8c4643c5e9a53867302aa00e9fa81146.ppt
- Количество слайдов: 24
 Conditional Random Fields for the Prediction of Signal Peptide Cleavage Sites M. W. Mak The Hong Kong Polytechnic University S. Y. Kung Princeton University 1 M. W. Mak and S. Y. Kung, ICASSP’ 09
Conditional Random Fields for the Prediction of Signal Peptide Cleavage Sites M. W. Mak The Hong Kong Polytechnic University S. Y. Kung Princeton University 1 M. W. Mak and S. Y. Kung, ICASSP’ 09
 Contents 1. Introduction Proteins and Their Subcellular Locations Importance of Protein Cleavage-Site Prediction Information in Amino Acid Sequences Existing Approaches to Cleavage Site Prediction 2. Conditional Random Field (CRF) CRF for Cleavage Site Prediction 3. Experiments and Results Effectiveness of Different Feature Functions Effect of Varying Window Size Fusion with Signal. P 2 M. W. Mak and S. Y. Kung, ICASSP’ 09
Contents 1. Introduction Proteins and Their Subcellular Locations Importance of Protein Cleavage-Site Prediction Information in Amino Acid Sequences Existing Approaches to Cleavage Site Prediction 2. Conditional Random Field (CRF) CRF for Cleavage Site Prediction 3. Experiments and Results Effectiveness of Different Feature Functions Effect of Varying Window Size Fusion with Signal. P 2 M. W. Mak and S. Y. Kung, ICASSP’ 09
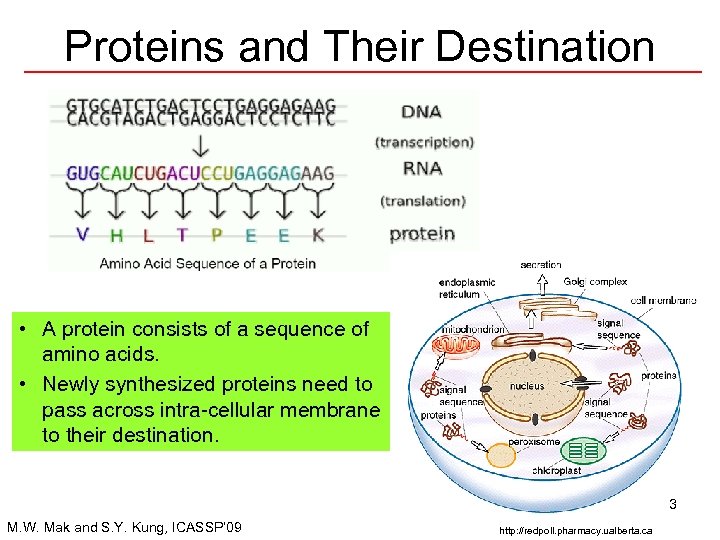 Proteins and Their Destination • A protein consists of a sequence of amino acids. • Newly synthesized proteins need to pass across intra-cellular membrane to their destination. 3 M. W. Mak and S. Y. Kung, ICASSP’ 09 http: //redpoll. pharmacy. ualberta. ca
