved to maintain homeostasis – regulate body as a whole • specialization – turn on & off large number of genes • must coordinate the body as a whole rather than serve the needs of individual cells
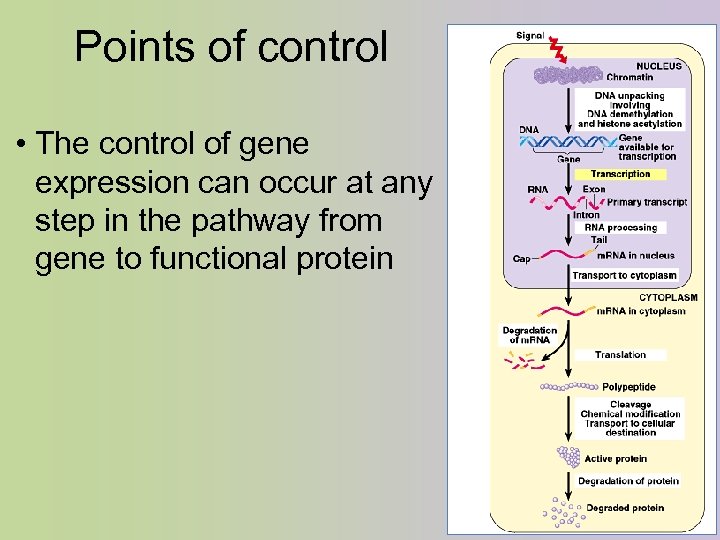 Points of control • The control of gene expression can occur at any step in the pathway from gene to functional protein
Points of control • The control of gene expression can occur at any step in the pathway from gene to functional protein
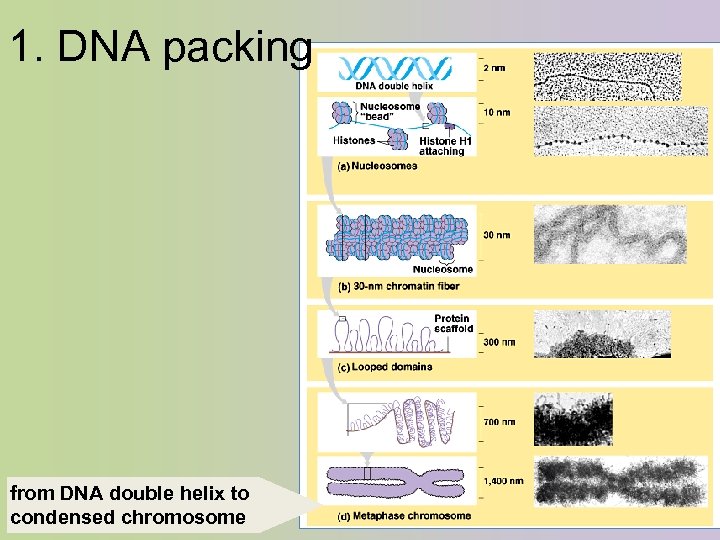 1. DNA packing from DNA double helix to condensed chromosome
1. DNA packing from DNA double helix to condensed chromosome
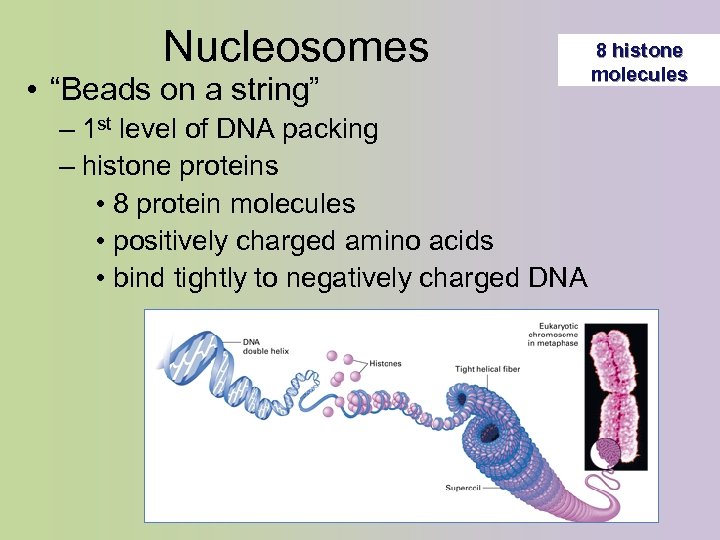 Nucleosomes • “Beads on a string” – 1 st level of DNA packing – histone proteins • 8 protein molecules • positively charged amino acids • bind tightly to negatively charged DNA 8 histone molecules
Nucleosomes • “Beads on a string” – 1 st level of DNA packing – histone proteins • 8 protein molecules • positively charged amino acids • bind tightly to negatively charged DNA 8 histone molecules
 DNA packing as gene control • Degree of packing of DNA regulates transcription – tightly wrapped around histones • no transcription • genes turned off § heterochromatin darker DNA (H) = tightly packed § euchromatin lighter DNA (E) = loosely packed
DNA packing as gene control • Degree of packing of DNA regulates transcription – tightly wrapped around histones • no transcription • genes turned off § heterochromatin darker DNA (H) = tightly packed § euchromatin lighter DNA (E) = loosely packed
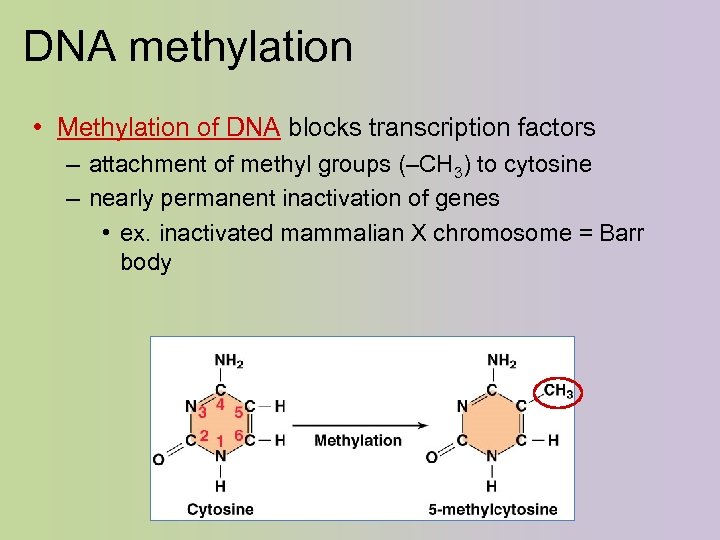 DNA methylation • Methylation of DNA blocks transcription factors – attachment of methyl groups (–CH 3) to cytosine – nearly permanent inactivation of genes • ex. inactivated mammalian X chromosome = Barr body
DNA methylation • Methylation of DNA blocks transcription factors – attachment of methyl groups (–CH 3) to cytosine – nearly permanent inactivation of genes • ex. inactivated mammalian X chromosome = Barr body
 Histone acetylation Chromatin changes ■ u Acetylation of histones unwinds DNA Loose histone wrapping ■ ■ u ■ RNA processing m. RNA degradation Translation Protein processing and degradation Histone tails enables transcription genes turned on attachment of acetyl groups ( –COCH 3) to histones ■ Transcription conformational change in histone proteins transcription factors have easier access to genes DNA double helix Amino acids available for chemical modification (a) Histone tails protrude outward from a nucleosome Unacetylated histones Acetylated histones (b) Acetylation of histone tails promotes loose chromatin structure that permits transcription
