92e71c5cea6c83d330550adf9f5eac70.ppt
- Количество слайдов: 23
 Computer Aided Drug Design Hanoch Senderowitz Department of Chemistry Bar Ilan University BIU-Valencia Workshop April 2010
Computer Aided Drug Design Hanoch Senderowitz Department of Chemistry Bar Ilan University BIU-Valencia Workshop April 2010
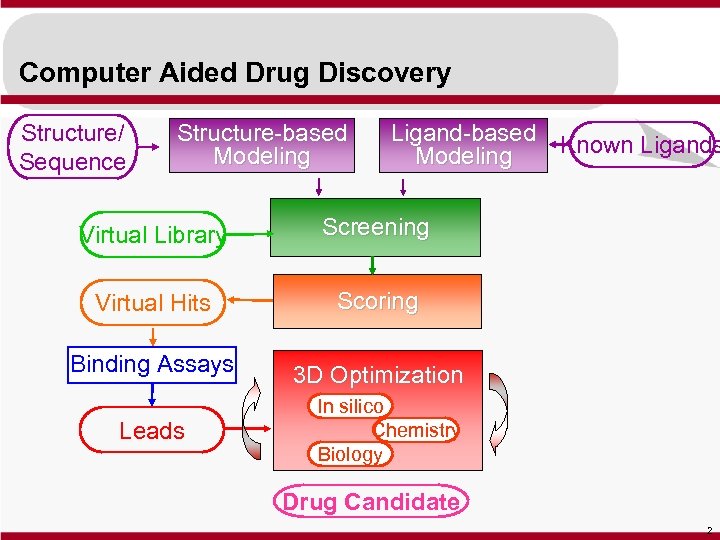 Computer Aided Drug Discovery Structure/ Sequence Structure-based Modeling Ligand-based Known Ligands Modeling Virtual Library Screening Virtual Hits Scoring Binding Assays 3 D Optimization Leads In silico Chemistry Biology Drug Candidate 2
Computer Aided Drug Discovery Structure/ Sequence Structure-based Modeling Ligand-based Known Ligands Modeling Virtual Library Screening Virtual Hits Scoring Binding Assays 3 D Optimization Leads In silico Chemistry Biology Drug Candidate 2
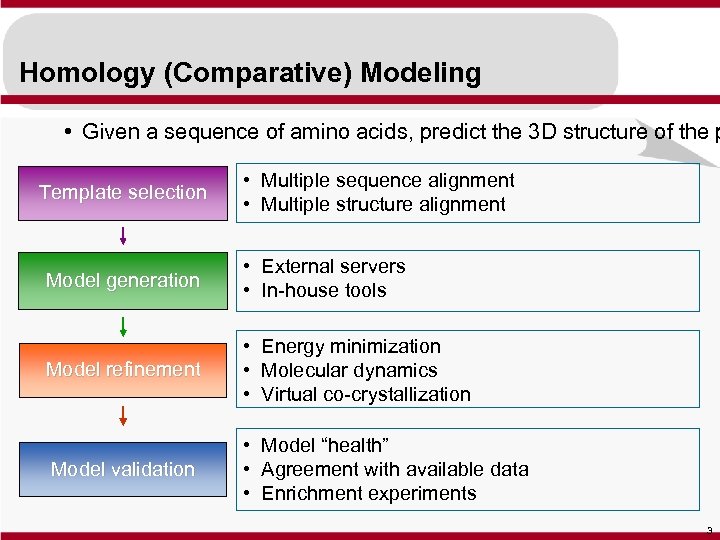 Homology (Comparative) Modeling • Given a sequence of amino acids, predict the 3 D structure of the p Template selection • Multiple sequence alignment • Multiple structure alignment Model generation • External servers • In-house tools Model refinement • Energy minimization • Molecular dynamics • Virtual co-crystallization Model validation • Model “health” • Agreement with available data • Enrichment experiments 3
Homology (Comparative) Modeling • Given a sequence of amino acids, predict the 3 D structure of the p Template selection • Multiple sequence alignment • Multiple structure alignment Model generation • External servers • In-house tools Model refinement • Energy minimization • Molecular dynamics • Virtual co-crystallization Model validation • Model “health” • Agreement with available data • Enrichment experiments 3
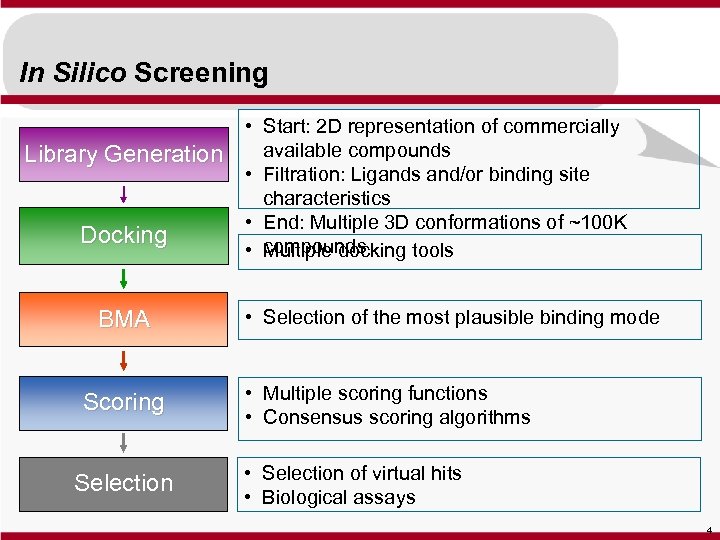 In Silico Screening Library Generation Docking BMA Scoring Selection • Start: 2 D representation of commercially available compounds • Filtration: Ligands and/or binding site characteristics • End: Multiple 3 D conformations of ~100 K • compounds Multiple docking tools • Selection of the most plausible binding mode • Multiple scoring functions • Consensus scoring algorithms • Selection of virtual hits • Biological assays 4