Proteins and Their Destination • A protein consists of a sequence of amino acids. • Newly synthesized proteins need to pass across intra-cellular membrane to their destination. 3 M. W. Mak and S. Y. Kung, ICASSP’ 09 http: //redpoll. pharmacy. ualberta. ca
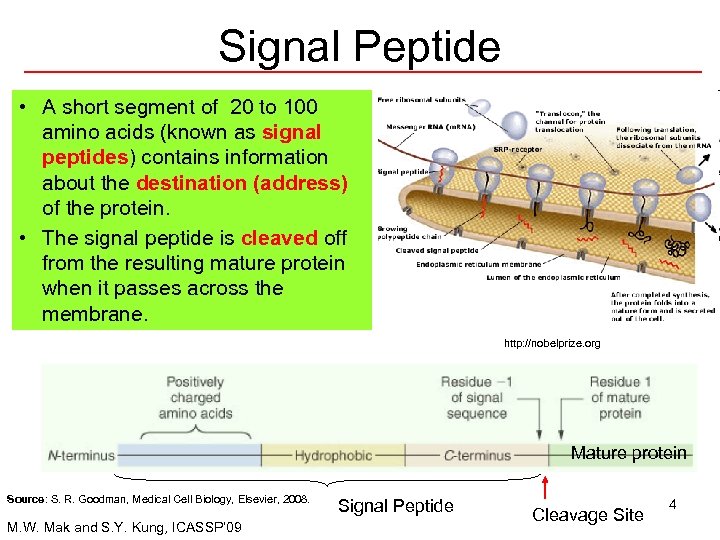 Signal Peptide • A short segment of 20 to 100 amino acids (known as signal peptides) contains information about the destination (address) of the protein. • The signal peptide is cleaved off from the resulting mature protein when it passes across the membrane. http: //nobelprize. org Mature protein Source: S. R. Goodman, Medical Cell Biology, Elsevier, 2008. M. W. Mak and S. Y. Kung, ICASSP’ 09 Signal Peptide Cleavage Site 4
Signal Peptide • A short segment of 20 to 100 amino acids (known as signal peptides) contains information about the destination (address) of the protein. • The signal peptide is cleaved off from the resulting mature protein when it passes across the membrane. http: //nobelprize. org Mature protein Source: S. R. Goodman, Medical Cell Biology, Elsevier, 2008. M. W. Mak and S. Y. Kung, ICASSP’ 09 Signal Peptide Cleavage Site 4
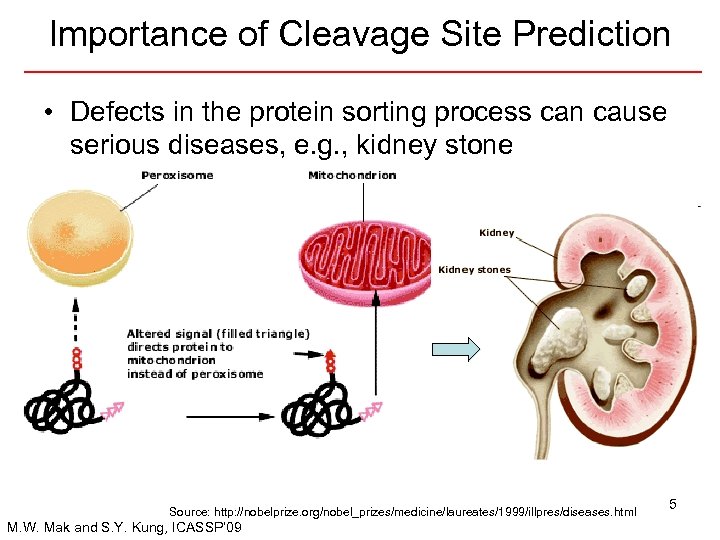 Importance of Cleavage Site Prediction • Defects in the protein sorting process can cause serious diseases, e. g. , kidney stone Source: http: //nobelprize. org/nobel_prizes/medicine/laureates/1999/illpres/diseases. html M. W. Mak and S. Y. Kung, ICASSP’ 09 5
Importance of Cleavage Site Prediction • Defects in the protein sorting process can cause serious diseases, e. g. , kidney stone Source: http: //nobelprize. org/nobel_prizes/medicine/laureates/1999/illpres/diseases. html M. W. Mak and S. Y. Kung, ICASSP’ 09 5