Histone acetylation Chromatin changes ■ u Acetylation of histones unwinds DNA Loose histone wrapping ■ ■ u ■ RNA processing m. RNA degradation Translation Protein processing and degradation Histone tails enables transcription genes turned on attachment of acetyl groups ( –COCH 3) to histones ■ Transcription conformational change in histone proteins transcription factors have easier access to genes DNA double helix Amino acids available for chemical modification (a) Histone tails protrude outward from a nucleosome Unacetylated histones Acetylated histones (b) Acetylation of histone tails promotes loose chromatin structure that permits transcription
 2. Transcription initiation • Control regions on DNA – promoter • nearby control sequence on DNA • binding of RNA polymerase & transcription factors – enhancer • distant control sequences on DNA • binding of activator proteins
2. Transcription initiation • Control regions on DNA – promoter • nearby control sequence on DNA • binding of RNA polymerase & transcription factors – enhancer • distant control sequences on DNA • binding of activator proteins
 Model for Enhancer action • Enhancer DNA sequences – distant control sequences • Activator proteins – bind to enhancer sequence & stimulates transcription • Silencer proteins – bind to enhancer sequence & block gene transcription
Model for Enhancer action • Enhancer DNA sequences – distant control sequences • Activator proteins – bind to enhancer sequence & stimulates transcription • Silencer proteins – bind to enhancer sequence & block gene transcription
 Transcription complex Activator Proteins • regulatory proteins bind to DNA at Enhancer Sites distant enhancer sites • increase the rate of transcription regulatory sites on DNA distant from gene Enhancer Activator Coactivator A E F B TFIID H RNA polymerase II Coding r eg ion T A Core promoter and initiation complex Initiation Complex at Promoter Site binding site of RNA polymerase
Transcription complex Activator Proteins • regulatory proteins bind to DNA at Enhancer Sites distant enhancer sites • increase the rate of transcription regulatory sites on DNA distant from gene Enhancer Activator Coactivator A E F B TFIID H RNA polymerase II Coding r eg ion T A Core promoter and initiation complex Initiation Complex at Promoter Site binding site of RNA polymerase
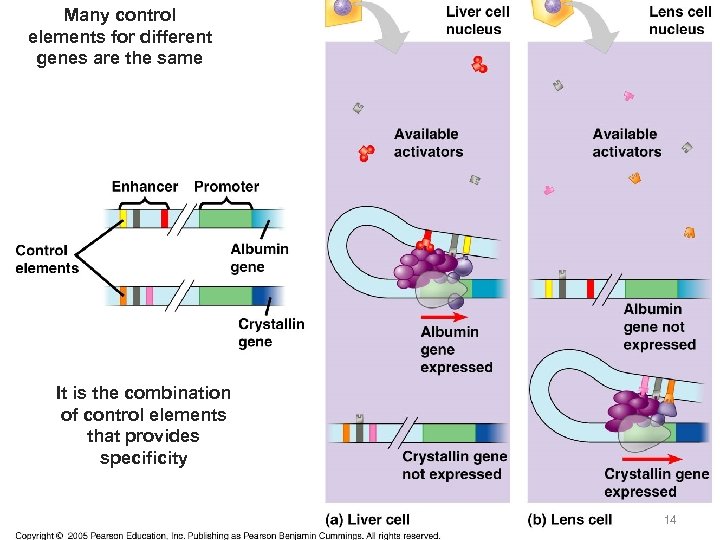 Many control elements for different genes are the same It is the combination of control elements that provides specificity 14
Many control elements for different genes are the same It is the combination of control elements that provides specificity 14
