79f004e5cdc013fc05804002b9422956.ppt
- Количество слайдов: 46
 Consolidating Software Tools for DNA Microarray Design and Manufacturing Mourad Atlas Nisar Hundewale Ludmila Perelygina Alex Zelikovsky
Consolidating Software Tools for DNA Microarray Design and Manufacturing Mourad Atlas Nisar Hundewale Ludmila Perelygina Alex Zelikovsky
 Agenda n n n Introduction DNA Array Flow (DAF) Benchmarks: Herpes B virus Experiments and Results Conclusion and Future Work
Agenda n n n Introduction DNA Array Flow (DAF) Benchmarks: Herpes B virus Experiments and Results Conclusion and Future Work
 Motivation n Microarrays provide a tool for answering a wide variety of questions about the dynamics of cells: q In which cells is each gene active? q Under what environmental conditions is each gene active? q How does the activity level of a gene change under different conditions? n Stage of a cell cycle? n Environmental conditions? n Diseases? q What genes seem to be regulated together?
Motivation n Microarrays provide a tool for answering a wide variety of questions about the dynamics of cells: q In which cells is each gene active? q Under what environmental conditions is each gene active? q How does the activity level of a gene change under different conditions? n Stage of a cell cycle? n Environmental conditions? n Diseases? q What genes seem to be regulated together?
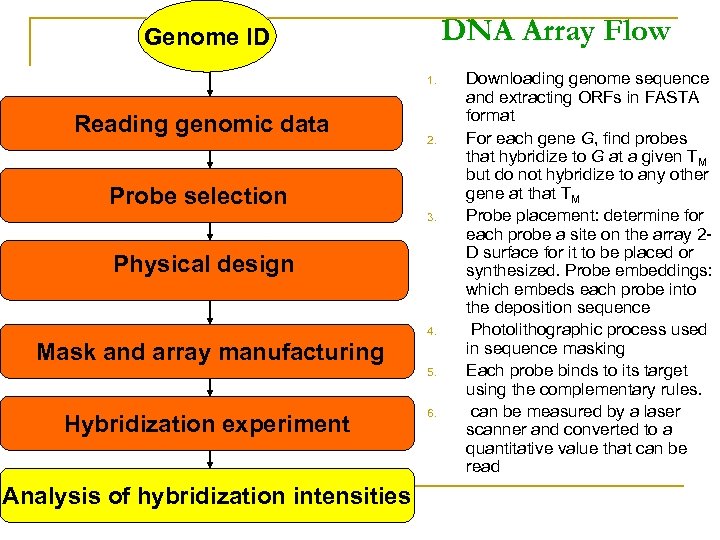 DNA Array Flow Genome ID 1. Reading genomic data 2. Probe selection 3. Physical design 4. Mask and array manufacturing 5. Hybridization experiment Analysis of hybridization intensities 6. Downloading genome sequence and extracting ORFs in FASTA format For each gene G, find probes that hybridize to G at a given TM but do not hybridize to any other gene at that TM Probe placement: determine for each probe a site on the array 2 D surface for it to be placed or synthesized. Probe embeddings: which embeds each probe into the deposition sequence Photolithographic process used in sequence masking Each probe binds to its target using the complementary rules. can be measured by a laser scanner and converted to a quantitative value that can be read
DNA Array Flow Genome ID 1. Reading genomic data 2. Probe selection 3. Physical design 4. Mask and array manufacturing 5. Hybridization experiment Analysis of hybridization intensities 6. Downloading genome sequence and extracting ORFs in FASTA format For each gene G, find probes that hybridize to G at a given TM but do not hybridize to any other gene at that TM Probe placement: determine for each probe a site on the array 2 D surface for it to be placed or synthesized. Probe embeddings: which embeds each probe into the deposition sequence Photolithographic process used in sequence masking Each probe binds to its target using the complementary rules. can be measured by a laser scanner and converted to a quantitative value that can be read
 Genome ID Reading genomic data Probe selection Physical design Mask and array manufacturing Hybridization experiment Analysis of hybridization intensities
Genome ID Reading genomic data Probe selection Physical design Mask and array manufacturing Hybridization experiment Analysis of hybridization intensities
