040d425b8ae148bb6ec6d120b335f1ec.ppt
- Количество слайдов: 40
 CONSERVATION GENETICS: studying genetic Maria Eugenia D’Amato
CONSERVATION GENETICS: studying genetic Maria Eugenia D’Amato
 METHODOLOGICAL APPROACHES TO THE STUDY OF GENETIC DIVERSITY • Molecular genetics techniques • Types and properties of molecular makers • Factors that determine the patterns of genetic variation
METHODOLOGICAL APPROACHES TO THE STUDY OF GENETIC DIVERSITY • Molecular genetics techniques • Types and properties of molecular makers • Factors that determine the patterns of genetic variation
 MOLECULAR TECHNIQUES 1. Southern blot 2. PCR 3. DNA sequencing
MOLECULAR TECHNIQUES 1. Southern blot 2. PCR 3. DNA sequencing
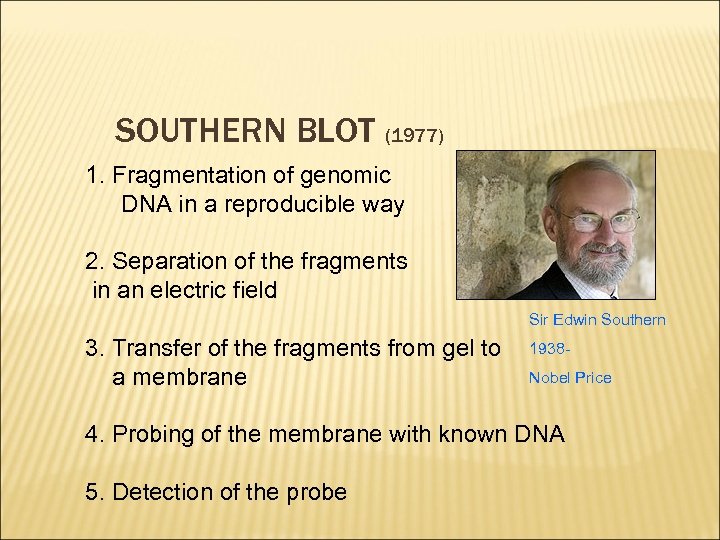 SOUTHERN BLOT (1977) 1. Fragmentation of genomic DNA in a reproducible way 2. Separation of the fragments in an electric field Sir Edwin Southern 3. Transfer of the fragments from gel to a membrane 1938 Nobel Price 4. Probing of the membrane with known DNA 5. Detection of the probe
SOUTHERN BLOT (1977) 1. Fragmentation of genomic DNA in a reproducible way 2. Separation of the fragments in an electric field Sir Edwin Southern 3. Transfer of the fragments from gel to a membrane 1938 Nobel Price 4. Probing of the membrane with known DNA 5. Detection of the probe
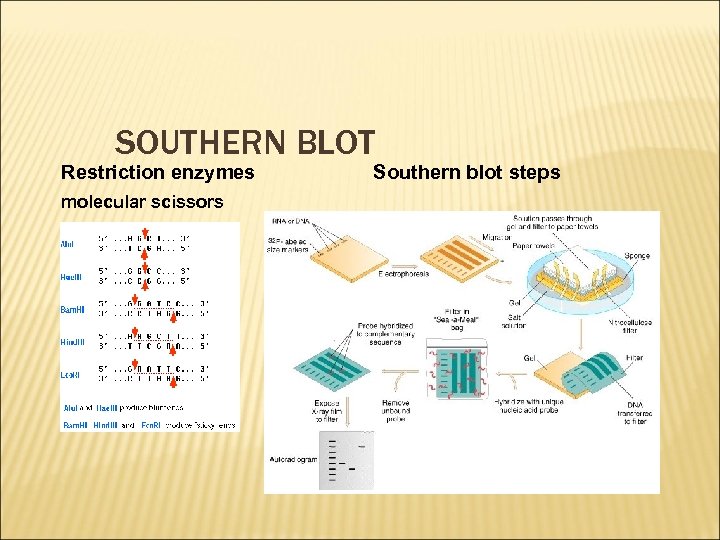 SOUTHERN BLOT Restriction enzymes molecular scissors Southern blot steps
SOUTHERN BLOT Restriction enzymes molecular scissors Southern blot steps
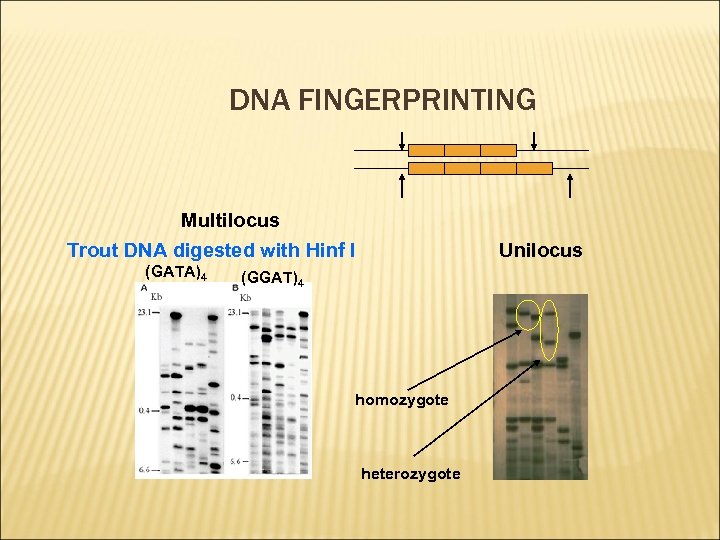 DNA FINGERPRINTING Multilocus Trout DNA digested with Hinf I (GATA)4 Unilocus (GGAT)4 homozygote heterozygote
DNA FINGERPRINTING Multilocus Trout DNA digested with Hinf I (GATA)4 Unilocus (GGAT)4 homozygote heterozygote
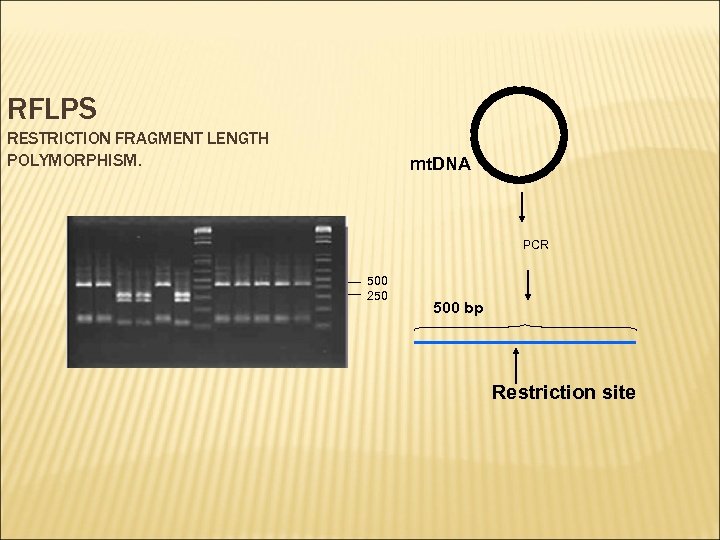 RFLPS RESTRICTION FRAGMENT LENGTH POLYMORPHISM. mt. DNA PCR 500 250 500 bp Restriction site
RFLPS RESTRICTION FRAGMENT LENGTH POLYMORPHISM. mt. DNA PCR 500 250 500 bp Restriction site