In Silico Screening Library Generation Docking BMA Scoring Selection • Start: 2 D representation of commercially available compounds • Filtration: Ligands and/or binding site characteristics • End: Multiple 3 D conformations of ~100 K • compounds Multiple docking tools • Selection of the most plausible binding mode • Multiple scoring functions • Consensus scoring algorithms • Selection of virtual hits • Biological assays 4
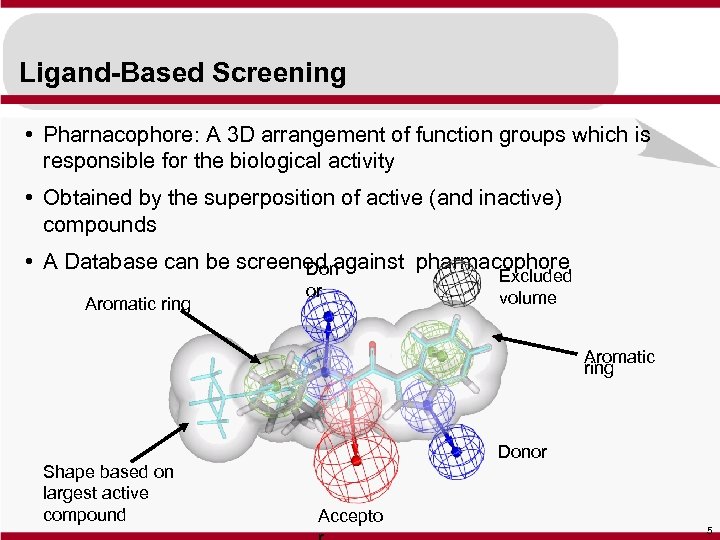 Ligand-Based Screening • Pharnacophore: A 3 D arrangement of function groups which is responsible for the biological activity • Obtained by the superposition of active (and inactive) compounds • A Database can be screened against pharmacophore Don Aromatic ring or Excluded volume Aromatic ring Shape based on largest active compound Donor Accepto 5
Ligand-Based Screening • Pharnacophore: A 3 D arrangement of function groups which is responsible for the biological activity • Obtained by the superposition of active (and inactive) compounds • A Database can be screened against pharmacophore Don Aromatic ring or Excluded volume Aromatic ring Shape based on largest active compound Donor Accepto 5
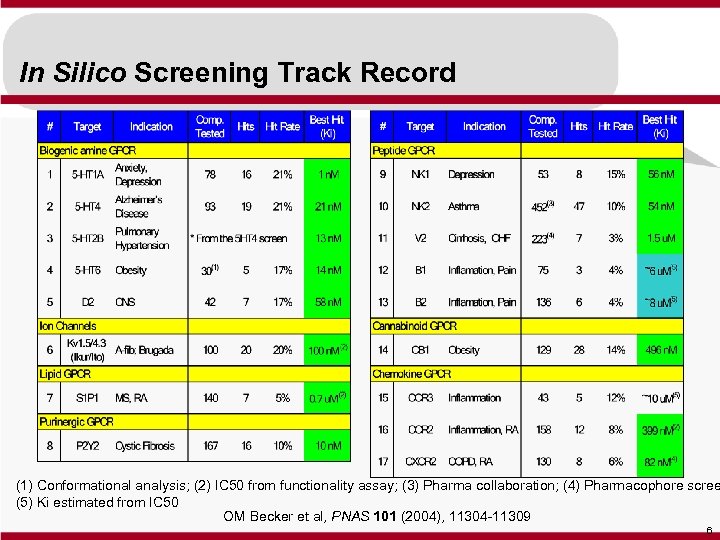 In Silico Screening Track Record (1) Conformational analysis; (2) IC 50 from functionality assay; (3) Pharma collaboration; (4) Pharmacophore scree (5) Ki estimated from IC 50 OM Becker et al, PNAS 101 (2004), 11304 -11309 6
In Silico Screening Track Record (1) Conformational analysis; (2) IC 50 from functionality assay; (3) Pharma collaboration; (4) Pharmacophore scree (5) Ki estimated from IC 50 OM Becker et al, PNAS 101 (2004), 11304 -11309 6
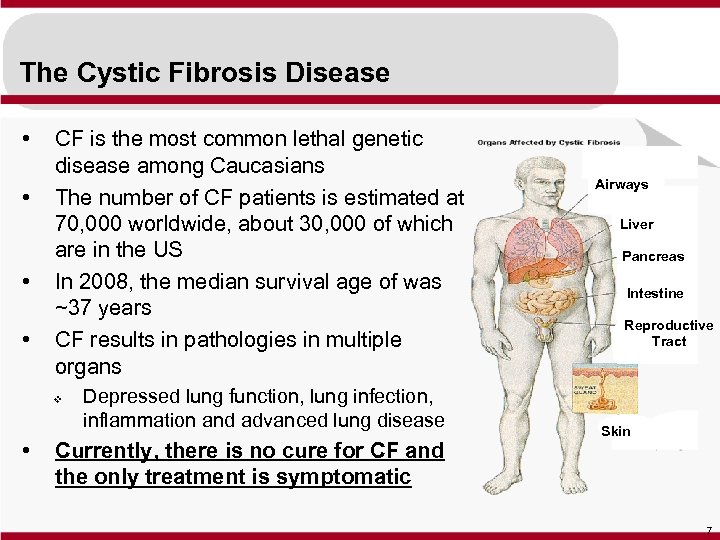 The Cystic Fibrosis Disease • • CF is the most common lethal genetic disease among Caucasians The number of CF patients is estimated at 70, 000 worldwide, about 30, 000 of which are in the US In 2008, the median survival age of was ~37 years CF results in pathologies in multiple organs v • Depressed lung function, lung infection, inflammation and advanced lung disease Currently, there is no cure for CF and the only treatment is symptomatic Airways Liver Pancreas Intestine Reproductive Tract Skin 7
The Cystic Fibrosis Disease • • CF is the most common lethal genetic disease among Caucasians The number of CF patients is estimated at 70, 000 worldwide, about 30, 000 of which are in the US In 2008, the median survival age of was ~37 years CF results in pathologies in multiple organs v • Depressed lung function, lung infection, inflammation and advanced lung disease Currently, there is no cure for CF and the only treatment is symptomatic Airways Liver Pancreas Intestine Reproductive Tract Skin 7