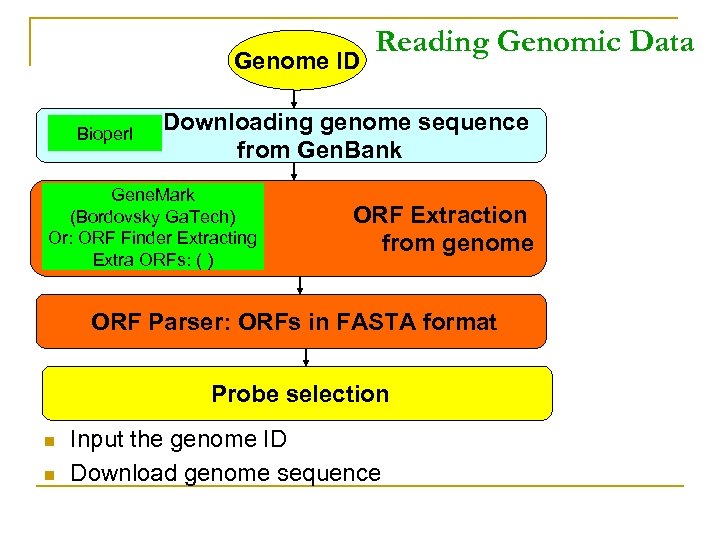 Genome ID Bioperl Reading Genomic Data Downloading genome sequence from Gen. Bank Gene. Mark (Bordovsky Ga. Tech) Or: ORF Finder Extracting Extra ORFs: ( ) ORF Extraction from genome ORF Parser: ORFs in FASTA format Probe selection n n Input the genome ID Download genome sequence
Genome ID Bioperl Reading Genomic Data Downloading genome sequence from Gen. Bank Gene. Mark (Bordovsky Ga. Tech) Or: ORF Finder Extracting Extra ORFs: ( ) ORF Extraction from genome ORF Parser: ORFs in FASTA format Probe selection n n Input the genome ID Download genome sequence
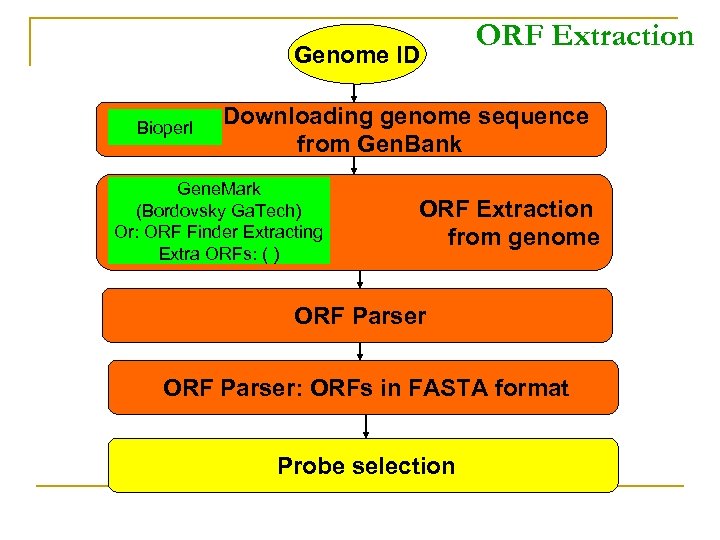 Genome ID Bioperl ORF Extraction Downloading genome sequence from Gen. Bank Gene. Mark (Bordovsky Ga. Tech) Or: ORF Finder Extracting Extra ORFs: ( ) ORF Extraction from genome ORF Parser: ORFs in FASTA format Probe selection
Genome ID Bioperl ORF Extraction Downloading genome sequence from Gen. Bank Gene. Mark (Bordovsky Ga. Tech) Or: ORF Finder Extracting Extra ORFs: ( ) ORF Extraction from genome ORF Parser: ORFs in FASTA format Probe selection
 What is ORF? n n 1. 2. Open reading frame (ORF) is a subsequence of DNA that could potentially be transcribed into messenger RNA (m. RNA) Because of the differences between prokaryotic and eukaryotic transcription systems there are two types of ORF: Prokaryotes: start and stop codon Eukaryotic: stop codon
What is ORF? n n 1. 2. Open reading frame (ORF) is a subsequence of DNA that could potentially be transcribed into messenger RNA (m. RNA) Because of the differences between prokaryotic and eukaryotic transcription systems there are two types of ORF: Prokaryotes: start and stop codon Eukaryotic: stop codon
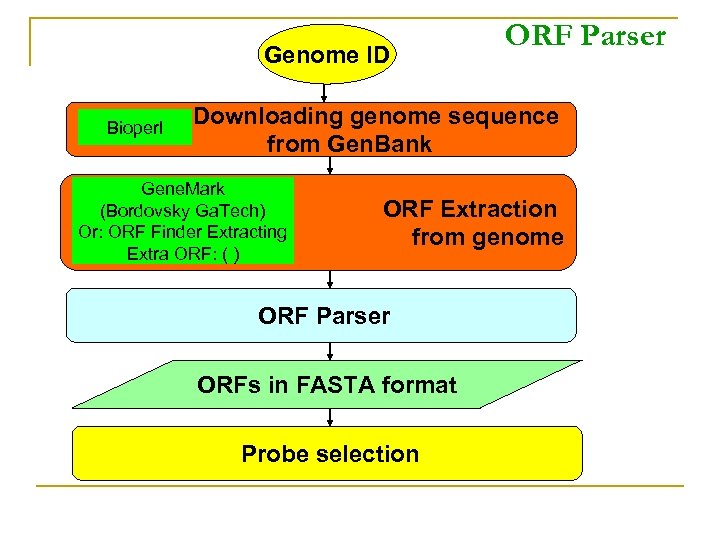 Genome ID Bioperl ORF Parser Downloading genome sequence from Gen. Bank Gene. Mark (Bordovsky Ga. Tech) Or: ORF Finder Extracting Extra ORF: ( ) ORF Extraction from genome ORF Parser ORFs in FASTA format Probe selection
Genome ID Bioperl ORF Parser Downloading genome sequence from Gen. Bank Gene. Mark (Bordovsky Ga. Tech) Or: ORF Finder Extracting Extra ORF: ( ) ORF Extraction from genome ORF Parser ORFs in FASTA format Probe selection
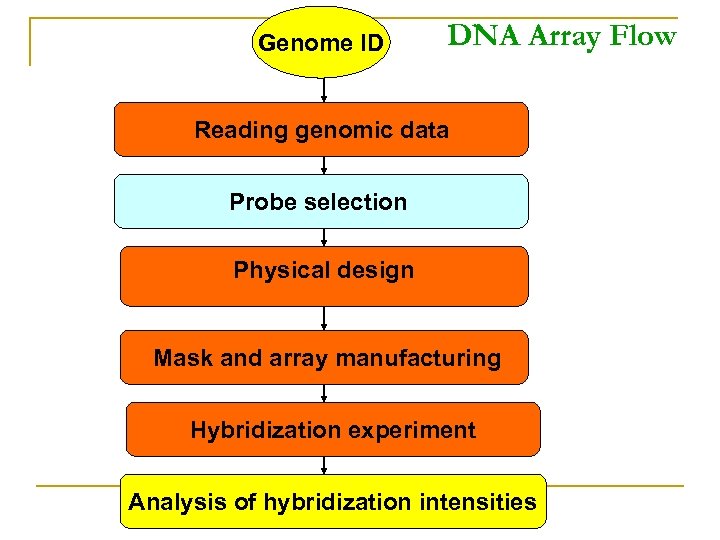 Genome ID DNA Array Flow Reading genomic data Probe selection Physical design Mask and array manufacturing Hybridization experiment Analysis of hybridization intensities
Genome ID DNA Array Flow Reading genomic data Probe selection Physical design Mask and array manufacturing Hybridization experiment Analysis of hybridization intensities
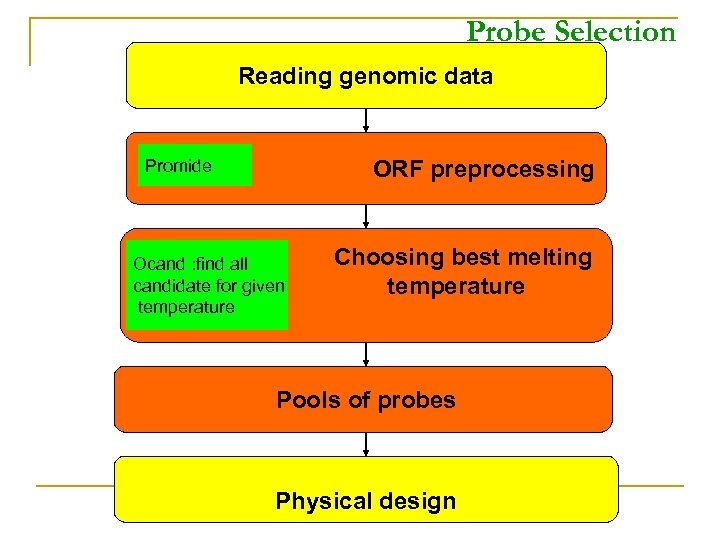 Probe Selection Reading genomic data ORF preprocessing Promide Ocand : find all candidate for given temperature Choosing best melting temperature Pools of probes Physical design
Probe Selection Reading genomic data ORF preprocessing Promide Ocand : find all candidate for given temperature Choosing best melting temperature Pools of probes Physical design