 PCR (1981) Polymerase Chain Reaction • In vitro replication of DNA Kary Mullis 1938 Nobel Price 1993
PCR (1981) Polymerase Chain Reaction • In vitro replication of DNA Kary Mullis 1938 Nobel Price 1993
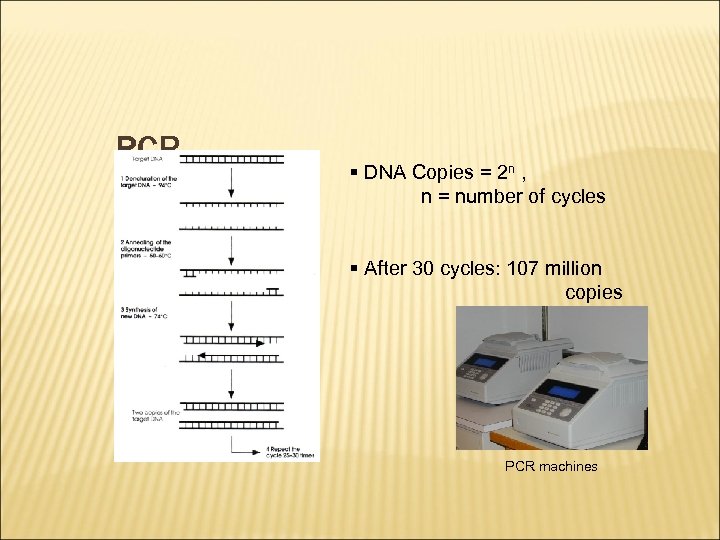 PCR § DNA Copies = 2 n , n = number of cycles § After 30 cycles: 107 million copies PCR machines
PCR § DNA Copies = 2 n , n = number of cycles § After 30 cycles: 107 million copies PCR machines
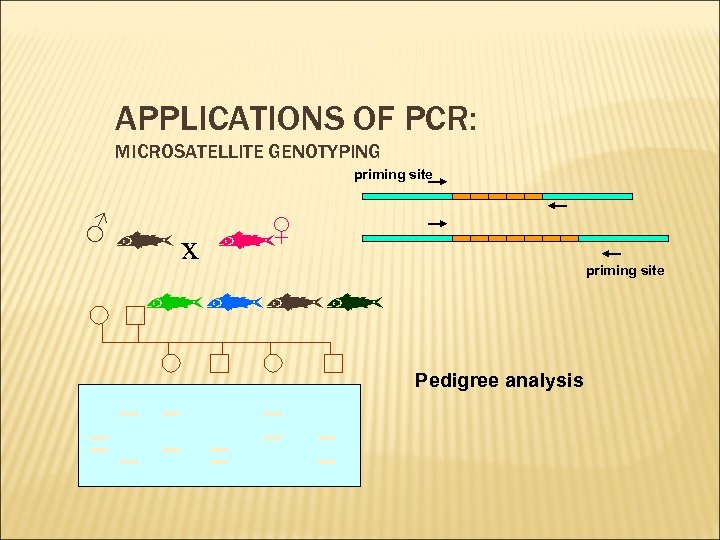 APPLICATIONS OF PCR: MICROSATELLITE GENOTYPING priming site x ♂ ♀ priming site Pedigree analysis
APPLICATIONS OF PCR: MICROSATELLITE GENOTYPING priming site x ♂ ♀ priming site Pedigree analysis
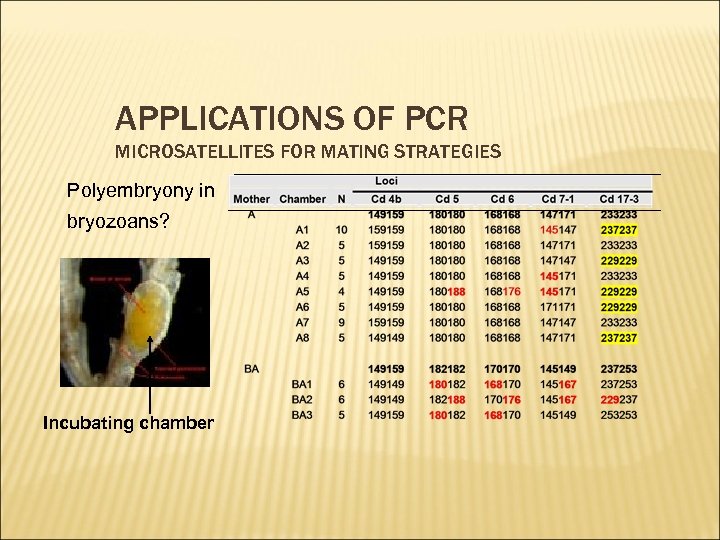 APPLICATIONS OF PCR MICROSATELLITES FOR MATING STRATEGIES Polyembryony in bryozoans? Incubating chamber
APPLICATIONS OF PCR MICROSATELLITES FOR MATING STRATEGIES Polyembryony in bryozoans? Incubating chamber
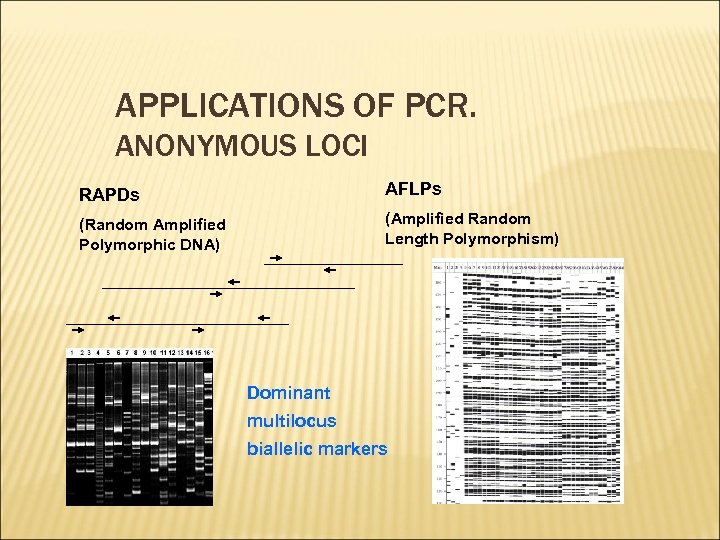 APPLICATIONS OF PCR. ANONYMOUS LOCI RAPDs AFLPs (Random Amplified Polymorphic DNA) (Amplified Random Length Polymorphism) Dominant multilocus biallelic markers
APPLICATIONS OF PCR. ANONYMOUS LOCI RAPDs AFLPs (Random Amplified Polymorphic DNA) (Amplified Random Length Polymorphism) Dominant multilocus biallelic markers
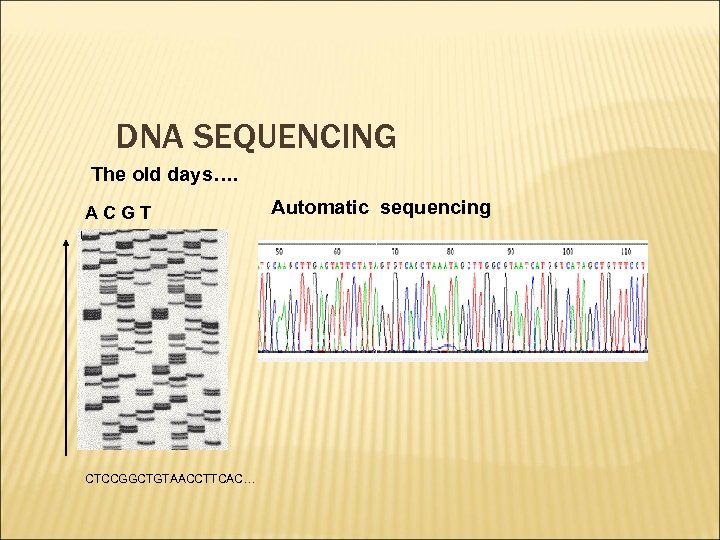 DNA SEQUENCING The old days…. ACGT CTCCGGCTGTAACCTTCAC… Automatic sequencing
DNA SEQUENCING The old days…. ACGT CTCCGGCTGTAACCTTCAC… Automatic sequencing
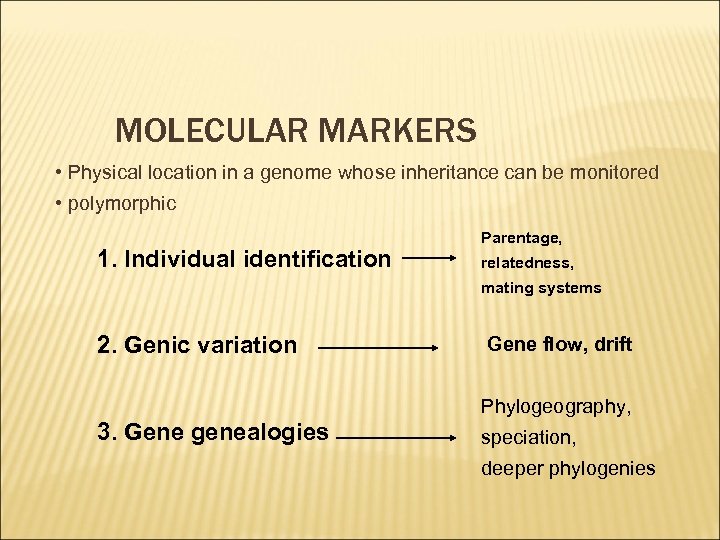 MOLECULAR MARKERS • Physical location in a genome whose inheritance can be monitored • polymorphic 1. Individual identification Parentage, relatedness, mating systems 2. Genic variation Gene flow, drift Phylogeography, 3. Gene genealogies speciation, deeper phylogenies