 Importance of Cleavage Site Prediction • Many proteins (e. g. insulin) are produced in living cells. To cause the proteins to be secreted out of the cell, they are provided with a signal peptide. Bioreactor Source: http: //nobelprize. org/nobel_prizes/medicine /laureates/1999/illpres/diseases. html 6 M. W. Mak and S. Y. Kung, ICASSP’ 09
Importance of Cleavage Site Prediction • Many proteins (e. g. insulin) are produced in living cells. To cause the proteins to be secreted out of the cell, they are provided with a signal peptide. Bioreactor Source: http: //nobelprize. org/nobel_prizes/medicine /laureates/1999/illpres/diseases. html 6 M. W. Mak and S. Y. Kung, ICASSP’ 09
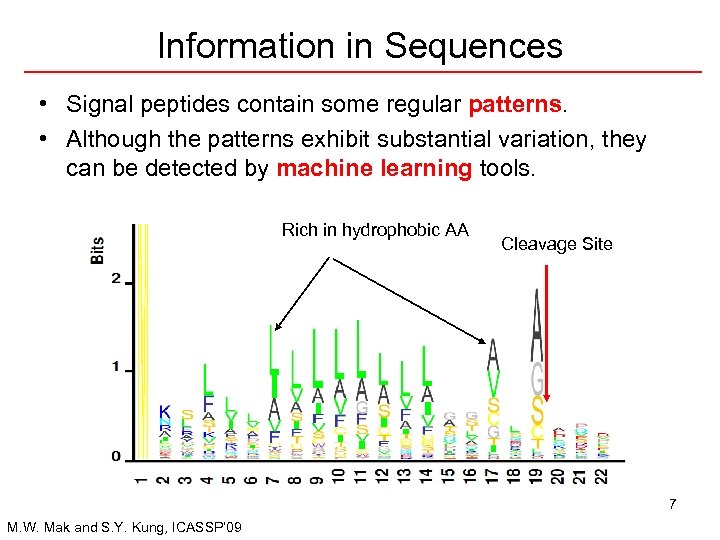 Information in Sequences • Signal peptides contain some regular patterns. • Although the patterns exhibit substantial variation, they can be detected by machine learning tools. Rich in hydrophobic AA Cleavage Site 7 M. W. Mak and S. Y. Kung, ICASSP’ 09
Information in Sequences • Signal peptides contain some regular patterns. • Although the patterns exhibit substantial variation, they can be detected by machine learning tools. Rich in hydrophobic AA Cleavage Site 7 M. W. Mak and S. Y. Kung, ICASSP’ 09
 Existing Methods • Weight matrices (Predi. Si) • Neural Networks (Signal. P 1. 1) • HMMs (Signal. P 3. 0) 8 M. W. Mak and S. Y. Kung, ICASSP’ 09
Existing Methods • Weight matrices (Predi. Si) • Neural Networks (Signal. P 1. 1) • HMMs (Signal. P 3. 0) 8 M. W. Mak and S. Y. Kung, ICASSP’ 09