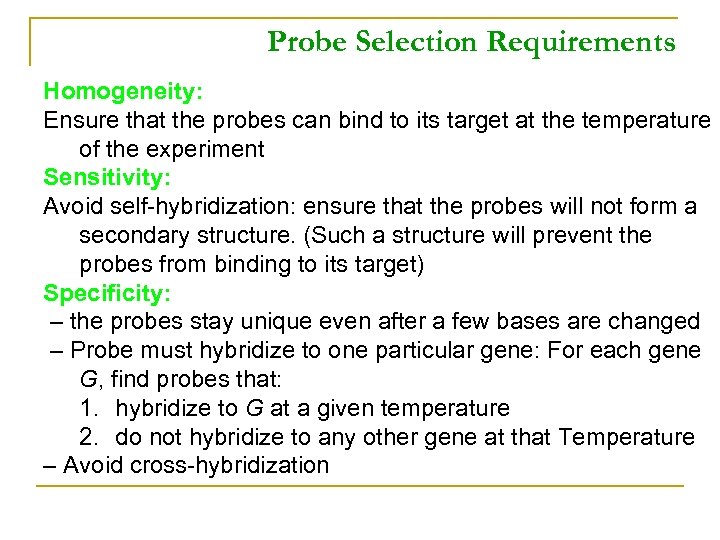 Probe Selection Requirements Homogeneity: Ensure that the probes can bind to its target at the temperature of the experiment Sensitivity: Avoid self-hybridization: ensure that the probes will not form a secondary structure. (Such a structure will prevent the probes from binding to its target) Specificity: – the probes stay unique even after a few bases are changed – Probe must hybridize to one particular gene: For each gene G, find probes that: 1. hybridize to G at a given temperature 2. do not hybridize to any other gene at that Temperature – Avoid cross-hybridization
Probe Selection Requirements Homogeneity: Ensure that the probes can bind to its target at the temperature of the experiment Sensitivity: Avoid self-hybridization: ensure that the probes will not form a secondary structure. (Such a structure will prevent the probes from binding to its target) Specificity: – the probes stay unique even after a few bases are changed – Probe must hybridize to one particular gene: For each gene G, find probes that: 1. hybridize to G at a given temperature 2. do not hybridize to any other gene at that Temperature – Avoid cross-hybridization
 Why Promide? Possible solutions: n Li and Stormo 2001 n Kaderali and Schliep 2002 n Rahmann (Promide) 2003 n They use the same data structure: Suffix array n Promide handles truly large scale datasets in a reasonable amount of time q n Human Gene. Nest clusters: in 50 hours Neurospora Crassa: q q Promide: few hours Li and Stormo: 1 week
Why Promide? Possible solutions: n Li and Stormo 2001 n Kaderali and Schliep 2002 n Rahmann (Promide) 2003 n They use the same data structure: Suffix array n Promide handles truly large scale datasets in a reasonable amount of time q n Human Gene. Nest clusters: in 50 hours Neurospora Crassa: q q Promide: few hours Li and Stormo: 1 week
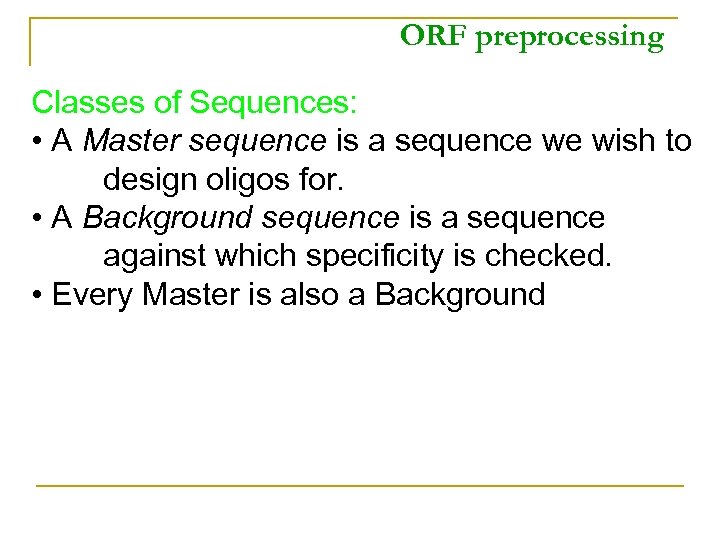 ORF preprocessing Classes of Sequences: • A Master sequence is a sequence we wish to design oligos for. • A Background sequence is a sequence against which specificity is checked. • Every Master is also a Background
ORF preprocessing Classes of Sequences: • A Master sequence is a sequence we wish to design oligos for. • A Background sequence is a sequence against which specificity is checked. • Every Master is also a Background
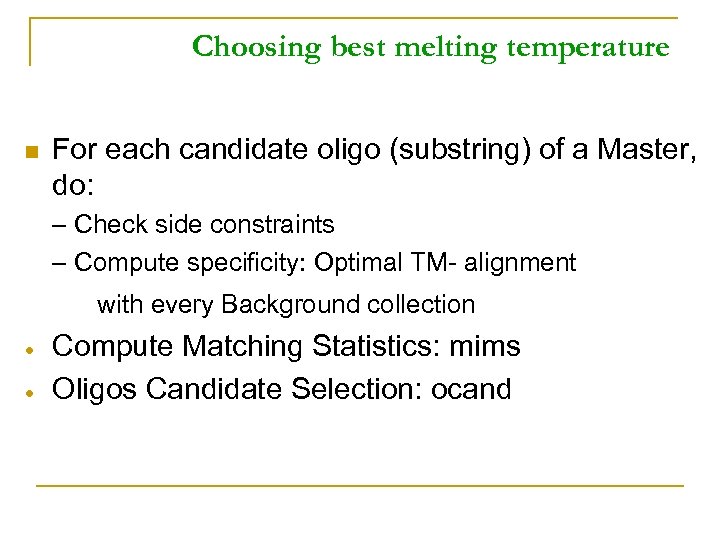 Choosing best melting temperature n For each candidate oligo (substring) of a Master, do: – Check side constraints – Compute specificity: Optimal TM- alignment with every Background collection Compute Matching Statistics: mims Oligos Candidate Selection: ocand
Choosing best melting temperature n For each candidate oligo (substring) of a Master, do: – Check side constraints – Compute specificity: Optimal TM- alignment with every Background collection Compute Matching Statistics: mims Oligos Candidate Selection: ocand
 Genome ID Mask and array manufacturing Reading genomic data Probe selection Physical design Mask and array manufacturing Hybridization experiment Analysis of hybridization intensities
Genome ID Mask and array manufacturing Reading genomic data Probe selection Physical design Mask and array manufacturing Hybridization experiment Analysis of hybridization intensities
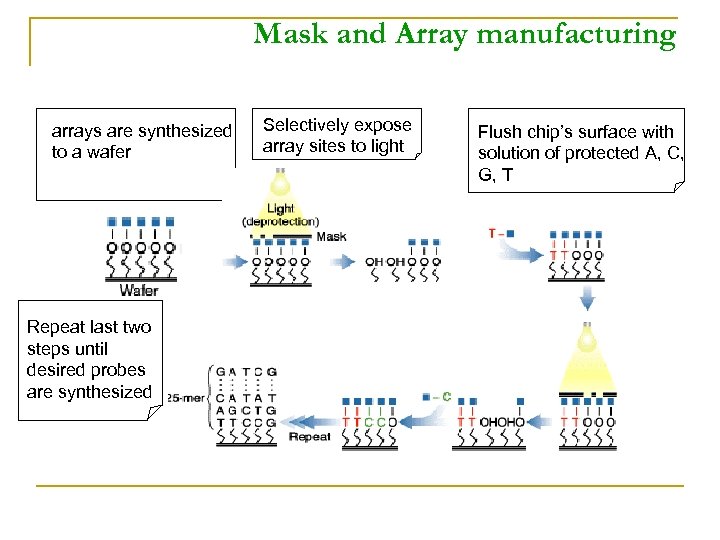 Mask and Array manufacturing arrays are synthesized to a wafer Repeat last two steps until desired probes are synthesized Selectively expose array sites to light Flush chip’s surface with solution of protected A, C, G, T