MOLECULAR MARKERS • Physical location in a genome whose inheritance can be monitored • polymorphic 1. Individual identification Parentage, relatedness, mating systems 2. Genic variation Gene flow, drift Phylogeography, 3. Gene genealogies speciation, deeper phylogenies
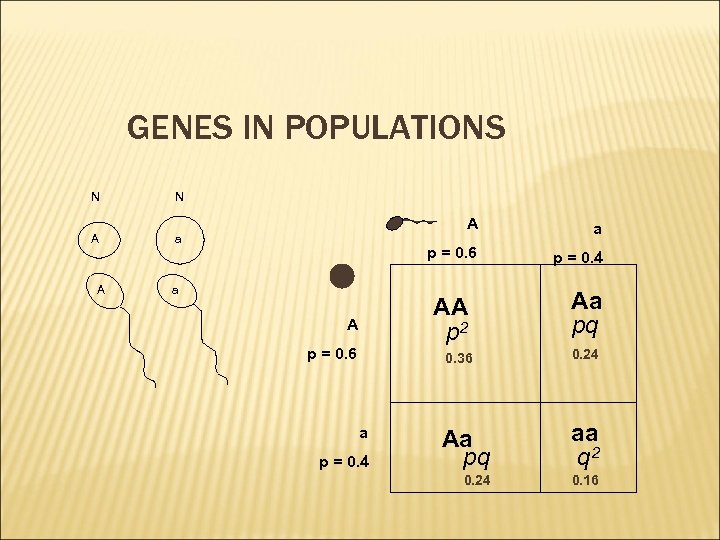 GENES IN POPULATIONS N N A a A A p = 0. 6 a a p = 0. 4 AA p 2 A p = 0. 6 Aa pq 0. 36 a p = 0. 4 0. 24 Aa pq aa q 2 0. 24 0. 16
GENES IN POPULATIONS N N A a A A p = 0. 6 a a p = 0. 4 AA p 2 A p = 0. 6 Aa pq 0. 36 a p = 0. 4 0. 24 Aa pq aa q 2 0. 24 0. 16
 GENES IN POPULATIONS: EQUILIBRIUM OF HARDY WEINBERG (p + q) 2 = p 2 + 2 pq + q 2 p = freq A q = freq a the organism is diploid with sexual reproduction generations are non overlapping Assumptions loci are biallelic allele frequencies are identical in males and females random mating population size is infinite no migration, no mutation, no selection
GENES IN POPULATIONS: EQUILIBRIUM OF HARDY WEINBERG (p + q) 2 = p 2 + 2 pq + q 2 p = freq A q = freq a the organism is diploid with sexual reproduction generations are non overlapping Assumptions loci are biallelic allele frequencies are identical in males and females random mating population size is infinite no migration, no mutation, no selection
 HARDY WEINBERG EQUILIBRIUM Consequences of the model • Allele frequencies remain constant, generation after generation • Genotype frequencies can be determined from allele frequencies
HARDY WEINBERG EQUILIBRIUM Consequences of the model • Allele frequencies remain constant, generation after generation • Genotype frequencies can be determined from allele frequencies
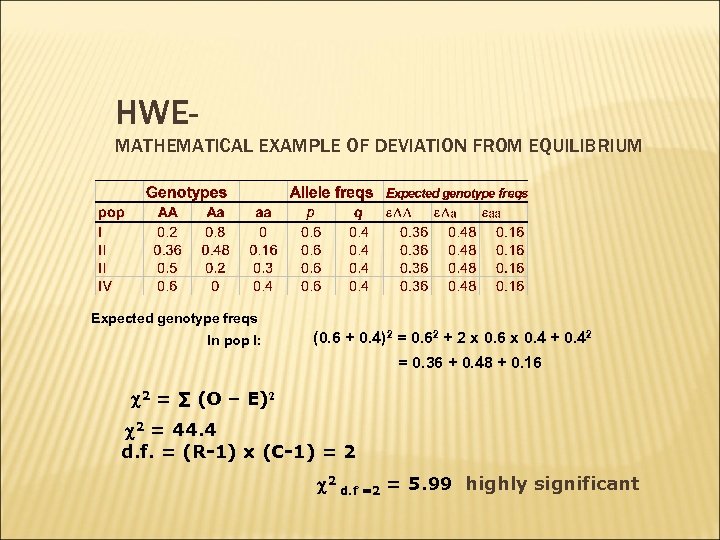 HWEMATHEMATICAL EXAMPLE OF DEVIATION FROM EQUILIBRIUM Expected genotype freqs In pop I: (0. 6 + 0. 4)2 = 0. 62 + 2 x 0. 6 x 0. 4 + 0. 42 = 0. 36 + 0. 48 + 0. 16 2 = ∑ (O – E)2 2 = 44. 4 d. f. = (R-1) x (C-1) = 2 2 d. f =2 = 5. 99 highly significant
HWEMATHEMATICAL EXAMPLE OF DEVIATION FROM EQUILIBRIUM Expected genotype freqs In pop I: (0. 6 + 0. 4)2 = 0. 62 + 2 x 0. 6 x 0. 4 + 0. 42 = 0. 36 + 0. 48 + 0. 16 2 = ∑ (O – E)2 2 = 44. 4 d. f. = (R-1) x (C-1) = 2 2 d. f =2 = 5. 99 highly significant