 3. Post-transcriptional control • Alternative RNA splicing – variable processing of exons creates a family of proteins
3. Post-transcriptional control • Alternative RNA splicing – variable processing of exons creates a family of proteins
 4. Regulation of m. RNA degradation • Life span of m. RNA determines amount of protein synthesis – m. RNA can last from hours to weeks
4. Regulation of m. RNA degradation • Life span of m. RNA determines amount of protein synthesis – m. RNA can last from hours to weeks
 RNA interference 1 The micro. RNA (mi. RNA) precursor folds back on itself, held together by hydrogen bonds. 2 2 An enzyme called Dicer moves along the doublestranded RNA, cutting it into shorter segments. One strand of 3 each short doublestranded RNA is degraded; the other strand (mi. RNA) then associates with a complex of proteins. 4 The bound mi. RNA can base-pair with any target m. RNA that contains the complementary sequence. NEW ! The mi. RNA-protein 5 complex prevents gene expression either by degrading the target m. RNA or by blocking its translation. Chromatin changes Transcription RNA processing m. RNA degradation Protein complex Translation Protein processing and degradation Dicer Degradation of m. RNA OR mi. RNA Target m. RNA Hydrogen bond Blockage of translation
RNA interference 1 The micro. RNA (mi. RNA) precursor folds back on itself, held together by hydrogen bonds. 2 2 An enzyme called Dicer moves along the doublestranded RNA, cutting it into shorter segments. One strand of 3 each short doublestranded RNA is degraded; the other strand (mi. RNA) then associates with a complex of proteins. 4 The bound mi. RNA can base-pair with any target m. RNA that contains the complementary sequence. NEW ! The mi. RNA-protein 5 complex prevents gene expression either by degrading the target m. RNA or by blocking its translation. Chromatin changes Transcription RNA processing m. RNA degradation Protein complex Translation Protein processing and degradation Dicer Degradation of m. RNA OR mi. RNA Target m. RNA Hydrogen bond Blockage of translation
 A Brief, RNAi Cartoon
A Brief, RNAi Cartoon
 mi. RNA’s • Scientists discovery video • Animation of mi. RNA function 19
mi. RNA’s • Scientists discovery video • Animation of mi. RNA function 19
 RNA interference 1990 s | 2006 “for their discovery of RNA interference — gene silencing by double-stranded RNA” Andrew Fire Stanford Craig Mello U Mass
RNA interference 1990 s | 2006 “for their discovery of RNA interference — gene silencing by double-stranded RNA” Andrew Fire Stanford Craig Mello U Mass