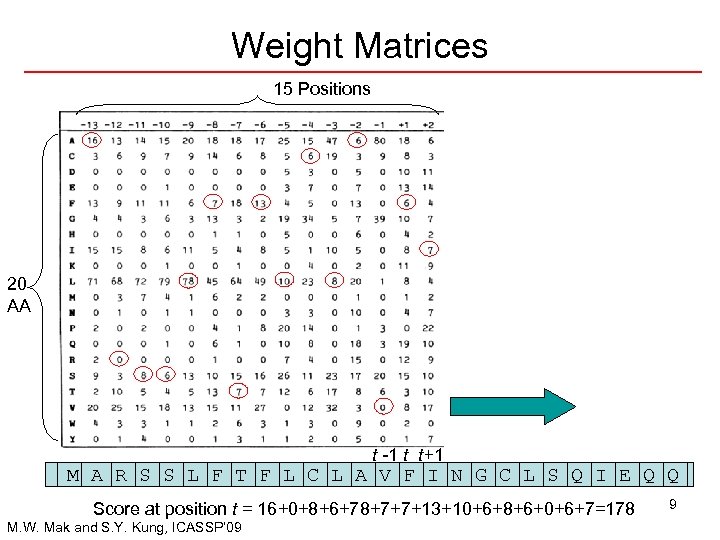 Weight Matrices 15 Positions 20 AA t -1 t t+1 M A R S S L F T F L C L A V F I N G C L S Q I E Q Q Score at position t = 16+0+8+6+78+7+7+13+10+6+8+6+0+6+7=178 M. W. Mak and S. Y. Kung, ICASSP’ 09 9
Weight Matrices 15 Positions 20 AA t -1 t t+1 M A R S S L F T F L C L A V F I N G C L S Q I E Q Q Score at position t = 16+0+8+6+78+7+7+13+10+6+8+6+0+6+7=178 M. W. Mak and S. Y. Kung, ICASSP’ 09 9
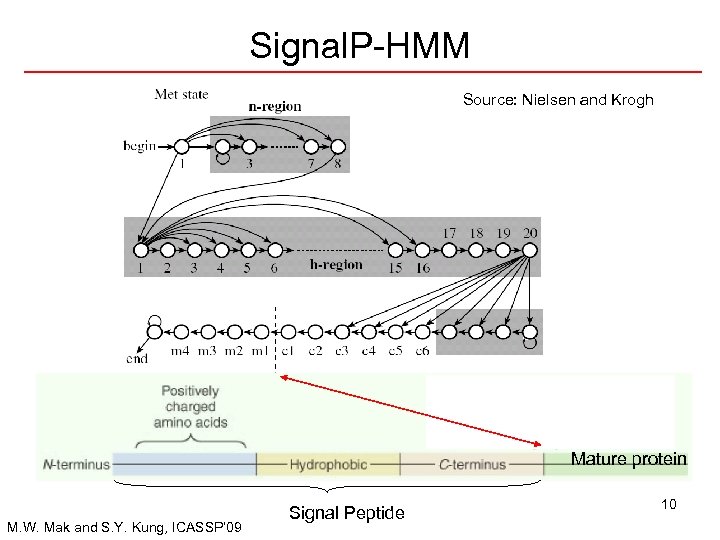 Signal. P-HMM Source: Nielsen and Krogh Mature protein M. W. Mak and S. Y. Kung, ICASSP’ 09 Signal Peptide 10
Signal. P-HMM Source: Nielsen and Krogh Mature protein M. W. Mak and S. Y. Kung, ICASSP’ 09 Signal Peptide 10
 Contents 1. Introduction Proteins and Their Subcellular Locations Importance of Protein Cleavage-Site Prediction Information in Amino Acid Sequences Existing Approaches to Cleavage Site Prediction 2. Conditional Random Field (CRF) CRF for Cleavage Site Prediction 3. Experiments and Results Effectiveness of Amino Acid Properties Effectiveness of Different Feature Functions Fusion with Signal. P 11 M. W. Mak and S. Y. Kung, ICASSP’ 09
Contents 1. Introduction Proteins and Their Subcellular Locations Importance of Protein Cleavage-Site Prediction Information in Amino Acid Sequences Existing Approaches to Cleavage Site Prediction 2. Conditional Random Field (CRF) CRF for Cleavage Site Prediction 3. Experiments and Results Effectiveness of Amino Acid Properties Effectiveness of Different Feature Functions Fusion with Signal. P 11 M. W. Mak and S. Y. Kung, ICASSP’ 09