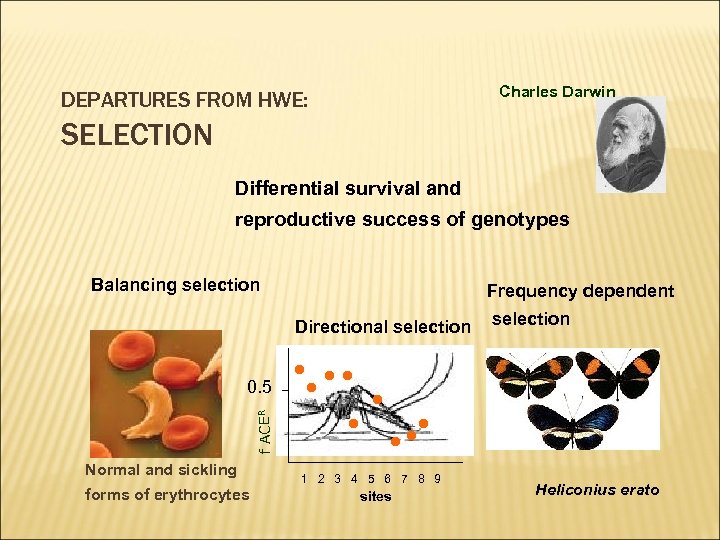 Charles Darwin DEPARTURES FROM HWE: SELECTION Differential survival and reproductive success of genotypes Balancing selection Frequency dependent Directional selection f ACER 0. 5 Normal and sickling forms of erythrocytes selection 1 2 3 4 5 6 7 8 9 sites Heliconius erato
Charles Darwin DEPARTURES FROM HWE: SELECTION Differential survival and reproductive success of genotypes Balancing selection Frequency dependent Directional selection f ACER 0. 5 Normal and sickling forms of erythrocytes selection 1 2 3 4 5 6 7 8 9 sites Heliconius erato
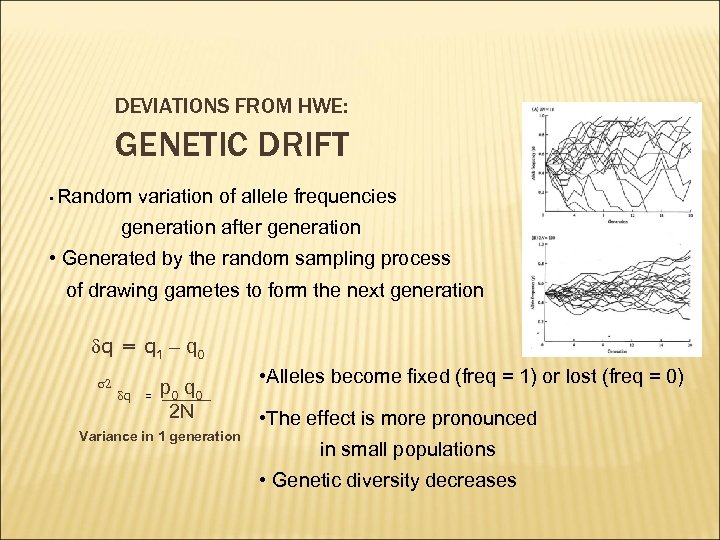 DEVIATIONS FROM HWE: GENETIC DRIFT • Random variation of allele frequencies generation after generation • Generated by the random sampling process of drawing gametes to form the next generation dq = q 1 – q 0 s 2 dq = p 0 q 0 2 N Variance in 1 generation • Alleles become fixed (freq = 1) or lost (freq = 0) • The effect is more pronounced in small populations • Genetic diversity decreases
DEVIATIONS FROM HWE: GENETIC DRIFT • Random variation of allele frequencies generation after generation • Generated by the random sampling process of drawing gametes to form the next generation dq = q 1 – q 0 s 2 dq = p 0 q 0 2 N Variance in 1 generation • Alleles become fixed (freq = 1) or lost (freq = 0) • The effect is more pronounced in small populations • Genetic diversity decreases
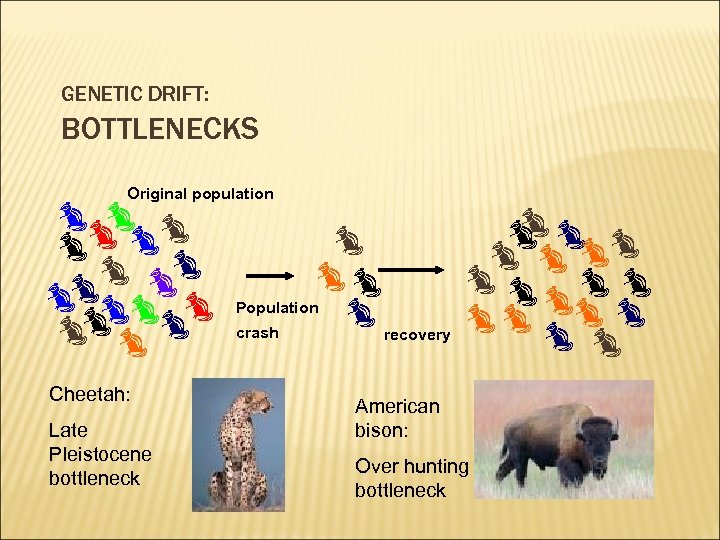 GENETIC DRIFT: BOTTLENECKS Original population Cheetah: Late Pleistocene bottleneck Population crash recovery American bison: Over hunting bottleneck
GENETIC DRIFT: BOTTLENECKS Original population Cheetah: Late Pleistocene bottleneck Population crash recovery American bison: Over hunting bottleneck