 5. Control of translation • Block initiation of translation stage – regulatory proteins attach to 5' end of m. RNA • prevent attachment of ribosomal subunits & initiator t. RNA • block translation of m. RNA to protein
5. Control of translation • Block initiation of translation stage – regulatory proteins attach to 5' end of m. RNA • prevent attachment of ribosomal subunits & initiator t. RNA • block translation of m. RNA to protein
 6 -7. Protein processing & degradation • Protein processing – folding, cleaving, adding sugar groups, targeting for transport • Protein degradation – ubiquitin tagging – proteasome degradation
6 -7. Protein processing & degradation • Protein processing – folding, cleaving, adding sugar groups, targeting for transport • Protein degradation – ubiquitin tagging – proteasome degradation
 Ubiquitin 1980 s | 2004 • “Death tag” – mark unwanted proteins with a label – 76 amino acid polypeptide, ubiquitin – labeled proteins are broken down rapidly in "waste disposers" • proteasomes Aaron Ciechanover Israel Avram Hershko Israel Irwin Rose UC Riverside
Ubiquitin 1980 s | 2004 • “Death tag” – mark unwanted proteins with a label – 76 amino acid polypeptide, ubiquitin – labeled proteins are broken down rapidly in "waste disposers" • proteasomes Aaron Ciechanover Israel Avram Hershko Israel Irwin Rose UC Riverside
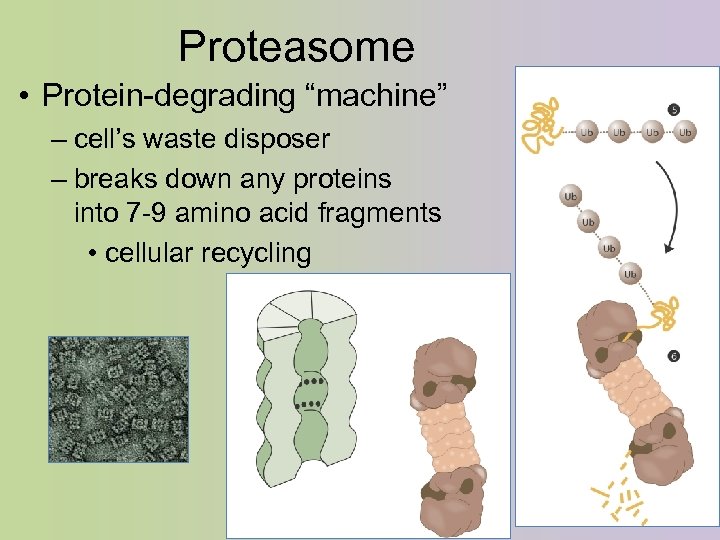 Proteasome • Protein-degrading “machine” – cell’s waste disposer – breaks down any proteins into 7 -9 amino acid fragments • cellular recycling
Proteasome • Protein-degrading “machine” – cell’s waste disposer – breaks down any proteins into 7 -9 amino acid fragments • cellular recycling
 6 7 Gene Regulation protein processing & degradation 1 & 2. transcription - DNA packing - transcription factors 3 & 4. post-transcription - m. RNA processing - splicing 4 - 5’ cap & poly-A tail m. RNA - breakdown by si. RNA processing 5 initiation of translation 5. translation - block start of translation 1 2 initiation of transcription 3 m. RNA splicing 6 & 7. post-translation - protein processing - protein degradation 4 m. RNA protection
6 7 Gene Regulation protein processing & degradation 1 & 2. transcription - DNA packing - transcription factors 3 & 4. post-transcription - m. RNA processing - splicing 4 - 5’ cap & poly-A tail m. RNA - breakdown by si. RNA processing 5 initiation of translation 5. translation - block start of translation 1 2 initiation of transcription 3 m. RNA splicing 6 & 7. post-translation - protein processing - protein degradation 4 m. RNA protection
 Concept Check 1. Do mi. RNA’s increase or decrease gene expression? 2. Does histone de-acetylation increase or decrease gene expression? 26