Mask and Array manufacturing arrays are synthesized to a wafer Repeat last two steps until desired probes are synthesized Selectively expose array sites to light Flush chip’s surface with solution of protected A, C, G, T
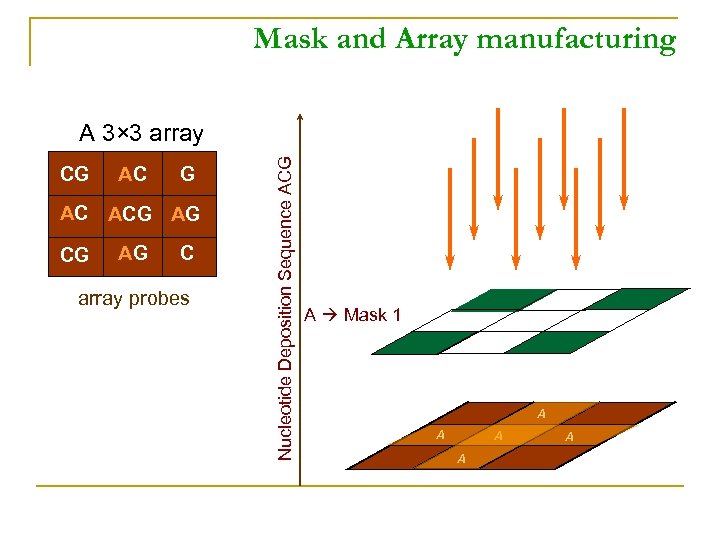 Mask and Array manufacturing CG AC ACG AG C array probes Nucleotide Deposition Sequence ACG A 3× 3 array A Mask 1 A A A
Mask and Array manufacturing CG AC ACG AG C array probes Nucleotide Deposition Sequence ACG A 3× 3 array A Mask 1 A A A
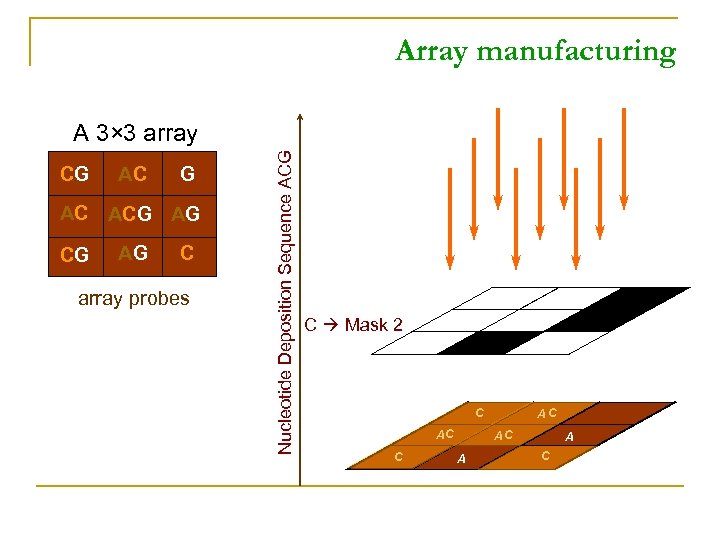 Array manufacturing CG AC ACG AG C array probes Nucleotide Deposition Sequence ACG A 3× 3 array C Mask 2 C AC AC A A C
Array manufacturing CG AC ACG AG C array probes Nucleotide Deposition Sequence ACG A 3× 3 array C Mask 2 C AC AC A A C
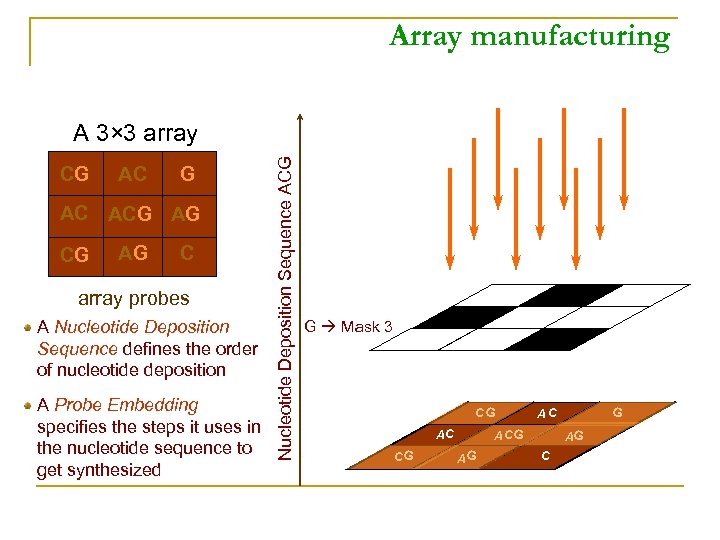 Array manufacturing CG AC ACG AG C array probes A Nucleotide Deposition Sequence defines the order of nucleotide deposition A Probe Embedding specifies the steps it uses in the nucleotide sequence to get synthesized Nucleotide Deposition Sequence ACG A 3× 3 array G Mask 3 CG AC G AG G AC AG C
Array manufacturing CG AC ACG AG C array probes A Nucleotide Deposition Sequence defines the order of nucleotide deposition A Probe Embedding specifies the steps it uses in the nucleotide sequence to get synthesized Nucleotide Deposition Sequence ACG A 3× 3 array G Mask 3 CG AC G AG G AC AG C
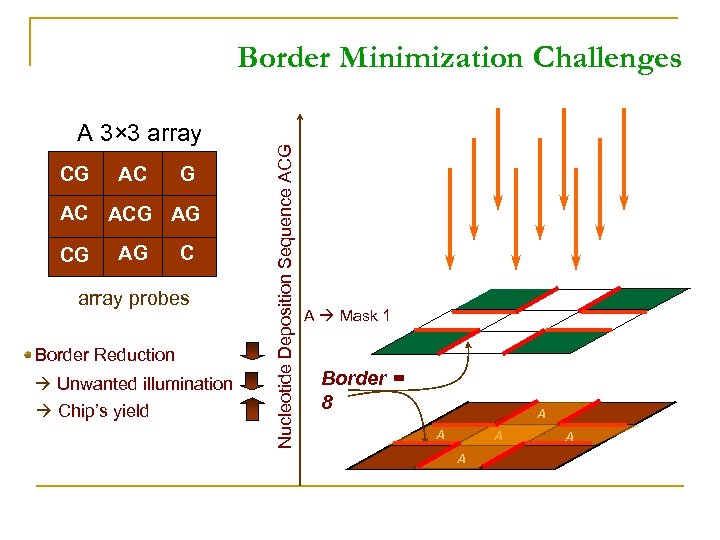 A 3× 3 array CG AC ACG AG C array probes Border Reduction Unwanted illumination Chip’s yield Nucleotide Deposition Sequence ACG Border Minimization Challenges A Mask 1 Border = 8 A A A
A 3× 3 array CG AC ACG AG C array probes Border Reduction Unwanted illumination Chip’s yield Nucleotide Deposition Sequence ACG Border Minimization Challenges A Mask 1 Border = 8 A A A
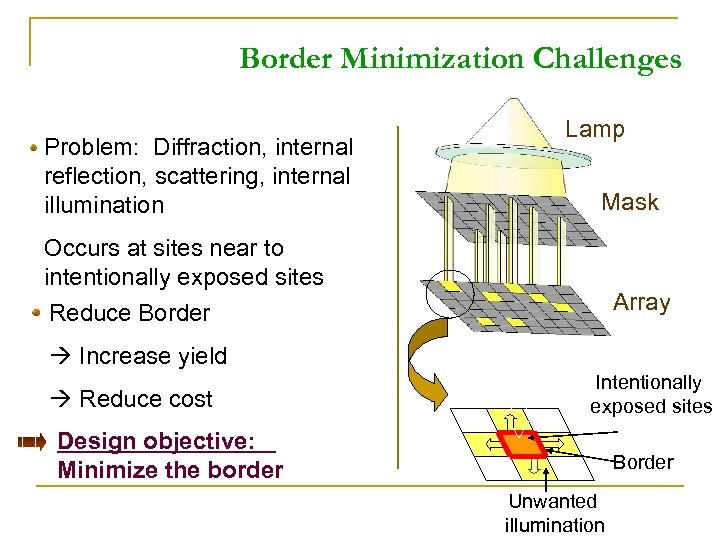 Border Minimization Challenges Problem: Diffraction, internal reflection, scattering, internal illumination Lamp Mask Occurs at sites near to intentionally exposed sites Array Reduce Border Increase yield Reduce cost Intentionally exposed sites Design objective: Minimize the border Border Unwanted illumination
Border Minimization Challenges Problem: Diffraction, internal reflection, scattering, internal illumination Lamp Mask Occurs at sites near to intentionally exposed sites Array Reduce Border Increase yield Reduce cost Intentionally exposed sites Design objective: Minimize the border Border Unwanted illumination
 Genome ID Physical design Reading genomic data Probe selection Physical design Mask and array manufacturing Hybridization experiment Analysis of hybridization intensities
Genome ID Physical design Reading genomic data Probe selection Physical design Mask and array manufacturing Hybridization experiment Analysis of hybridization intensities
 Physical Design Probe Selection Deposition sequence design Test control 2 D-probe placement 3 D-probe embedding Mask and array manufacturing
Physical Design Probe Selection Deposition sequence design Test control 2 D-probe placement 3 D-probe embedding Mask and array manufacturing