 Conditional Random Fields • Conditional Random Fields (CRFs) were originally designed for sequence labeling tasks such as Part-of. Speech (POS) tagging • Given a sequence of observations (e. g. , words), a CRF attempts to find the most likely label sequence, i. e. , it gives a label for each of the observations. 12 M. W. Mak and S. Y. Kung, ICASSP’ 09
Conditional Random Fields • Conditional Random Fields (CRFs) were originally designed for sequence labeling tasks such as Part-of. Speech (POS) tagging • Given a sequence of observations (e. g. , words), a CRF attempts to find the most likely label sequence, i. e. , it gives a label for each of the observations. 12 M. W. Mak and S. Y. Kung, ICASSP’ 09
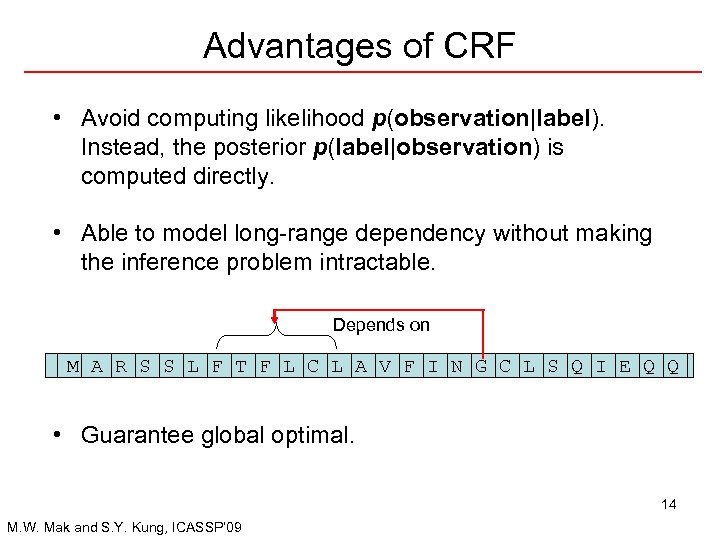 Advantages of CRF • Avoid computing likelihood p(observation|label). Instead, the posterior p(label|observation) is computed directly. • Able to model long-range dependency without making the inference problem intractable. Depends on M A R S S L F T F L C L A V F I N G C L S Q I E Q Q • Guarantee global optimal. 14 M. W. Mak and S. Y. Kung, ICASSP’ 09
Advantages of CRF • Avoid computing likelihood p(observation|label). Instead, the posterior p(label|observation) is computed directly. • Able to model long-range dependency without making the inference problem intractable. Depends on M A R S S L F T F L C L A V F I N G C L S Q I E Q Q • Guarantee global optimal. 14 M. W. Mak and S. Y. Kung, ICASSP’ 09
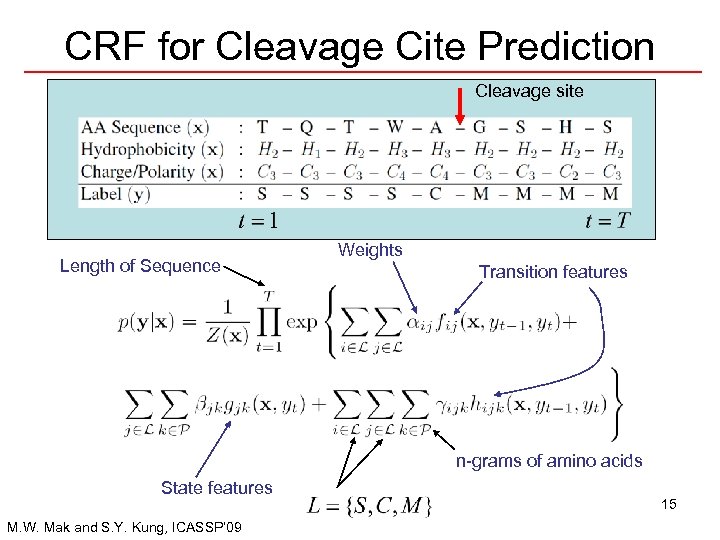 CRF for Cleavage Cite Prediction Cleavage site Length of Sequence Weights Transition features n-grams of amino acids State features M. W. Mak and S. Y. Kung, ICASSP’ 09 15