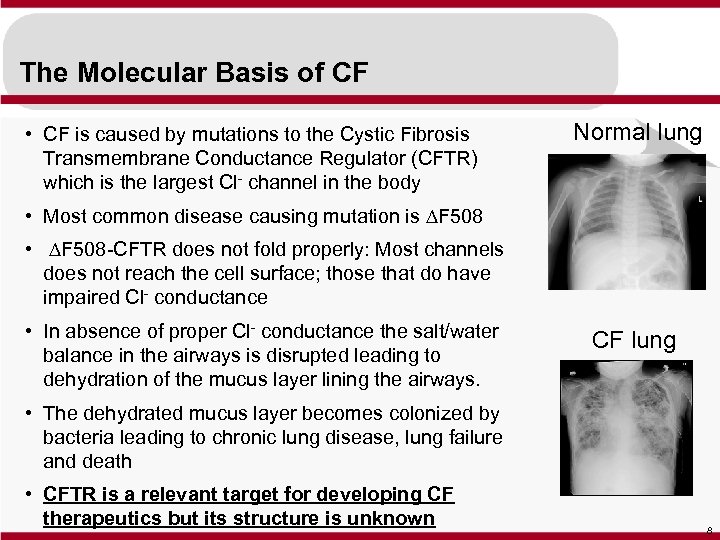 The Molecular Basis of CF • CF is caused by mutations to the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) which is the largest Cl- channel in the body Normal lung • Most common disease causing mutation is DF 508 • DF 508 -CFTR does not fold properly: Most channels does not reach the cell surface; those that do have impaired Cl- conductance • In absence of proper Cl- conductance the salt/water balance in the airways is disrupted leading to dehydration of the mucus layer lining the airways. CF lung • The dehydrated mucus layer becomes colonized by bacteria leading to chronic lung disease, lung failure and death • CFTR is a relevant target for developing CF therapeutics but its structure is unknown 8
The Molecular Basis of CF • CF is caused by mutations to the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) which is the largest Cl- channel in the body Normal lung • Most common disease causing mutation is DF 508 • DF 508 -CFTR does not fold properly: Most channels does not reach the cell surface; those that do have impaired Cl- conductance • In absence of proper Cl- conductance the salt/water balance in the airways is disrupted leading to dehydration of the mucus layer lining the airways. CF lung • The dehydrated mucus layer becomes colonized by bacteria leading to chronic lung disease, lung failure and death • CFTR is a relevant target for developing CF therapeutics but its structure is unknown 8
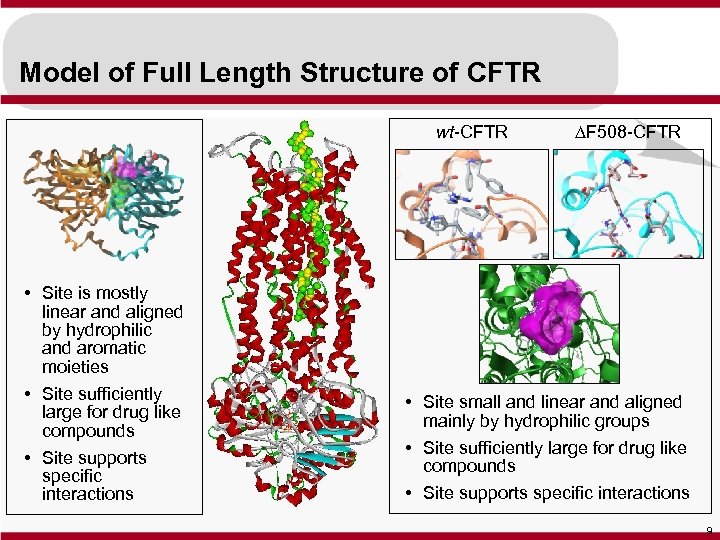 Model of Full Length Structure of CFTR wt-CFTR • Site is mostly linear and aligned by hydrophilic and aromatic moieties • Site sufficiently large for drug like compounds • Site supports specific interactions DF 508 -CFTR • Site small and linear and aligned mainly by hydrophilic groups • Site sufficiently large for drug like compounds • Site supports specific interactions 9
Model of Full Length Structure of CFTR wt-CFTR • Site is mostly linear and aligned by hydrophilic and aromatic moieties • Site sufficiently large for drug like compounds • Site supports specific interactions DF 508 -CFTR • Site small and linear and aligned mainly by hydrophilic groups • Site sufficiently large for drug like compounds • Site supports specific interactions 9