Concept Check 1. Do mi. RNA’s increase or decrease gene expression? 2. Does histone de-acetylation increase or decrease gene expression? 26
 Genome Sizes and Estimated Numbers of Genes*
Genome Sizes and Estimated Numbers of Genes*
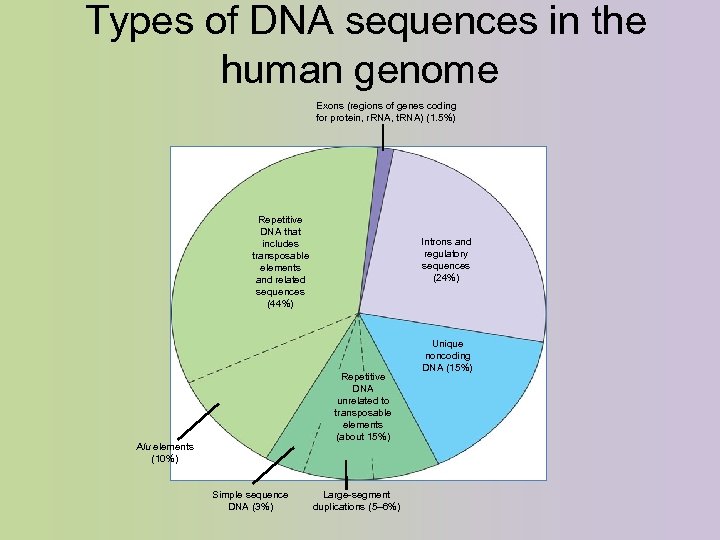 Types of DNA sequences in the human genome Exons (regions of genes coding for protein, r. RNA, t. RNA) (1. 5%) Repetitive DNA that includes transposable elements and related sequences (44%) Introns and regulatory sequences (24%) Repetitive DNA unrelated to transposable elements (about 15%) Alu elements (10%) Simple sequence DNA (3%) Large-segment duplications (5– 6%) Unique noncoding DNA (15%)
Types of DNA sequences in the human genome Exons (regions of genes coding for protein, r. RNA, t. RNA) (1. 5%) Repetitive DNA that includes transposable elements and related sequences (44%) Introns and regulatory sequences (24%) Repetitive DNA unrelated to transposable elements (about 15%) Alu elements (10%) Simple sequence DNA (3%) Large-segment duplications (5– 6%) Unique noncoding DNA (15%)
 Just how little of out Genome Codes for Genes?
Just how little of out Genome Codes for Genes?
 Cancer results from genetic changes in cell cycle control 30
Cancer results from genetic changes in cell cycle control 30
 Proto-oncogene --> oncogene 31
Proto-oncogene --> oncogene 31
 32
32
 Multistep Model of cancer 33
Multistep Model of cancer 33
 Genome Evolution • How did our genome get so big? 34
Genome Evolution • How did our genome get so big? 34
 35
35
 36
36
 • The similarity in the amino acid sequences of the various globin proteins – Supports this model of gene duplication and mutation Table 19. 1 37
• The similarity in the amino acid sequences of the various globin proteins – Supports this model of gene duplication and mutation Table 19. 1 37
 Evolution of Genes with Novel Functions • The copies of some duplicated genes – Have diverged so much during evolutionary time that the functions of their encoded proteins are now substantially different 38
Evolution of Genes with Novel Functions • The copies of some duplicated genes – Have diverged so much during evolutionary time that the functions of their encoded proteins are now substantially different 38
 39
39
 Concept check 1. How much of the human genome consists of exons? 2. How can exon shuffling lead to the evolution of a new gene 40
Concept check 1. How much of the human genome consists of exons? 2. How can exon shuffling lead to the evolution of a new gene 40
 Turn your Question Genes on!
Turn your Question Genes on!