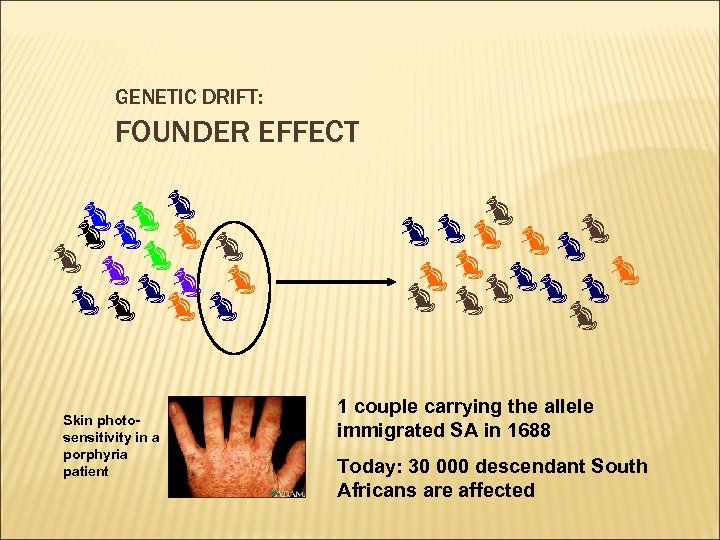 GENETIC DRIFT: FOUNDER EFFECT Skin photosensitivity in a porphyria patient 1 couple carrying the allele immigrated SA in 1688 Today: 30 000 descendant South Africans are affected
GENETIC DRIFT: FOUNDER EFFECT Skin photosensitivity in a porphyria patient 1 couple carrying the allele immigrated SA in 1688 Today: 30 000 descendant South Africans are affected
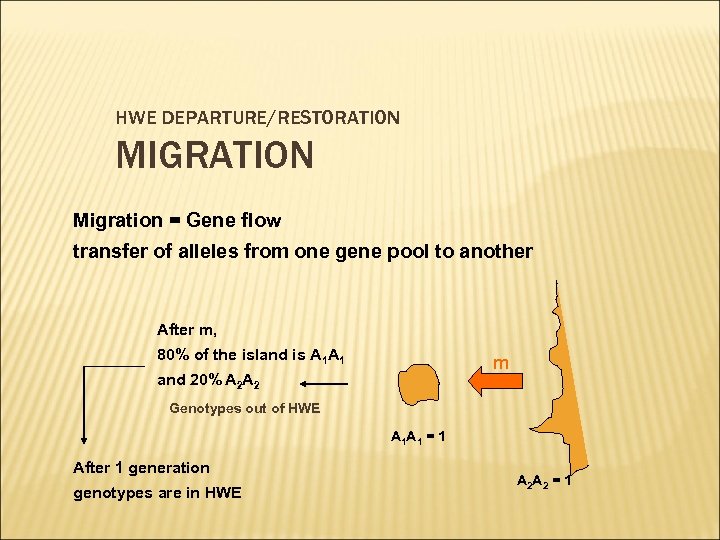 HWE DEPARTURE/RESTORATION MIGRATION Migration = Gene flow transfer of alleles from one gene pool to another After m, 80% of the island is A 1 A 1 m and 20% A 2 A 2 Genotypes out of HWE A 1 A 1 = 1 After 1 generation genotypes are in HWE A 2 A 2 = 1
HWE DEPARTURE/RESTORATION MIGRATION Migration = Gene flow transfer of alleles from one gene pool to another After m, 80% of the island is A 1 A 1 m and 20% A 2 A 2 Genotypes out of HWE A 1 A 1 = 1 After 1 generation genotypes are in HWE A 2 A 2 = 1
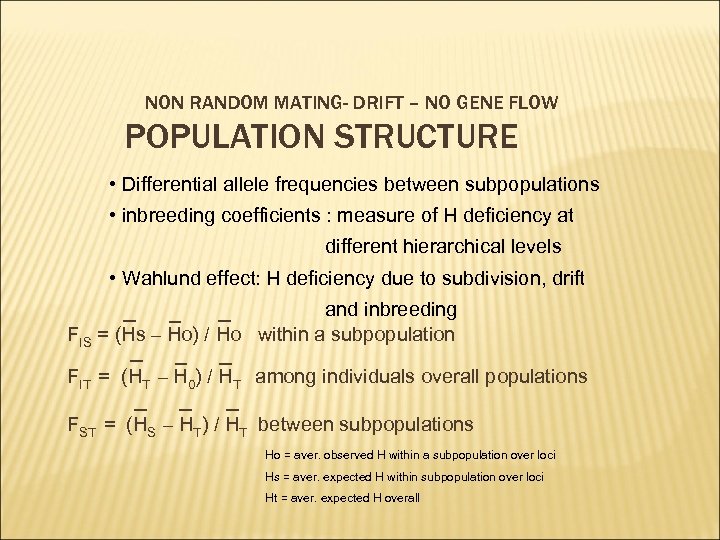 NON RANDOM MATING- DRIFT – NO GENE FLOW POPULATION STRUCTURE • Differential allele frequencies between subpopulations • inbreeding coefficients : measure of H deficiency at different hierarchical levels • Wahlund effect: H deficiency due to subdivision, drift and inbreeding FIS = (Hs – Ho) / Ho within a subpopulation FIT = (HT – H 0) / HT among individuals overall populations FST = (HS – HT) / HT between subpopulations Ho = aver. observed H within a subpopulation over loci Hs = aver. expected H within subpopulation over loci Ht = aver. expected H overall
NON RANDOM MATING- DRIFT – NO GENE FLOW POPULATION STRUCTURE • Differential allele frequencies between subpopulations • inbreeding coefficients : measure of H deficiency at different hierarchical levels • Wahlund effect: H deficiency due to subdivision, drift and inbreeding FIS = (Hs – Ho) / Ho within a subpopulation FIT = (HT – H 0) / HT among individuals overall populations FST = (HS – HT) / HT between subpopulations Ho = aver. observed H within a subpopulation over loci Hs = aver. expected H within subpopulation over loci Ht = aver. expected H overall