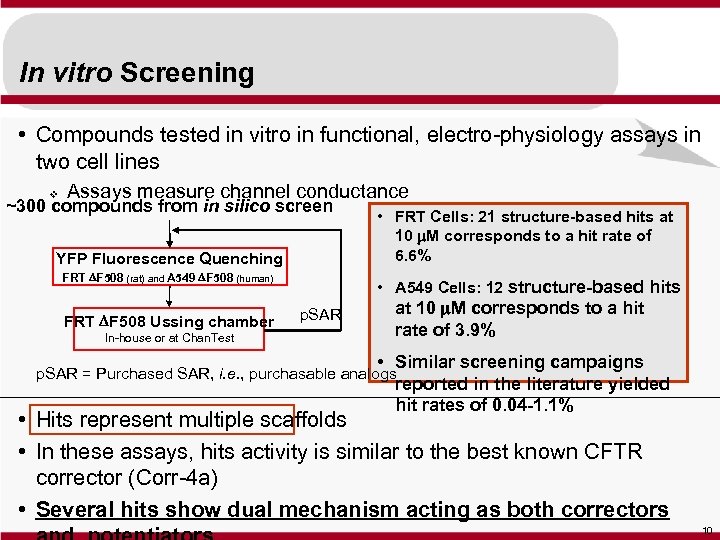 In vitro Screening • Compounds tested in vitro in functional, electro-physiology assays in two cell lines v Assays measure channel conductance ~300 compounds from in silico screen YFP Fluorescence Quenching FRT DF 508 (rat) and A 549 DF 508 (human) FRT DF 508 Ussing chamber In-house or at Chan. Test • FRT Cells: 21 structure-based hits at 10 m. M corresponds to a hit rate of 6. 6% • A 549 Cells: 12 structure-based hits p. SAR at 10 m. M corresponds to a hit rate of 3. 9% • Similar screening campaigns reported in the literature yielded hit rates of 0. 04 -1. 1% p. SAR = Purchased SAR, i. e. , purchasable analogs • Hits represent multiple scaffolds • In these assays, hits activity is similar to the best known CFTR corrector (Corr-4 a) • Several hits show dual mechanism acting as both correctors 10
In vitro Screening • Compounds tested in vitro in functional, electro-physiology assays in two cell lines v Assays measure channel conductance ~300 compounds from in silico screen YFP Fluorescence Quenching FRT DF 508 (rat) and A 549 DF 508 (human) FRT DF 508 Ussing chamber In-house or at Chan. Test • FRT Cells: 21 structure-based hits at 10 m. M corresponds to a hit rate of 6. 6% • A 549 Cells: 12 structure-based hits p. SAR at 10 m. M corresponds to a hit rate of 3. 9% • Similar screening campaigns reported in the literature yielded hit rates of 0. 04 -1. 1% p. SAR = Purchased SAR, i. e. , purchasable analogs • Hits represent multiple scaffolds • In these assays, hits activity is similar to the best known CFTR corrector (Corr-4 a) • Several hits show dual mechanism acting as both correctors 10
 Lead Optimization: The Art of Balance BBB CYP Permeabilit h. ERG y Efficacy Solubility Binding 11
Lead Optimization: The Art of Balance BBB CYP Permeabilit h. ERG y Efficacy Solubility Binding 11
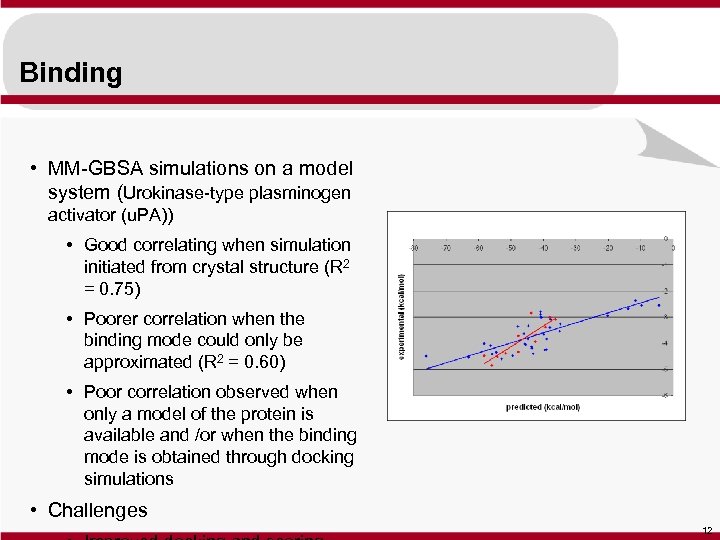 Binding • MM-GBSA simulations on a model system (Urokinase-type plasminogen activator (u. PA)) • Good correlating when simulation initiated from crystal structure (R 2 = 0. 75) • Poorer correlation when the binding mode could only be approximated (R 2 = 0. 60) • Poor correlation observed when only a model of the protein is available and /or when the binding mode is obtained through docking simulations • Challenges 12