 A larger portion of the DNA in a eukaryotic cell is transcribed than would be predicted by the proteins made by the cell. What is being transcribed and what is its function? 1. Multiple enhancer regions are being transcribed to amplify the transcription of protein-coding genes. 2. These transcriptions are of non-coding “junk” DNA that is a remnant of mutated protein coding segments. The transcripts are degraded by nuclear enzymes. 3. The additional DNA that is transcribed is introns that are excised fro the primary transcript in the produciton of m. RNA. 4. Many RNA coding genes are transcribed. Precursor RNAs fold into hairpin structures, which are cut and processed into mi. RNAs that regulate translation of m. RNAs. 42
A larger portion of the DNA in a eukaryotic cell is transcribed than would be predicted by the proteins made by the cell. What is being transcribed and what is its function? 1. Multiple enhancer regions are being transcribed to amplify the transcription of protein-coding genes. 2. These transcriptions are of non-coding “junk” DNA that is a remnant of mutated protein coding segments. The transcripts are degraded by nuclear enzymes. 3. The additional DNA that is transcribed is introns that are excised fro the primary transcript in the produciton of m. RNA. 4. Many RNA coding genes are transcribed. Precursor RNAs fold into hairpin structures, which are cut and processed into mi. RNAs that regulate translation of m. RNAs. 42
 How is the coordinated transcription of genes involved in the same pathway regulated? 1. The genes are transcribed in one transcription unit, although each gene has its own promoter. 2. The genes are located in the same region of the chromosome, and enzymes de-acetylate the entire region so that transcription may begin. 3. A steroid hormone selectively binds to the promoters for all the genes. 4. The genes have the same combination of control elements in the enhancer that bind with the particular activators present in the cell. 43
How is the coordinated transcription of genes involved in the same pathway regulated? 1. The genes are transcribed in one transcription unit, although each gene has its own promoter. 2. The genes are located in the same region of the chromosome, and enzymes de-acetylate the entire region so that transcription may begin. 3. A steroid hormone selectively binds to the promoters for all the genes. 4. The genes have the same combination of control elements in the enhancer that bind with the particular activators present in the cell. 43
 Histones are 1. Small positively charged proteins that bind tightly to DNA 2. Small bodies in the nucleus involved in r. RNA synthesis 3. Basic units of DNA packing consisting of DNA wound around a protein core. 4. DNA bending proteins that facilitate formation of transcription initiation complexes. 5. Proteins responsible for producing repeating sequences at telomeres. 44
Histones are 1. Small positively charged proteins that bind tightly to DNA 2. Small bodies in the nucleus involved in r. RNA synthesis 3. Basic units of DNA packing consisting of DNA wound around a protein core. 4. DNA bending proteins that facilitate formation of transcription initiation complexes. 5. Proteins responsible for producing repeating sequences at telomeres. 44
 Heterochromatin 1. Has a higher degree of packing than does euchromatin. 2. Is visible with a light microscope during interphase. 3. Is not actively involved in transcription 4. Makes up metaphase chromosomes 5. Is all of the above. 45
Heterochromatin 1. Has a higher degree of packing than does euchromatin. 2. Is visible with a light microscope during interphase. 3. Is not actively involved in transcription 4. Makes up metaphase chromosomes 5. Is all of the above. 45
 What is the main reason that prokaryotic genes average 1, 000 nucleotride base pairs whereas human genes average about 27. 000 base pairs? 1. Prokaryotes have smaller, bue many more individual genes. 2. Prokaryotes are more ancient organisms, longer genes arose later in evolution. 3. Prokaryotes are much simpler organism humans have many types of differentiated cells. 4. Prokaryotic genes do not have introns, human genes have many 5. Human proteins are much larger and more complex than prokaryotic proteins. 46
What is the main reason that prokaryotic genes average 1, 000 nucleotride base pairs whereas human genes average about 27. 000 base pairs? 1. Prokaryotes have smaller, bue many more individual genes. 2. Prokaryotes are more ancient organisms, longer genes arose later in evolution. 3. Prokaryotes are much simpler organism humans have many types of differentiated cells. 4. Prokaryotic genes do not have introns, human genes have many 5. Human proteins are much larger and more complex than prokaryotic proteins. 46
 A eukaryotic gene typically has all of the following associated with it except 1. A promoter 2. An operator 3. Enhancers 4. Introns and exons 5. Control elements. 47
A eukaryotic gene typically has all of the following associated with it except 1. A promoter 2. An operator 3. Enhancers 4. Introns and exons 5. Control elements. 47