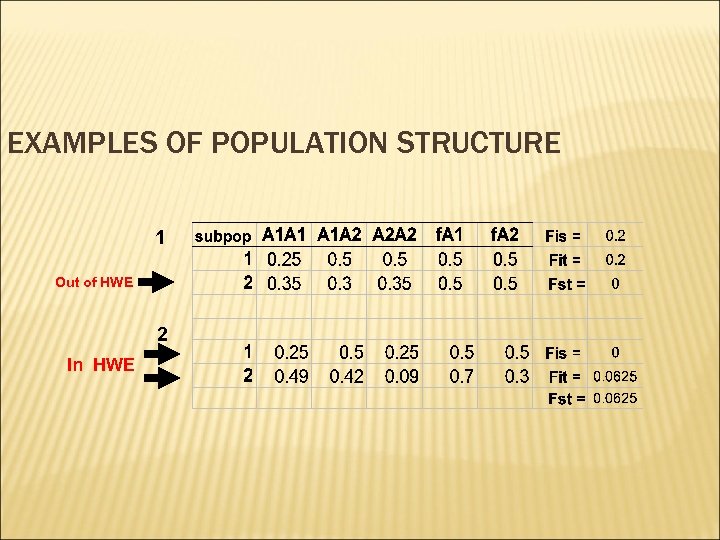 EXAMPLES OF POPULATION STRUCTURE 1 Out of HWE 2 In HWE
EXAMPLES OF POPULATION STRUCTURE 1 Out of HWE 2 In HWE
 GENEALOGIES: A HISTORICAL PERSPECTIVE Lineage: individuals or taxa related by a common ancestor Phylogenetic tree
GENEALOGIES: A HISTORICAL PERSPECTIVE Lineage: individuals or taxa related by a common ancestor Phylogenetic tree
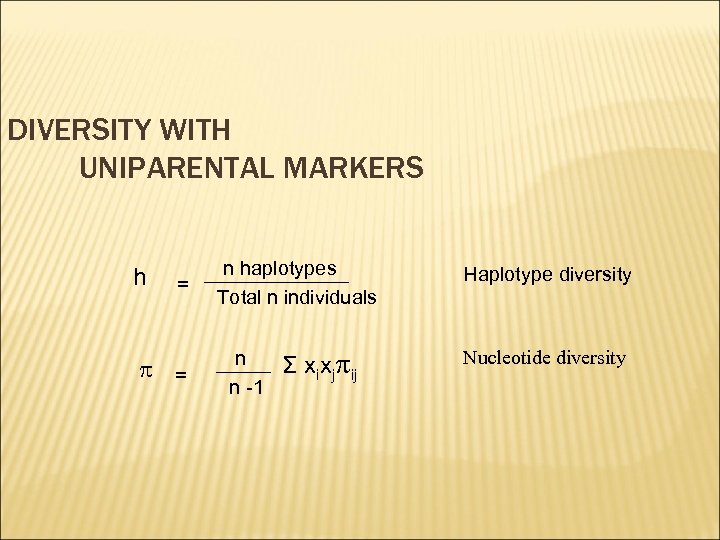 DIVERSITY WITH UNIPARENTAL MARKERS h = p = n haplotypes Total n individuals n n -1 Σ xixjpij Haplotype diversity Nucleotide diversity
DIVERSITY WITH UNIPARENTAL MARKERS h = p = n haplotypes Total n individuals n n -1 Σ xixjpij Haplotype diversity Nucleotide diversity
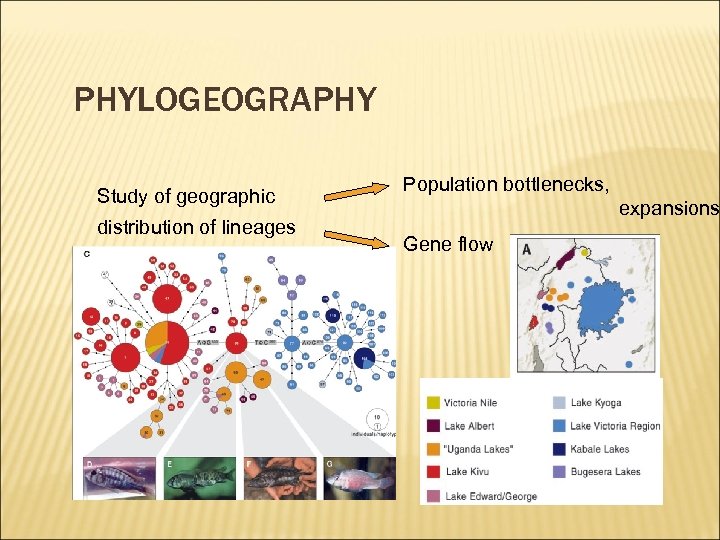 PHYLOGEOGRAPHY Study of geographic distribution of lineages Population bottlenecks, expansions Gene flow
PHYLOGEOGRAPHY Study of geographic distribution of lineages Population bottlenecks, expansions Gene flow
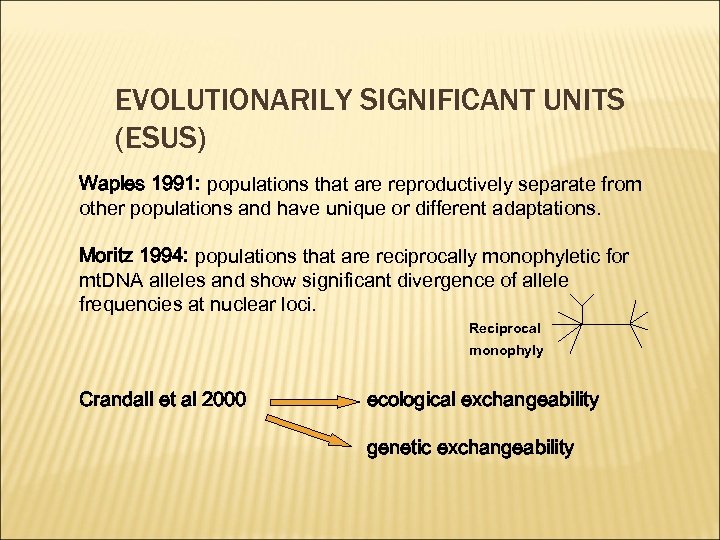 EVOLUTIONARILY SIGNIFICANT UNITS (ESUS) Waples 1991: populations that are reproductively separate from other populations and have unique or different adaptations. Moritz 1994: populations that are reciprocally monophyletic for mt. DNA alleles and show significant divergence of allele frequencies at nuclear loci. Reciprocal monophyly Crandall et al 2000 ecological exchangeability genetic exchangeability
EVOLUTIONARILY SIGNIFICANT UNITS (ESUS) Waples 1991: populations that are reproductively separate from other populations and have unique or different adaptations. Moritz 1994: populations that are reciprocally monophyletic for mt. DNA alleles and show significant divergence of allele frequencies at nuclear loci. Reciprocal monophyly Crandall et al 2000 ecological exchangeability genetic exchangeability