 Physical Design • Probe Placement • Similar probes should be placed close together • Constructive placement • Placement improvement operators • Probe Embedding • Degrees of freedom (DOF) in probe embedding • DOF exploitation for border conflict reduction
Physical Design • Probe Placement • Similar probes should be placed close together • Constructive placement • Placement improvement operators • Probe Embedding • Degrees of freedom (DOF) in probe embedding • DOF exploitation for border conflict reduction
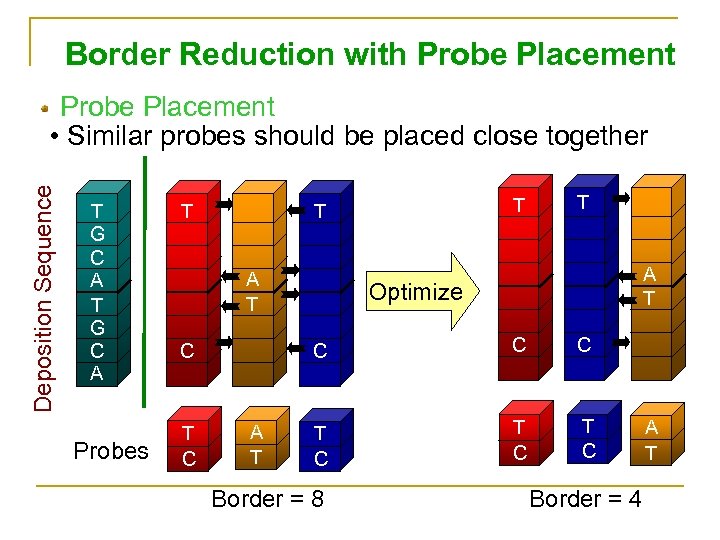 Border Reduction with Probe Placement Deposition Sequence Probe Placement • Similar probes should be placed close together T G C A Probes T A T Optimize C C T T A T C C T C T C Border = 8 Border = 4 A T
Border Reduction with Probe Placement Deposition Sequence Probe Placement • Similar probes should be placed close together T G C A Probes T A T Optimize C C T T A T C C T C T C Border = 8 Border = 4 A T
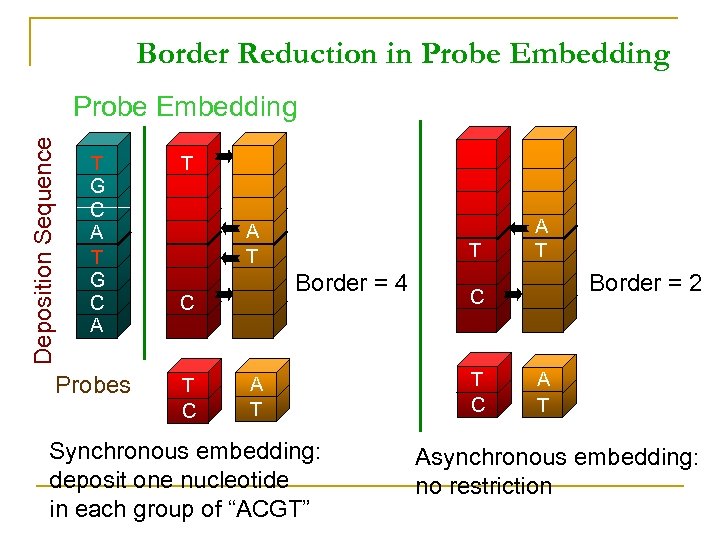 Border Reduction in Probe Embedding Deposition Sequence Probe Embedding T G C A Probes T A T Border = 4 C T A T Synchronous embedding: deposit one nucleotide in each group of “ACGT” A T Border = 2 C T C A T Asynchronous embedding: no restriction
Border Reduction in Probe Embedding Deposition Sequence Probe Embedding T G C A Probes T A T Border = 4 C T A T Synchronous embedding: deposit one nucleotide in each group of “ACGT” A T Border = 2 C T C A T Asynchronous embedding: no restriction
 Physical Design Problem Give: n 2 probes Find: Placement of probes in n x n sites Embedding of the probes Minimize: Total border cost
Physical Design Problem Give: n 2 probes Find: Placement of probes in n x n sites Embedding of the probes Minimize: Total border cost
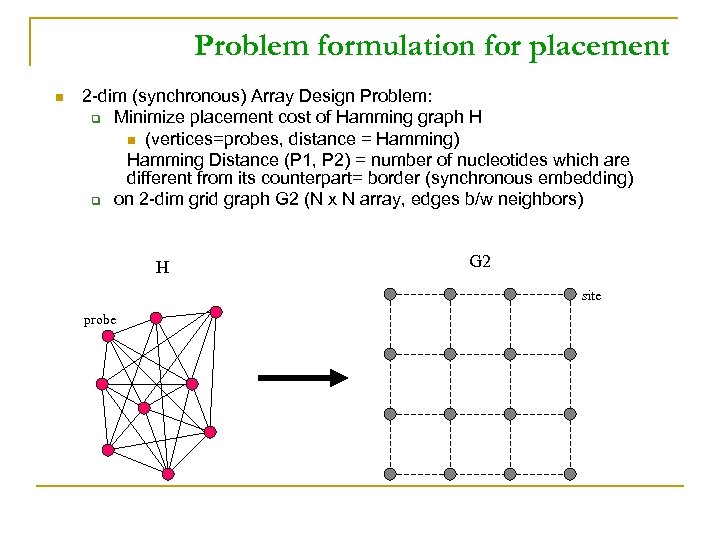 Problem formulation for placement n 2 -dim (synchronous) Array Design Problem: q Minimize placement cost of Hamming graph H n (vertices=probes, distance = Hamming) Hamming Distance (P 1, P 2) = number of nucleotides which are different from its counterpart= border (synchronous embedding) q on 2 -dim grid graph G 2 (N x N array, edges b/w neighbors) H G 2 site probe
Problem formulation for placement n 2 -dim (synchronous) Array Design Problem: q Minimize placement cost of Hamming graph H n (vertices=probes, distance = Hamming) Hamming Distance (P 1, P 2) = number of nucleotides which are different from its counterpart= border (synchronous embedding) q on 2 -dim grid graph G 2 (N x N array, edges b/w neighbors) H G 2 site probe
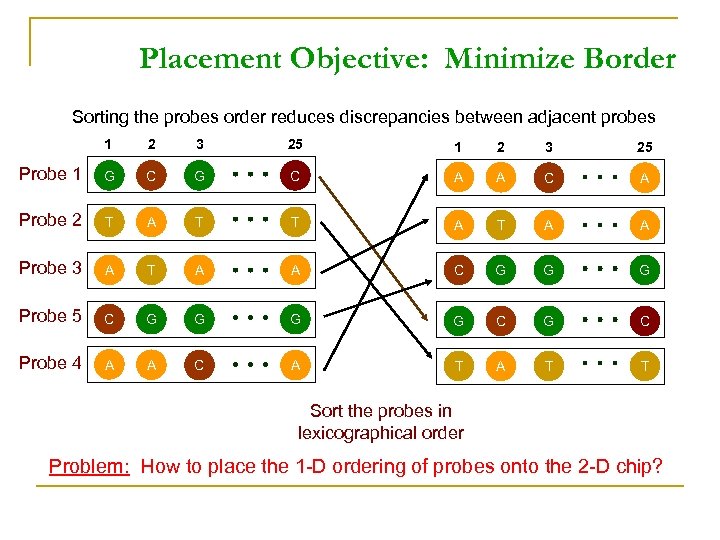 Placement Objective: Minimize Border Sorting the probes order reduces discrepancies between adjacent probes 1 2 3 25 Probe 1 G C A A C A Probe 2 T A T A A Probe 3 A T A A C G G G Probe 5 C G G C G C Probe 4 A A C A T T Sort the probes in lexicographical order Problem: How to place the 1 -D ordering of probes onto the 2 -D chip?
Placement Objective: Minimize Border Sorting the probes order reduces discrepancies between adjacent probes 1 2 3 25 Probe 1 G C A A C A Probe 2 T A T A A Probe 3 A T A A C G G G Probe 5 C G G C G C Probe 4 A A C A T T Sort the probes in lexicographical order Problem: How to place the 1 -D ordering of probes onto the 2 -D chip?