Binding • MM-GBSA simulations on a model system (Urokinase-type plasminogen activator (u. PA)) • Good correlating when simulation initiated from crystal structure (R 2 = 0. 75) • Poorer correlation when the binding mode could only be approximated (R 2 = 0. 60) • Poor correlation observed when only a model of the protein is available and /or when the binding mode is obtained through docking simulations • Challenges 12
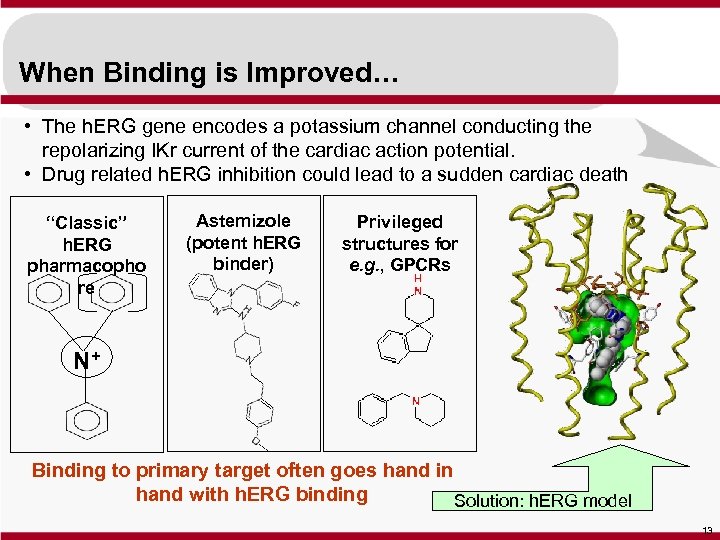 When Binding is Improved… • The h. ERG gene encodes a potassium channel conducting the repolarizing IKr current of the cardiac action potential. • Drug related h. ERG inhibition could lead to a sudden cardiac death “Classic” h. ERG pharmacopho re Astemizole (potent h. ERG binder) Privileged structures for e. g. , GPCRs N+ Binding to primary target often goes hand in hand with h. ERG binding Solution: h. ERG model 13
When Binding is Improved… • The h. ERG gene encodes a potassium channel conducting the repolarizing IKr current of the cardiac action potential. • Drug related h. ERG inhibition could lead to a sudden cardiac death “Classic” h. ERG pharmacopho re Astemizole (potent h. ERG binder) Privileged structures for e. g. , GPCRs N+ Binding to primary target often goes hand in hand with h. ERG binding Solution: h. ERG model 13
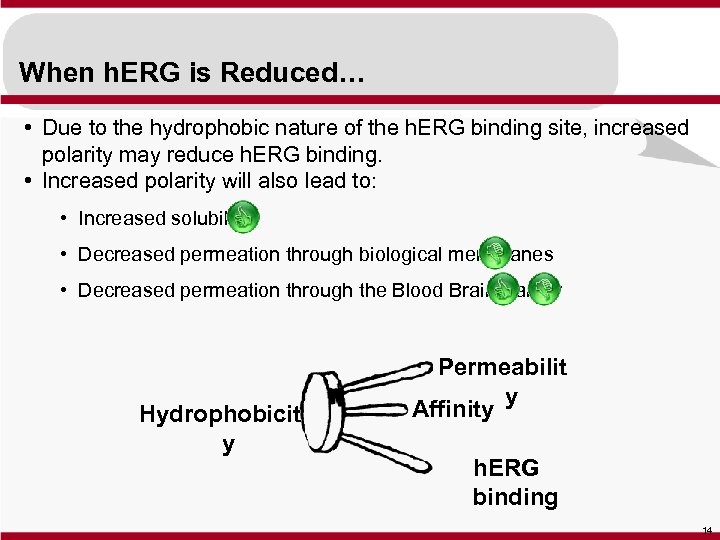 When h. ERG is Reduced… • Due to the hydrophobic nature of the h. ERG binding site, increased polarity may reduce h. ERG binding. • Increased polarity will also lead to: • Increased solubility • Decreased permeation through biological membranes • Decreased permeation through the Blood Brain Barrier Hydrophobicit y Permeabilit y Affinity h. ERG binding 14
When h. ERG is Reduced… • Due to the hydrophobic nature of the h. ERG binding site, increased polarity may reduce h. ERG binding. • Increased polarity will also lead to: • Increased solubility • Decreased permeation through biological membranes • Decreased permeation through the Blood Brain Barrier Hydrophobicit y Permeabilit y Affinity h. ERG binding 14