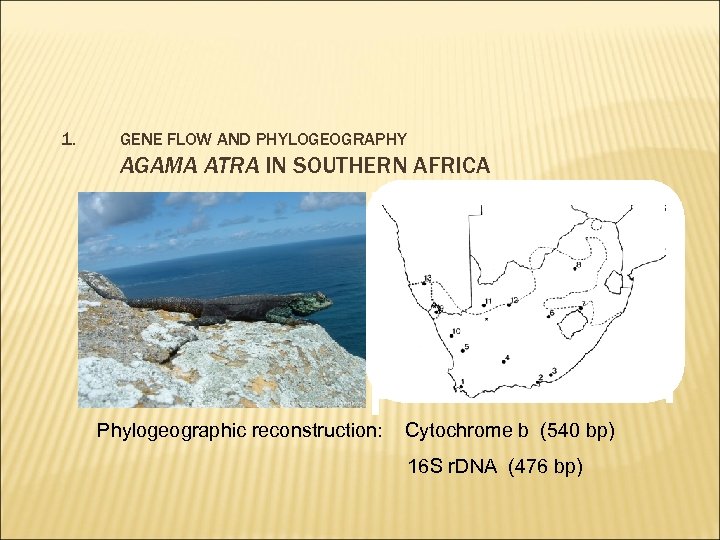 1. GENE FLOW AND PHYLOGEOGRAPHY AGAMA ATRA IN SOUTHERN AFRICA Phylogeographic reconstruction: Cytochrome b (540 bp) 16 S r. DNA (476 bp)
1. GENE FLOW AND PHYLOGEOGRAPHY AGAMA ATRA IN SOUTHERN AFRICA Phylogeographic reconstruction: Cytochrome b (540 bp) 16 S r. DNA (476 bp)
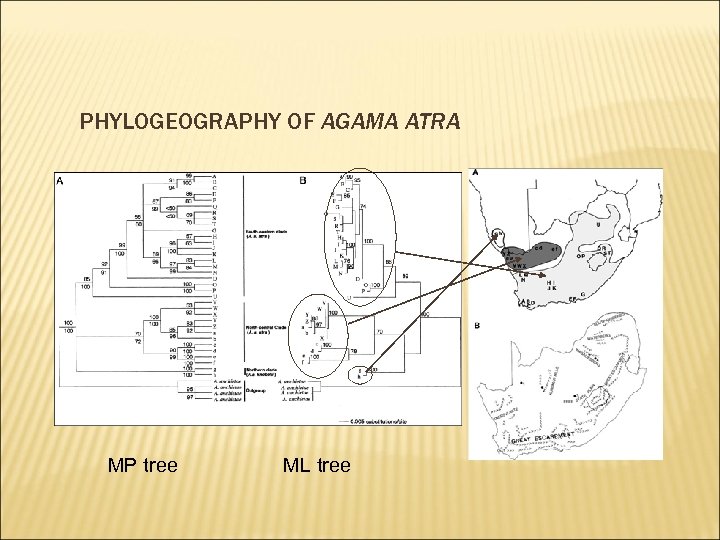 PHYLOGEOGRAPHY OF AGAMA ATRA MP tree ML tree
PHYLOGEOGRAPHY OF AGAMA ATRA MP tree ML tree
 PHYLOGEOGRAPHY OF AGAMA ATRA COINCIDENT PATTERN WITH OTHER ROCK-DWELLING SPECIES Pronolagus rupestris Pachydactilus rugosus Vicariant event cycles of dry-humid period during glacial –interglacial produced fragmentation of habitat
PHYLOGEOGRAPHY OF AGAMA ATRA COINCIDENT PATTERN WITH OTHER ROCK-DWELLING SPECIES Pronolagus rupestris Pachydactilus rugosus Vicariant event cycles of dry-humid period during glacial –interglacial produced fragmentation of habitat
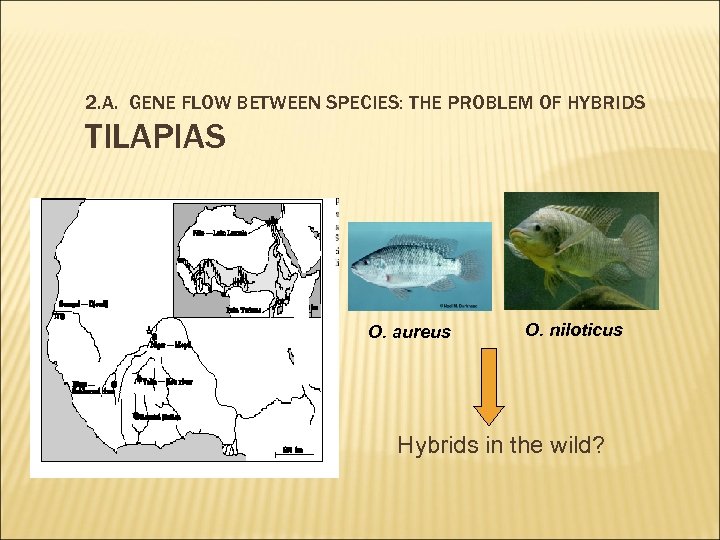 2. A. GENE FLOW BETWEEN SPECIES: THE PROBLEM OF HYBRIDS TILAPIAS O. aureus O. niloticus Hybrids in the wild?
2. A. GENE FLOW BETWEEN SPECIES: THE PROBLEM OF HYBRIDS TILAPIAS O. aureus O. niloticus Hybrids in the wild?
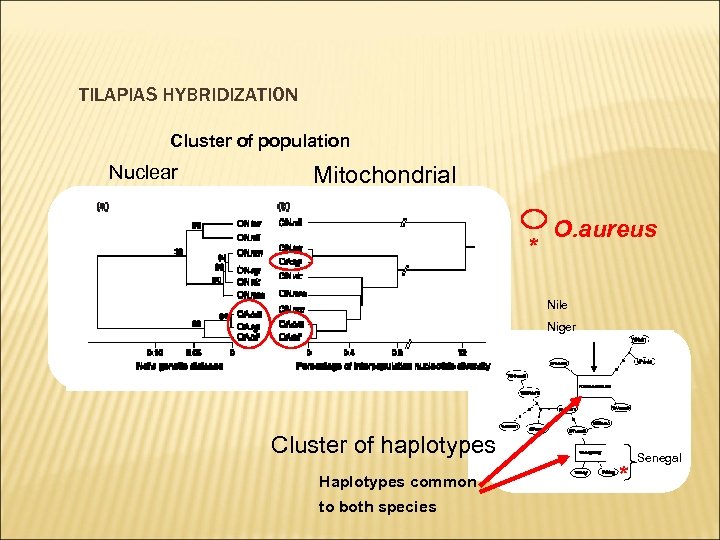 TILAPIAS HYBRIDIZATION Cluster of population Nuclear Mitochondrial * O. aureus Nile Niger Cluster of haplotypes Haplotypes common to both species * Senegal
TILAPIAS HYBRIDIZATION Cluster of population Nuclear Mitochondrial * O. aureus Nile Niger Cluster of haplotypes Haplotypes common to both species * Senegal
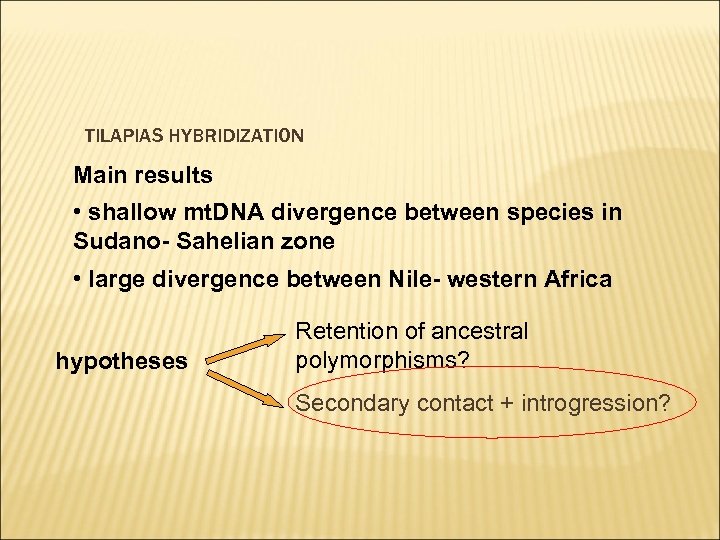 TILAPIAS HYBRIDIZATION Main results • shallow mt. DNA divergence between species in Sudano- Sahelian zone • large divergence between Nile- western Africa hypotheses Retention of ancestral polymorphisms? Secondary contact + introgression?
TILAPIAS HYBRIDIZATION Main results • shallow mt. DNA divergence between species in Sudano- Sahelian zone • large divergence between Nile- western Africa hypotheses Retention of ancestral polymorphisms? Secondary contact + introgression?