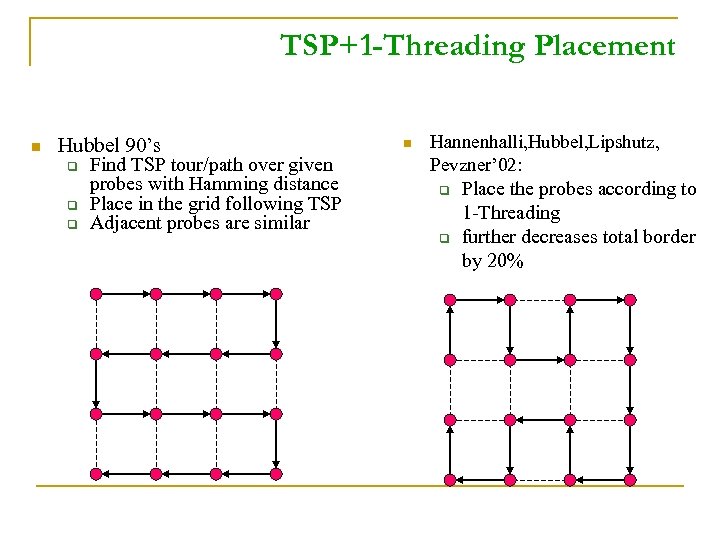 TSP+1 -Threading Placement n Hubbel 90’s q Find TSP tour/path over given probes with Hamming distance q Place in the grid following TSP q Adjacent probes are similar n Hannenhalli, Hubbel, Lipshutz, Pevzner’ 02: q q Place the probes according to 1 -Threading further decreases total border by 20%
TSP+1 -Threading Placement n Hubbel 90’s q Find TSP tour/path over given probes with Hamming distance q Place in the grid following TSP q Adjacent probes are similar n Hannenhalli, Hubbel, Lipshutz, Pevzner’ 02: q q Place the probes according to 1 -Threading further decreases total border by 20%
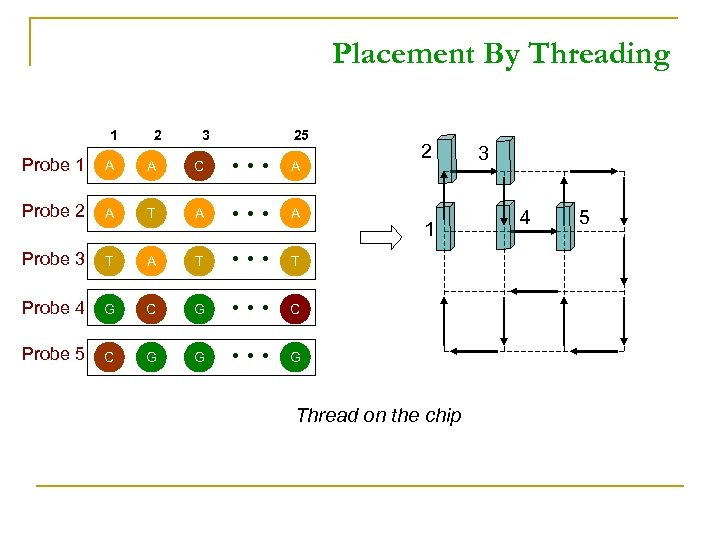 Placement By Threading 1 2 3 25 Probe 1 A A C A Probe 2 A T A A Probe 3 T A T T Probe 4 G C Probe 5 C G G 2 G 1 Thread on the chip 3 4 5
Placement By Threading 1 2 3 25 Probe 1 A A C A Probe 2 A T A A Probe 3 T A T T Probe 4 G C Probe 5 C G G 2 G 1 Thread on the chip 3 4 5
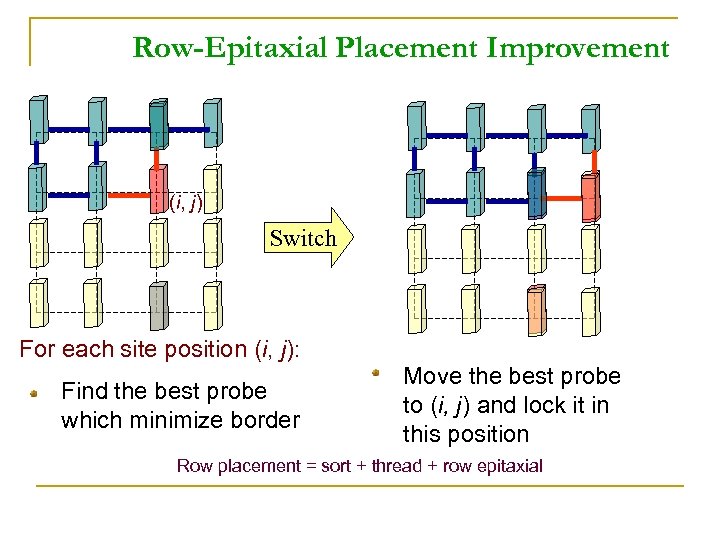 Row-Epitaxial Placement Improvement (i, j) Switch For each site position (i, j): Find the best probe which minimize border Move the best probe to (i, j) and lock it in this position Row placement = sort + thread + row epitaxial
Row-Epitaxial Placement Improvement (i, j) Switch For each site position (i, j): Find the best probe which minimize border Move the best probe to (i, j) and lock it in this position Row placement = sort + thread + row epitaxial
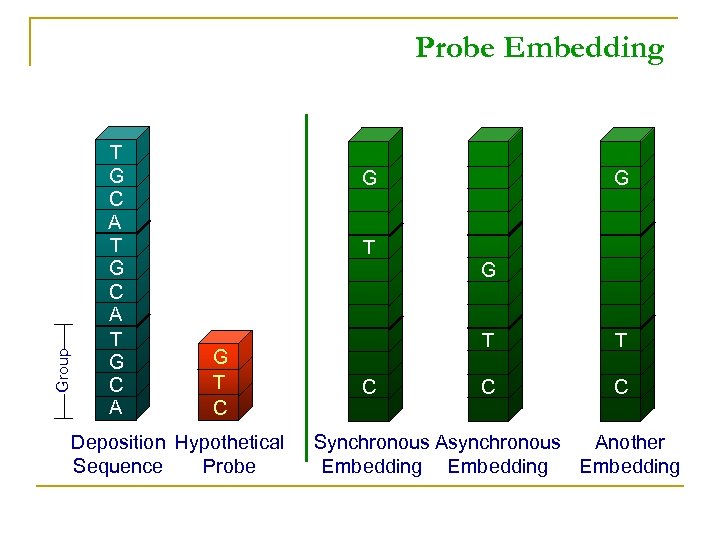 Group Probe Embedding T G C A G G T C Deposition Hypothetical Sequence Probe T C C Synchronous Asynchronous Another Embedding
Group Probe Embedding T G C A G G T C Deposition Hypothetical Sequence Probe T C C Synchronous Asynchronous Another Embedding
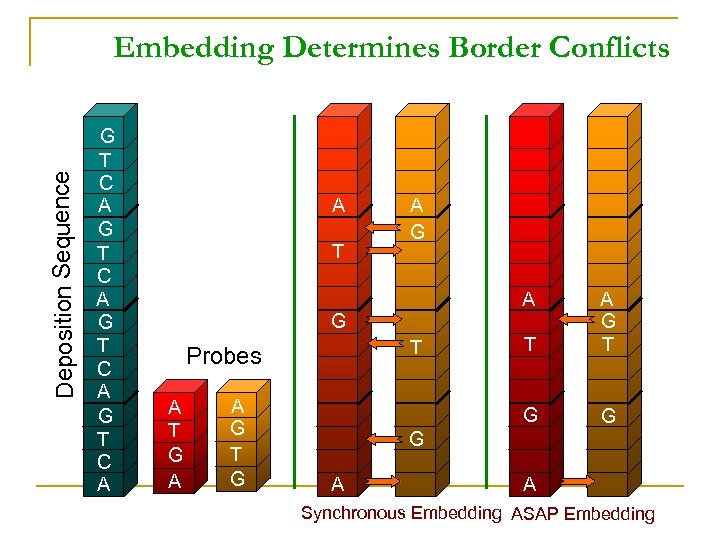 Deposition Sequence Embedding Determines Border Conflicts G T C A A T A G A T Probes A T G A A G T G G A A Synchronous Embedding ASAP Embedding
Deposition Sequence Embedding Determines Border Conflicts G T C A A T A G A T Probes A T G A A G T G G A A Synchronous Embedding ASAP Embedding
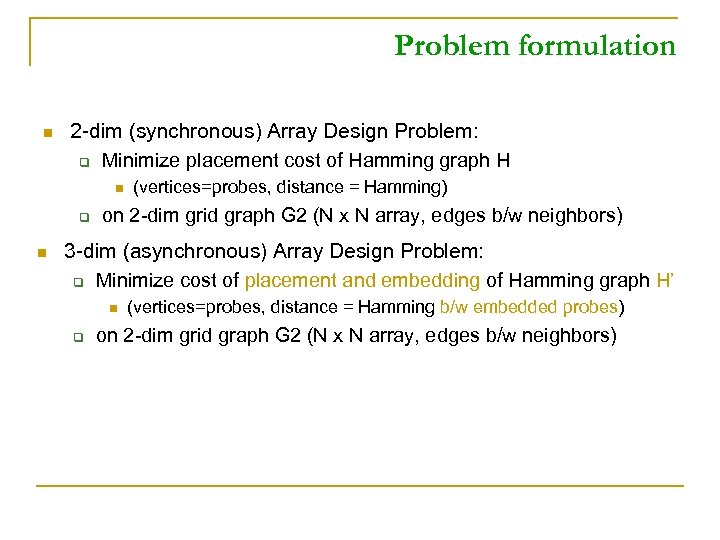 Problem formulation n 2 -dim (synchronous) Array Design Problem: q Minimize placement cost of Hamming graph H n q n (vertices=probes, distance = Hamming) on 2 -dim grid graph G 2 (N x N array, edges b/w neighbors) 3 -dim (asynchronous) Array Design Problem: q Minimize cost of placement and embedding of Hamming graph H’ n q (vertices=probes, distance = Hamming b/w embedded probes) on 2 -dim grid graph G 2 (N x N array, edges b/w neighbors)
Problem formulation n 2 -dim (synchronous) Array Design Problem: q Minimize placement cost of Hamming graph H n q n (vertices=probes, distance = Hamming) on 2 -dim grid graph G 2 (N x N array, edges b/w neighbors) 3 -dim (asynchronous) Array Design Problem: q Minimize cost of placement and embedding of Hamming graph H’ n q (vertices=probes, distance = Hamming b/w embedded probes) on 2 -dim grid graph G 2 (N x N array, edges b/w neighbors)
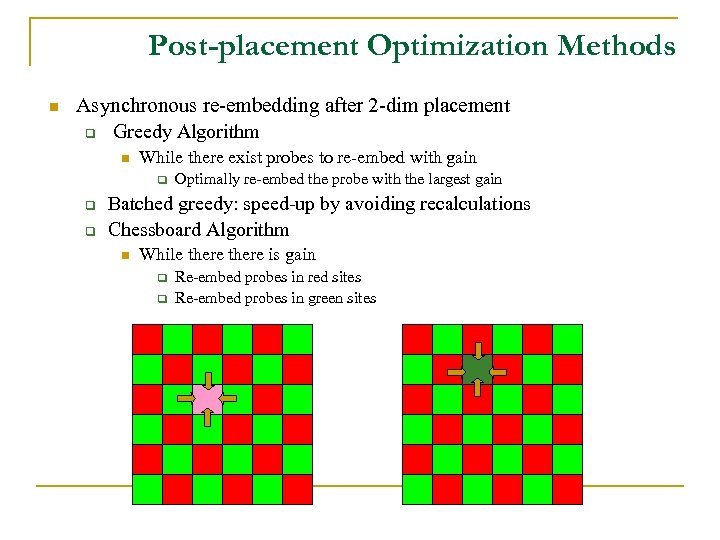 Post-placement Optimization Methods n Asynchronous re-embedding after 2 -dim placement q Greedy Algorithm n While there exist probes to re-embed with gain q q q Optimally re-embed the probe with the largest gain Batched greedy: speed-up by avoiding recalculations Chessboard Algorithm n While there is gain q q Re-embed probes in red sites Re-embed probes in green sites