CRF for Cleavage Cite Prediction Cleavage site Length of Sequence Weights Transition features n-grams of amino acids State features M. W. Mak and S. Y. Kung, ICASSP’ 09 15
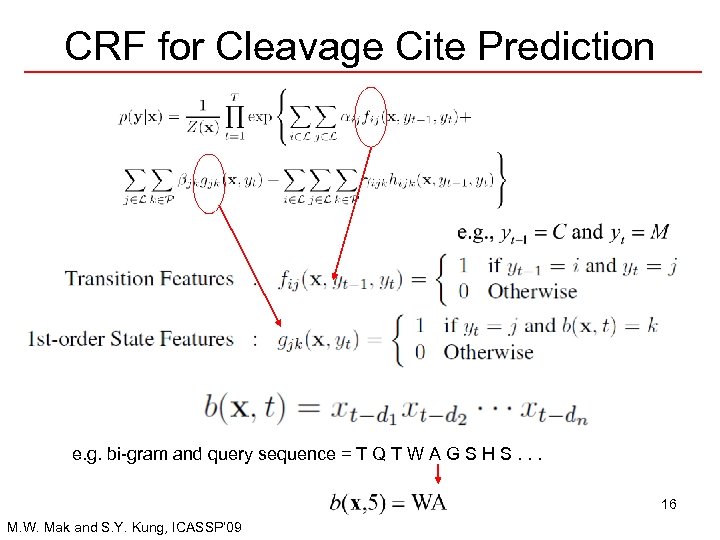 CRF for Cleavage Cite Prediction e. g. bi-gram and query sequence = T Q T W A G S H S. . . 16 M. W. Mak and S. Y. Kung, ICASSP’ 09
CRF for Cleavage Cite Prediction e. g. bi-gram and query sequence = T Q T W A G S H S. . . 16 M. W. Mak and S. Y. Kung, ICASSP’ 09
 Contents 1. Introduction Proteins and Their Subcellular Locations Importance of Protein Cleavage-Site Prediction Information in Amino Acid Sequences Existing Approaches to Cleavage Site Prediction 2. Conditional Random Field (CRF) CRF for Cleavage Site Prediction 3. Experiments and Results Effectiveness of Different Feature Functions Effect of Varying Window Size Fusion with Signal. P 18 M. W. Mak and S. Y. Kung, ICASSP’ 09
Contents 1. Introduction Proteins and Their Subcellular Locations Importance of Protein Cleavage-Site Prediction Information in Amino Acid Sequences Existing Approaches to Cleavage Site Prediction 2. Conditional Random Field (CRF) CRF for Cleavage Site Prediction 3. Experiments and Results Effectiveness of Different Feature Functions Effect of Varying Window Size Fusion with Signal. P 18 M. W. Mak and S. Y. Kung, ICASSP’ 09