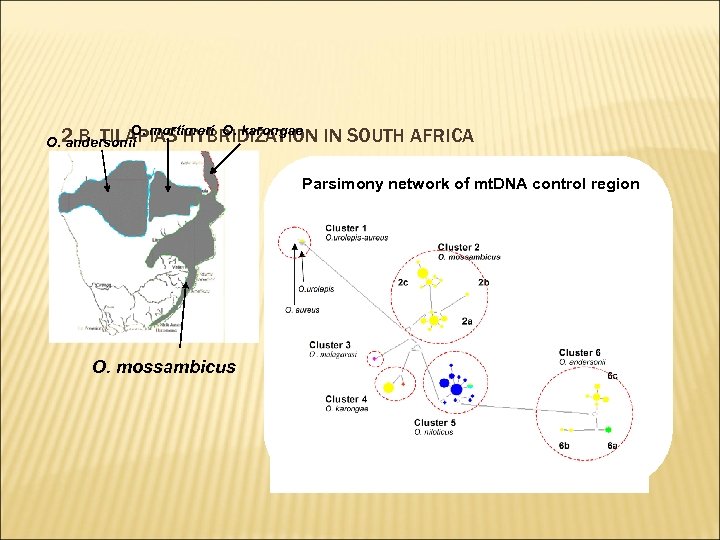 O. mortimeri O. karongae O. 2. B. TILAPIAS HYBRIDIZATION andersonii IN SOUTH AFRICA Parsimony network of mt. DNA control region O. mossambicus
O. mortimeri O. karongae O. 2. B. TILAPIAS HYBRIDIZATION andersonii IN SOUTH AFRICA Parsimony network of mt. DNA control region O. mossambicus
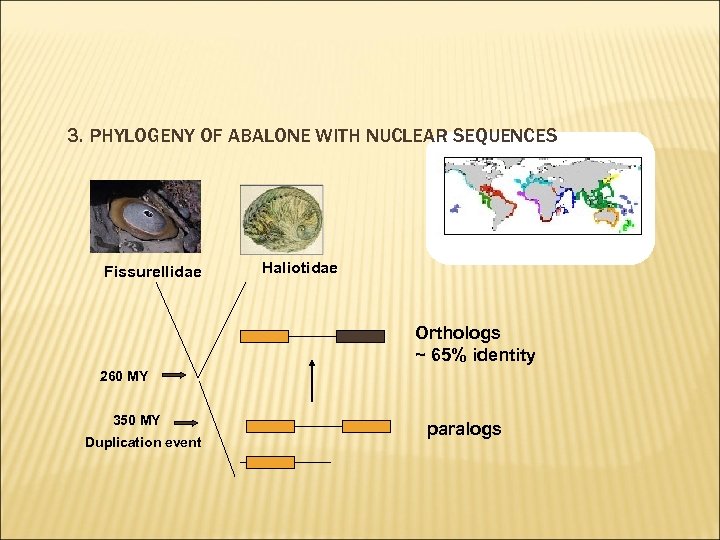 3. PHYLOGENY OF ABALONE WITH NUCLEAR SEQUENCES Fissurellidae Haliotidae Orthologs ~ 65% identity 260 MY 350 MY Duplication event paralogs
3. PHYLOGENY OF ABALONE WITH NUCLEAR SEQUENCES Fissurellidae Haliotidae Orthologs ~ 65% identity 260 MY 350 MY Duplication event paralogs
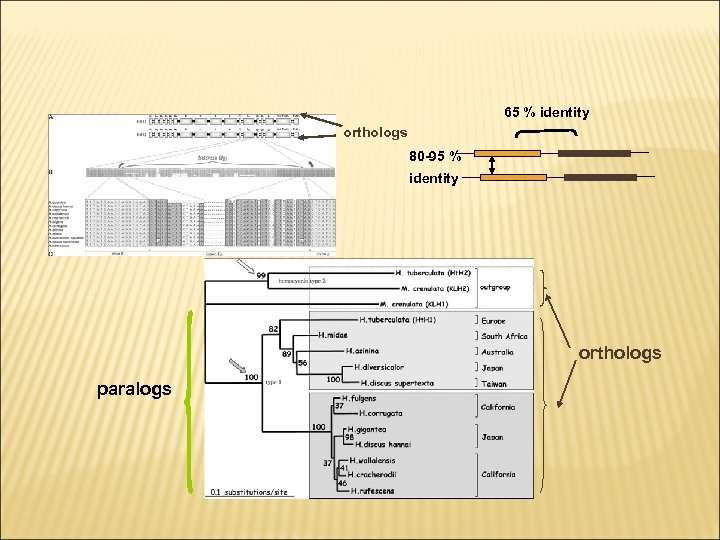 PHYLOGENY OF ABALONES 65 % identity orthologs 80 -95 % identity orthologs paralogs
PHYLOGENY OF ABALONES 65 % identity orthologs 80 -95 % identity orthologs paralogs
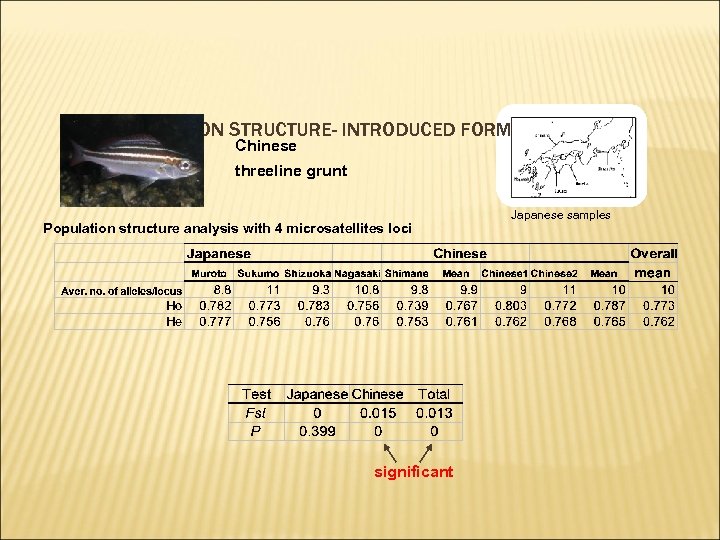 4. POPULATION STRUCTURE- INTRODUCED FORMS Chinese threeline grunt Population structure analysis with 4 microsatellites loci significant Japanese samples
4. POPULATION STRUCTURE- INTRODUCED FORMS Chinese threeline grunt Population structure analysis with 4 microsatellites loci significant Japanese samples
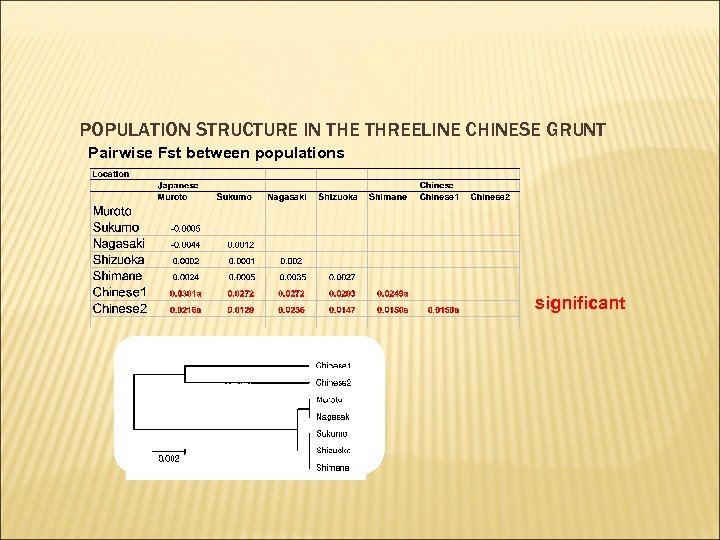 POPULATION STRUCTURE IN THE THREELINE CHINESE GRUNT Pairwise Fst between populations significant
POPULATION STRUCTURE IN THE THREELINE CHINESE GRUNT Pairwise Fst between populations significant