Post-placement Optimization Methods n Asynchronous re-embedding after 2 -dim placement q Greedy Algorithm n While there exist probes to re-embed with gain q q q Optimally re-embed the probe with the largest gain Batched greedy: speed-up by avoiding recalculations Chessboard Algorithm n While there is gain q q Re-embed probes in red sites Re-embed probes in green sites
 Genome ID Analysis of hybridization intensities Reading genomic data Probe selection Physical design Mask and array manufacturing Hybridization experiment Analysis of hybridization intensities
Genome ID Analysis of hybridization intensities Reading genomic data Probe selection Physical design Mask and array manufacturing Hybridization experiment Analysis of hybridization intensities
 Experimental Study n n n n In our experiment we have considered the following parameters and we measured the results for different values of these parameters. Melting Temperature: We choose the temperatures 60 C and 65 C as best melting temperatures for our DNA probe array. Number of Candidates: We experimented with different values of K (number of candidates) for each pools of probes: 1 and 2. Chip Size: We ran our Experiments with 2 different chip sizes. We experimented with 50 x 50 and 60 x 60. We give the number of conflict and runtime for each algorithm for the Herpes B virus and simulated data
Experimental Study n n n n In our experiment we have considered the following parameters and we measured the results for different values of these parameters. Melting Temperature: We choose the temperatures 60 C and 65 C as best melting temperatures for our DNA probe array. Number of Candidates: We experimented with different values of K (number of candidates) for each pools of probes: 1 and 2. Chip Size: We ran our Experiments with 2 different chip sizes. We experimented with 50 x 50 and 60 x 60. We give the number of conflict and runtime for each algorithm for the Herpes B virus and simulated data
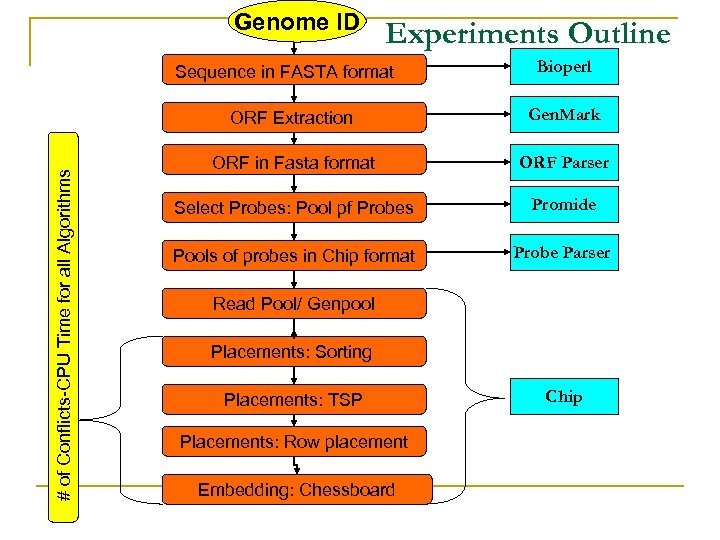 Genome ID Experiments Outline Sequence in FASTA format Bioperl # of Conflicts-CPU Time for all Algorithms ORF Extraction Gen. Mark ORF in Fasta format ORF Parser Select Probes: Pool pf Probes Promide Pools of probes in Chip format Probe Parser Read Pool/ Genpool Placements: Sorting Placements: TSP Placements: Row placement Embedding: Chessboard Chip
Genome ID Experiments Outline Sequence in FASTA format Bioperl # of Conflicts-CPU Time for all Algorithms ORF Extraction Gen. Mark ORF in Fasta format ORF Parser Select Probes: Pool pf Probes Promide Pools of probes in Chip format Probe Parser Read Pool/ Genpool Placements: Sorting Placements: TSP Placements: Row placement Embedding: Chessboard Chip
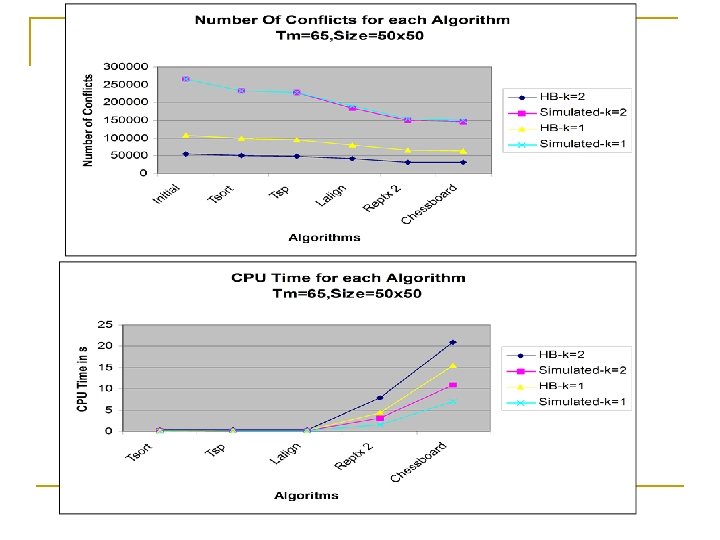
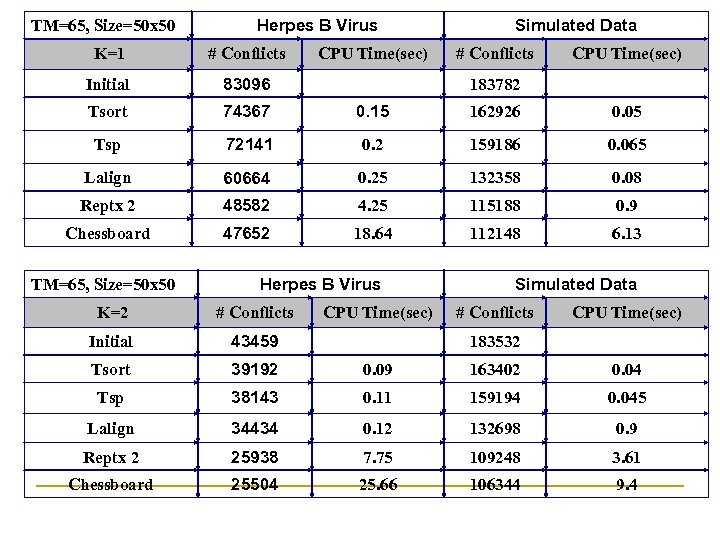 TM=65, Size=50 x 50 Herpes B Virus Simulated Data K=1 # Conflicts CPU Time(sec) Initial 83096 183782 Tsort 74367 0. 15 162926 0. 05 Tsp 72141 0. 2 159186 0. 065 Lalign 60664 0. 25 132358 0. 08 Reptx 2 48582 4. 25 115188 0. 9 Chessboard 47652 18. 64 112148 6. 13 TM=65, Size=50 x 50 Herpes B Virus Simulated Data K=2 # Conflicts CPU Time(sec) Initial 43459 183532 Tsort 39192 0. 09 163402 0. 04 Tsp 38143 0. 11 159194 0. 045 Lalign 34434 0. 12 132698 0. 9 Reptx 2 25938 7. 75 109248 3. 61 Chessboard 25504 25. 66 106344 9. 4