 Experiments • Data: 1937 protein sequences extracted from Swissprot 56. 5. The cleavage sites locations of these sequences were biologically determined • Ten-fold cross validation • For 1 st-order state features, up to 5 -grams of amino acids • For 2 nd-order state features, up to bi-grams of amino acids. • Use CRF++ software 19 M. W. Mak and S. Y. Kung, ICASSP’ 09
Experiments • Data: 1937 protein sequences extracted from Swissprot 56. 5. The cleavage sites locations of these sequences were biologically determined • Ten-fold cross validation • For 1 st-order state features, up to 5 -grams of amino acids • For 2 nd-order state features, up to bi-grams of amino acids. • Use CRF++ software 19 M. W. Mak and S. Y. Kung, ICASSP’ 09
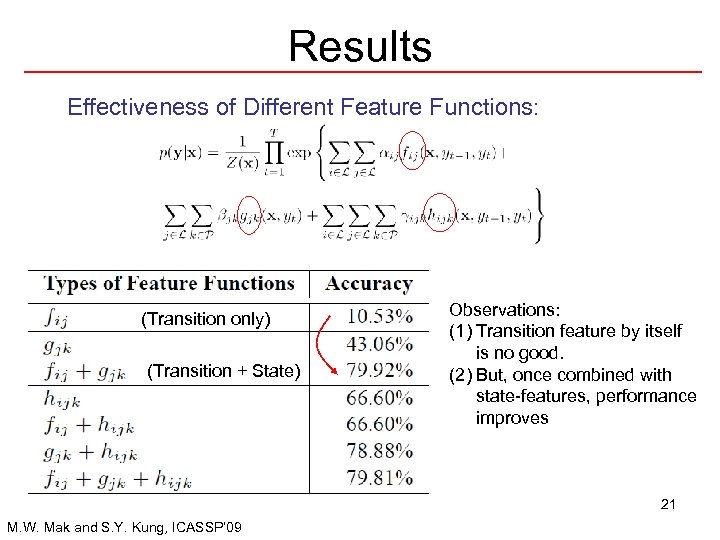 Results Effectiveness of Different Feature Functions: (Transition only) (Transition + State) Observations: (1) Transition feature by itself is no good. (2) But, once combined with state-features, performance improves 21 M. W. Mak and S. Y. Kung, ICASSP’ 09
Results Effectiveness of Different Feature Functions: (Transition only) (Transition + State) Observations: (1) Transition feature by itself is no good. (2) But, once combined with state-features, performance improves 21 M. W. Mak and S. Y. Kung, ICASSP’ 09
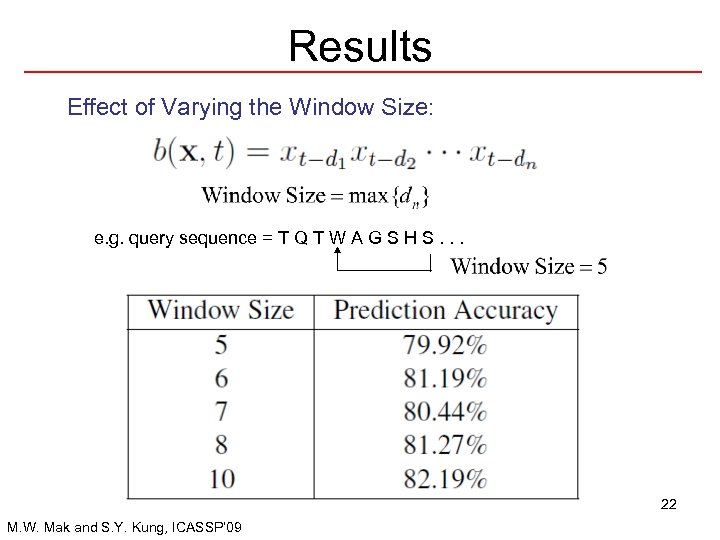 Results Effect of Varying the Window Size: e. g. query sequence = T Q T W A G S H S. . . 22 M. W. Mak and S. Y. Kung, ICASSP’ 09
Results Effect of Varying the Window Size: e. g. query sequence = T Q T W A G S H S. . . 22 M. W. Mak and S. Y. Kung, ICASSP’ 09
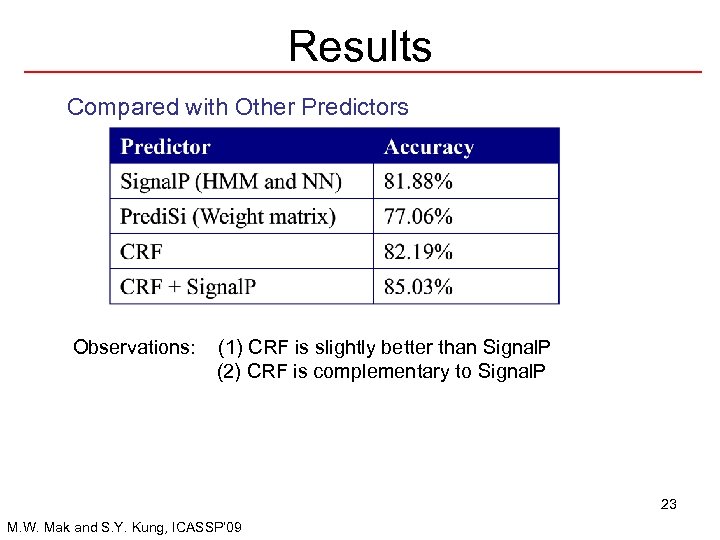 Results Compared with Other Predictors Observations: (1) CRF is slightly better than Signal. P (2) CRF is complementary to Signal. P 23 M. W. Mak and S. Y. Kung, ICASSP’ 09