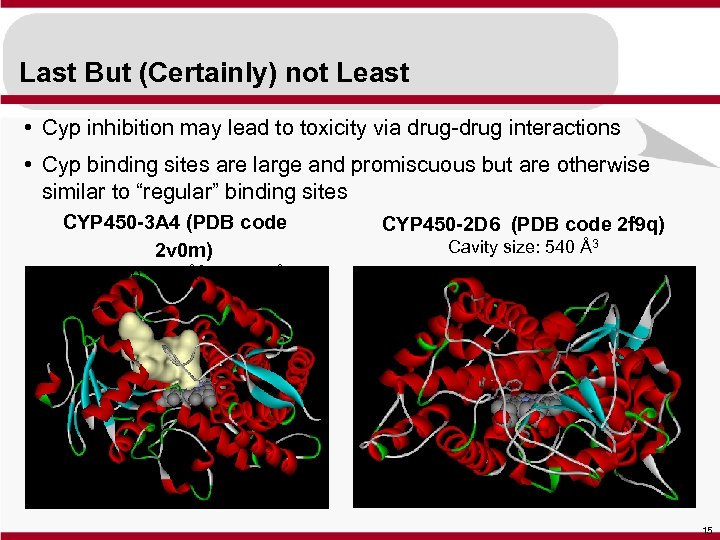 Last But (Certainly) not Least • Cyp inhibition may lead to toxicity via drug-drug interactions • Cyp binding sites are large and promiscuous but are otherwise similar to “regular” binding sites CYP 450 -3 A 4 (PDB code 2 v 0 m) CYP 450 -2 D 6 (PDB code 2 f 9 q) Cavity size: 540 Å3 Cavity size: 950 Å3 to 2000 Å3 15
Last But (Certainly) not Least • Cyp inhibition may lead to toxicity via drug-drug interactions • Cyp binding sites are large and promiscuous but are otherwise similar to “regular” binding sites CYP 450 -3 A 4 (PDB code 2 v 0 m) CYP 450 -2 D 6 (PDB code 2 f 9 q) Cavity size: 540 Å3 Cavity size: 950 Å3 to 2000 Å3 15
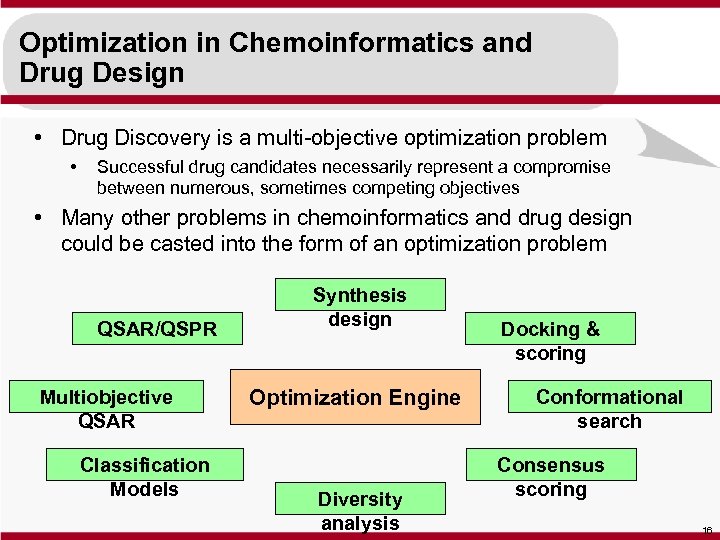 Optimization in Chemoinformatics and Drug Design • Drug Discovery is a multi-objective optimization problem • Successful drug candidates necessarily represent a compromise between numerous, sometimes competing objectives • Many other problems in chemoinformatics and drug design could be casted into the form of an optimization problem QSAR/QSPR Multiobjective QSAR Classification Models Synthesis design Optimization Engine Diversity analysis Docking & scoring Conformational search Consensus scoring 16
Optimization in Chemoinformatics and Drug Design • Drug Discovery is a multi-objective optimization problem • Successful drug candidates necessarily represent a compromise between numerous, sometimes competing objectives • Many other problems in chemoinformatics and drug design could be casted into the form of an optimization problem QSAR/QSPR Multiobjective QSAR Classification Models Synthesis design Optimization Engine Diversity analysis Docking & scoring Conformational search Consensus scoring 16
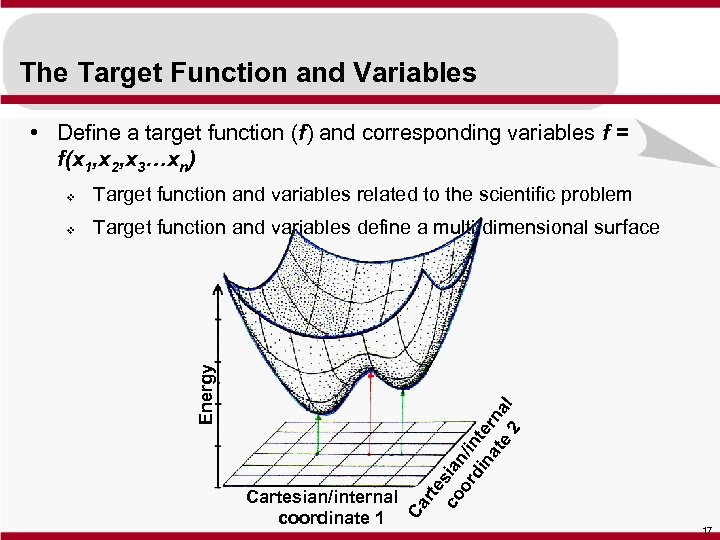 The Target Function and Variables • Define a target function (f) and corresponding variables f = f(x 1, x 2, x 3…xn) v Target function and variables define a multi-dimensional surface Ca Cartesian/internal coordinate 1 rte co sian or /in di te na rn te al 2 Target function and variables related to the scientific problem Energy v 17
The Target Function and Variables • Define a target function (f) and corresponding variables f = f(x 1, x 2, x 3…xn) v Target function and variables define a multi-dimensional surface Ca Cartesian/internal coordinate 1 rte co sian or /in di te na rn te al 2 Target function and variables related to the scientific problem Energy v 17