TM=65, Size=50 x 50 Herpes B Virus Simulated Data K=1 # Conflicts CPU Time(sec) Initial 83096 183782 Tsort 74367 0. 15 162926 0. 05 Tsp 72141 0. 2 159186 0. 065 Lalign 60664 0. 25 132358 0. 08 Reptx 2 48582 4. 25 115188 0. 9 Chessboard 47652 18. 64 112148 6. 13 TM=65, Size=50 x 50 Herpes B Virus Simulated Data K=2 # Conflicts CPU Time(sec) Initial 43459 183532 Tsort 39192 0. 09 163402 0. 04 Tsp 38143 0. 11 159194 0. 045 Lalign 34434 0. 12 132698 0. 9 Reptx 2 25938 7. 75 109248 3. 61 Chessboard 25504 25. 66 106344 9. 4
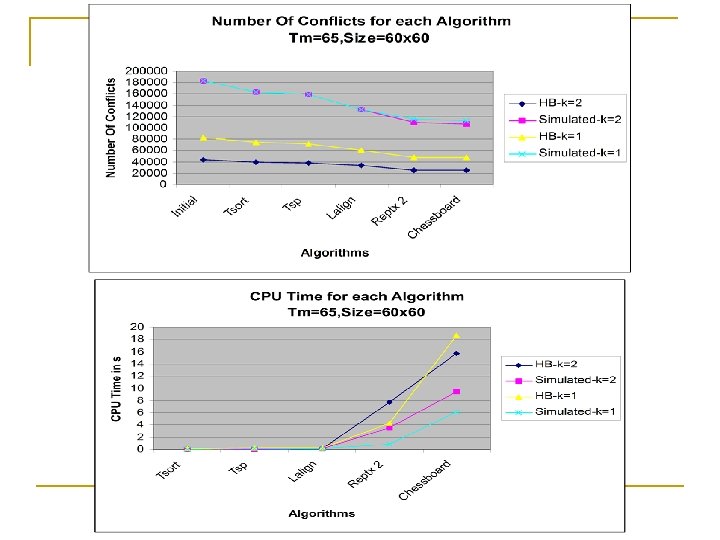
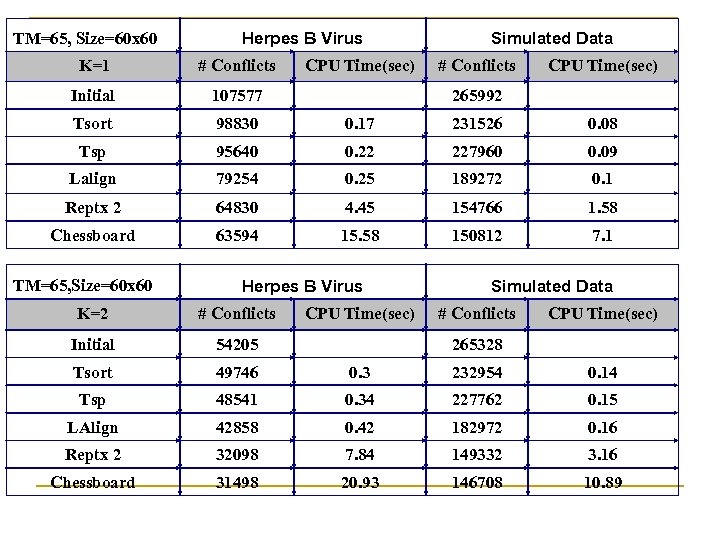 TM=65, Size=60 x 60 Herpes B Virus Simulated Data K=1 # Conflicts CPU Time(sec) Initial 107577 265992 Tsort 98830 0. 17 231526 0. 08 Tsp 95640 0. 22 227960 0. 09 Lalign 79254 0. 25 189272 0. 1 Reptx 2 64830 4. 45 154766 1. 58 Chessboard 63594 15. 58 150812 7. 1 TM=65, Size=60 x 60 Herpes B Virus Simulated Data K=2 # Conflicts CPU Time(sec) Initial 54205 265328 Tsort 49746 0. 3 232954 0. 14 Tsp 48541 0. 34 227762 0. 15 LAlign 42858 0. 42 182972 0. 16 Reptx 2 32098 7. 84 149332 3. 16 Chessboard 31498 20. 93 146708 10. 89
TM=65, Size=60 x 60 Herpes B Virus Simulated Data K=1 # Conflicts CPU Time(sec) Initial 107577 265992 Tsort 98830 0. 17 231526 0. 08 Tsp 95640 0. 22 227960 0. 09 Lalign 79254 0. 25 189272 0. 1 Reptx 2 64830 4. 45 154766 1. 58 Chessboard 63594 15. 58 150812 7. 1 TM=65, Size=60 x 60 Herpes B Virus Simulated Data K=2 # Conflicts CPU Time(sec) Initial 54205 265328 Tsort 49746 0. 3 232954 0. 14 Tsp 48541 0. 34 227762 0. 15 LAlign 42858 0. 42 182972 0. 16 Reptx 2 32098 7. 84 149332 3. 16 Chessboard 31498 20. 93 146708 10. 89
 n n n Conclusion and Future work Conclusion: Our experiments show: The genomic data follow the pattern predicted by simulated data In case of Herpes B virus, like simulated data, increasing number of candidates per probe (k) decreases number of border conflicts during the probe placement algorithms The number of border conflicts is several times smaller than for simulated data The trade-off between number of border conflicts and the CPU time taken for the various algorithms that are defined in the physical design We give a concatenate software solution for the entire DNA array flow We explore all steps in a single automated software suite of tools Future work: The entire software suite be made available through web services Users can enter name of organism or ID and with an option of choosing to set the required parameters the suite will produce the DNA probe micro-array chip layout
n n n Conclusion and Future work Conclusion: Our experiments show: The genomic data follow the pattern predicted by simulated data In case of Herpes B virus, like simulated data, increasing number of candidates per probe (k) decreases number of border conflicts during the probe placement algorithms The number of border conflicts is several times smaller than for simulated data The trade-off between number of border conflicts and the CPU time taken for the various algorithms that are defined in the physical design We give a concatenate software solution for the entire DNA array flow We explore all steps in a single automated software suite of tools Future work: The entire software suite be made available through web services Users can enter name of organism or ID and with an option of choosing to set the required parameters the suite will produce the DNA probe micro-array chip layout
 Thank you
Thank you