Results Compared with Other Predictors Observations: (1) CRF is slightly better than Signal. P (2) CRF is complementary to Signal. P 23 M. W. Mak and S. Y. Kung, ICASSP’ 09
 Web Server http: //158. 132. 148. 85: 8080/CSite. Pred/faces/Page 1. jsp 24 M. W. Mak and S. Y. Kung, ICASSP’ 09
Web Server http: //158. 132. 148. 85: 8080/CSite. Pred/faces/Page 1. jsp 24 M. W. Mak and S. Y. Kung, ICASSP’ 09
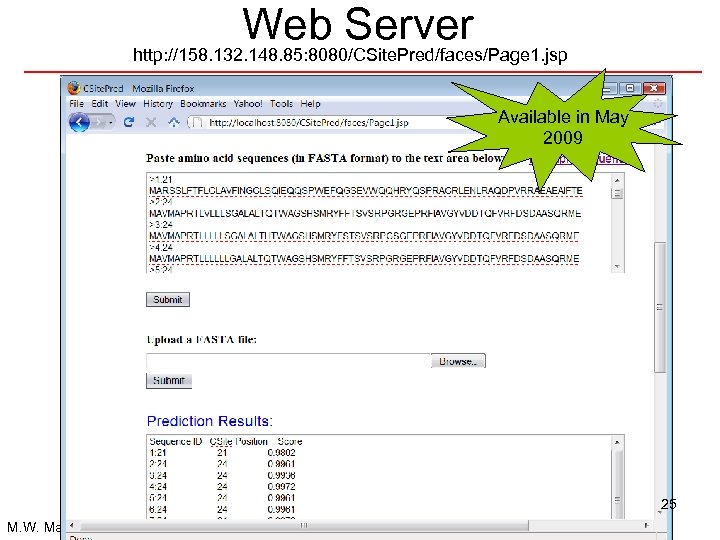 Web Server http: //158. 132. 148. 85: 8080/CSite. Pred/faces/Page 1. jsp Available in May 2009 25 M. W. Mak and S. Y. Kung, ICASSP’ 09
Web Server http: //158. 132. 148. 85: 8080/CSite. Pred/faces/Page 1. jsp Available in May 2009 25 M. W. Mak and S. Y. Kung, ICASSP’ 09
 26 M. W. Mak and S. Y. Kung, ICASSP’ 09
26 M. W. Mak and S. Y. Kung, ICASSP’ 09
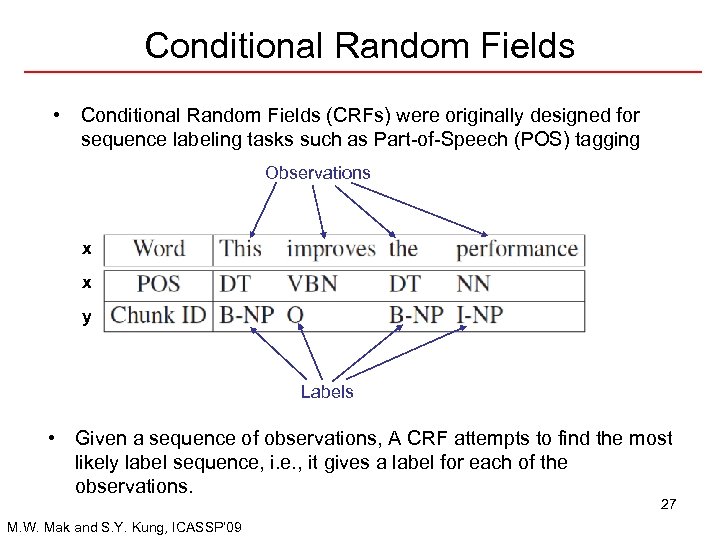 Conditional Random Fields • Conditional Random Fields (CRFs) were originally designed for sequence labeling tasks such as Part-of-Speech (POS) tagging Observations x x y Labels • Given a sequence of observations, A CRF attempts to find the most likely label sequence, i. e. , it gives a label for each of the observations. 27 M. W. Mak and S. Y. Kung, ICASSP’ 09
Conditional Random Fields • Conditional Random Fields (CRFs) were originally designed for sequence labeling tasks such as Part-of-Speech (POS) tagging Observations x x y Labels • Given a sequence of observations, A CRF attempts to find the most likely label sequence, i. e. , it gives a label for each of the observations. 27 M. W. Mak and S. Y. Kung, ICASSP’ 09