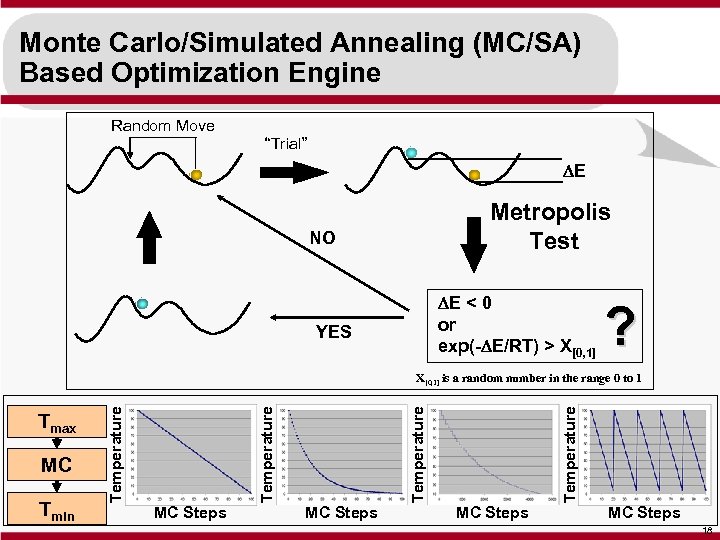 Monte Carlo/Simulated Annealing (MC/SA) Based Optimization Engine Random Move “Trial” DE Metropolis Test NO DE < 0 or exp(-DE/RT) > X[0, 1] YES ? MC Steps Temperature Tmin Temperature MC Temperature Tmax Temperature X[0, 1] is a random number in the range 0 to 1 MC Steps 18
Monte Carlo/Simulated Annealing (MC/SA) Based Optimization Engine Random Move “Trial” DE Metropolis Test NO DE < 0 or exp(-DE/RT) > X[0, 1] YES ? MC Steps Temperature Tmin Temperature MC Temperature Tmax Temperature X[0, 1] is a random number in the range 0 to 1 MC Steps 18
 Quantitative Structure Activity Relationship (QSAR) Quantitative Structure Property Relationship (QSPR) • Correlate specific biological activity for a set of compounds with their structure-derived molecular descriptors by means of a mathematical model • The nature of correlation, activity and descriptors are unlimited v BBB permeability = f (hydrophobicity, H-bonding potential) v Metabolic stability = f (presence/absence of specific fragments) v Protein crystallizability = f (amino acid composition, secondary structure) 19
Quantitative Structure Activity Relationship (QSAR) Quantitative Structure Property Relationship (QSPR) • Correlate specific biological activity for a set of compounds with their structure-derived molecular descriptors by means of a mathematical model • The nature of correlation, activity and descriptors are unlimited v BBB permeability = f (hydrophobicity, H-bonding potential) v Metabolic stability = f (presence/absence of specific fragments) v Protein crystallizability = f (amino acid composition, secondary structure) 19
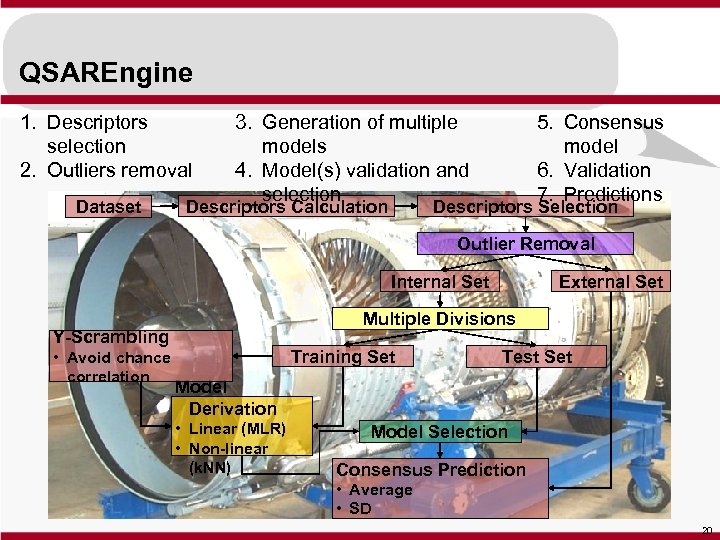 QSAREngine 1. Descriptors selection 2. Outliers removal Dataset 3. Generation of multiple models 4. Model(s) validation and selection Descriptors Calculation 5. Consensus model 6. Validation 7. Predictions Descriptors Selection Outlier Removal Internal Set Multiple Divisions Y-Scrambling • Avoid chance correlation External Set Training Set Test Set Model Derivation • Linear (MLR) • Non-linear (k. NN) Model Selection Consensus Prediction • Average • SD 20
QSAREngine 1. Descriptors selection 2. Outliers removal Dataset 3. Generation of multiple models 4. Model(s) validation and selection Descriptors Calculation 5. Consensus model 6. Validation 7. Predictions Descriptors Selection Outlier Removal Internal Set Multiple Divisions Y-Scrambling • Avoid chance correlation External Set Training Set Test Set Model Derivation • Linear (MLR) • Non-linear (k. NN) Model Selection Consensus Prediction • Average • SD 20
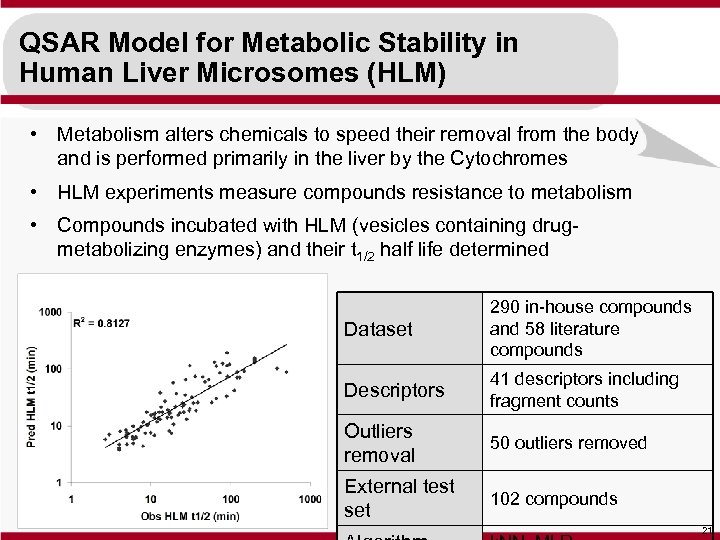 QSAR Model for Metabolic Stability in Human Liver Microsomes (HLM) • Metabolism alters chemicals to speed their removal from the body and is performed primarily in the liver by the Cytochromes • HLM experiments measure compounds resistance to metabolism • Compounds incubated with HLM (vesicles containing drugmetabolizing enzymes) and their t 1/2 half life determined Dataset 290 in-house compounds and 58 literature compounds Descriptors 41 descriptors including fragment counts Outliers removal 50 outliers removed External test set 102 compounds 21
QSAR Model for Metabolic Stability in Human Liver Microsomes (HLM) • Metabolism alters chemicals to speed their removal from the body and is performed primarily in the liver by the Cytochromes • HLM experiments measure compounds resistance to metabolism • Compounds incubated with HLM (vesicles containing drugmetabolizing enzymes) and their t 1/2 half life determined Dataset 290 in-house compounds and 58 literature compounds Descriptors 41 descriptors including fragment counts Outliers removal 50 outliers removed External test set 102 compounds 21
 The Grand Challenge • How can we reliably and consistently predict the pharmacological profile of bio-active compounds? v Basic scientific research v Practical applications in drug design • How can we make better drugs? ity CYP Ef fic B BB Binding Pe rm ea bil ac y h. ERG Solubility 22
The Grand Challenge • How can we reliably and consistently predict the pharmacological profile of bio-active compounds? v Basic scientific research v Practical applications in drug design • How can we make better drugs? ity CYP Ef fic B BB Binding Pe rm ea bil ac y h. ERG Solubility 22
 Acknowledgments • EPIX Pharmaceuticals • Lab members • Dr. Efrat Noy • Dr. Merav Fichman • Gal Fradin • Yocheved Beim • Funding • CFFT 23
Acknowledgments • EPIX Pharmaceuticals • Lab members • Dr. Efrat Noy • Dr. Merav Fichman • Gal Fradin • Yocheved Beim • Funding • CFFT 23


