2f539f5ecc5deca1ee1594d6ea6b7ca0.ppt
- Количество слайдов: 45

Computational Biology, Part 6 Sequence File Formats and Sequence Assembly Robert F. Murphy Copyright 1996 -2001. All rights reserved.
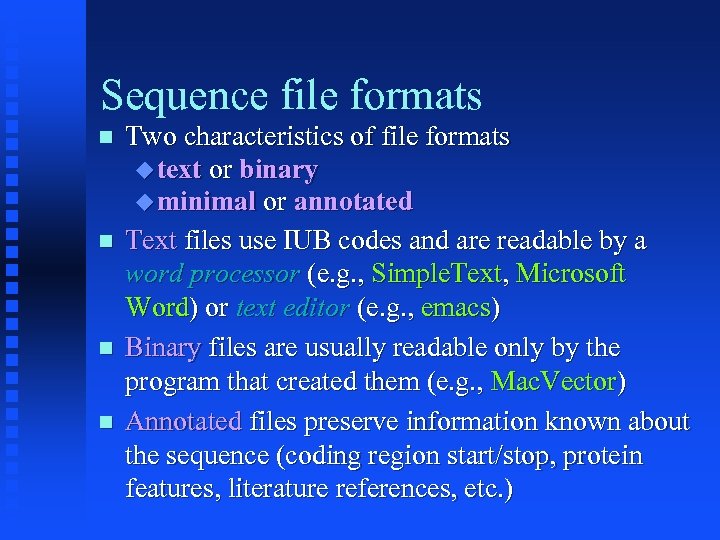
Sequence file formats Two characteristics of file formats text or binary minimal or annotated Text files use IUB codes and are readable by a word processor (e. g. , Simple. Text, Microsoft Word) or text editor (e. g. , emacs) Binary files are usually readable only by the program that created them (e. g. , Mac. Vector) Annotated files preserve information known about the sequence (coding region start/stop, protein features, literature references, etc. )
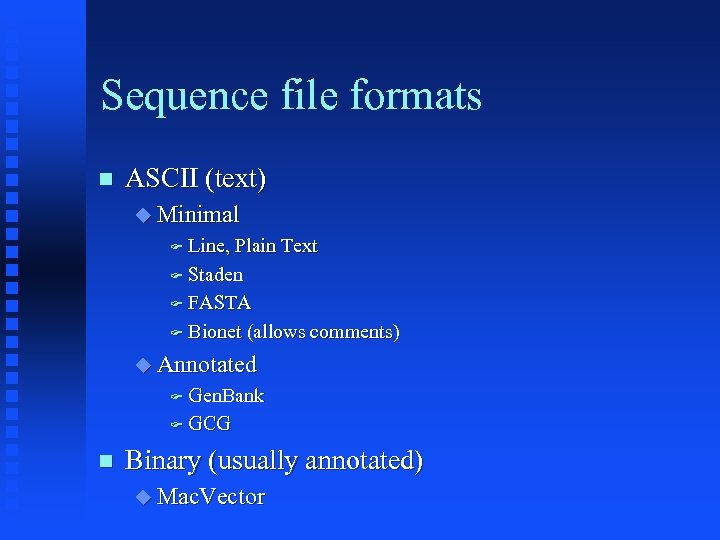
Sequence file formats ASCII (text) Minimal Line, Plain Text Staden FASTA Bionet (allows comments) Annotated Gen. Bank GCG Binary (usually annotated) Mac. Vector
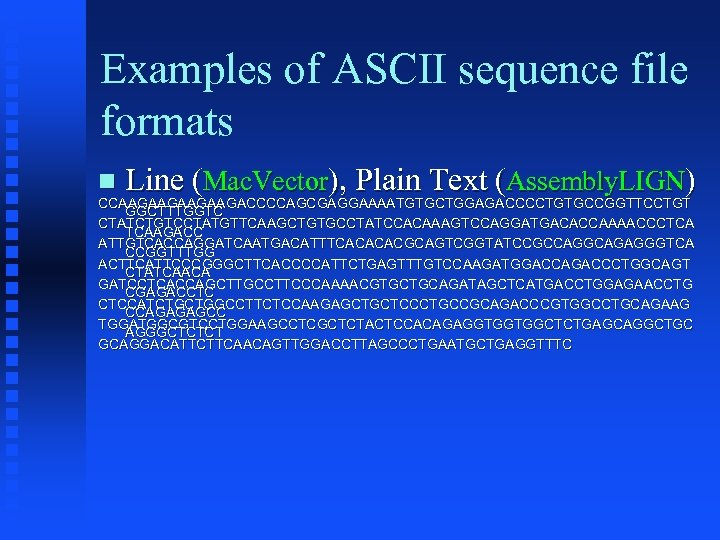
Examples of ASCII sequence file formats Line (Mac. Vector), Plain Text (Assembly. LIGN) CCAAGAAGACCCCAGCGAGGAAAATGTGCTGGAGACCCCTGTGCCGGTTCCTGT GGCTTTGGTC CTATCTGTCCTATGTTCAAGCTGTGCCTATCCACAAAGTCCAGGATGACACCAAAACCCTCA TCAAGACC ATTGTCACCAGGATCAATGACATTTCACACACGCAGTCGGTATCCGCCAGGCAGAGGGTCA CCGGTTTGG ACTTCATTCCCGGGCTTCACCCCATTCTGAGTTTGTCCAAGATGGACCAGACCCTGGCAGT CTATCAACA GATCCTCACCAGCTTGCCTTCCCAAAACGTGCTGCAGATAGCTCATGACCTGGAGAACCTG CGAGACCTC CTCCATCTGCTGGCCTTCTCCAAGAGCTGCTCCCTGCCGCAGACCCGTGGCCTGCAGAAG CCAGAGAGCC TGGATGGCGTCCTGGAAGCCTCGCTCTACTCCACAGAGGTGGTGGCTCTGAGCAGGCTGC AGGGCTCTCT GCAGGACATTCTTCAACAGTTGGACCTTAGCCCTGAATGCTGAGGTTTC

Examples of ASCII sequence file formats Fasta (Entrez) >gi|995614|dbj|D 49653|RATOBESE Rat m. RNA for obese. CCAAGAAGACCCCAGCGAGGAAAATGTGCTGGAGACCCCTGTGCCGGTTCCTGT GGCTTTGGTC CTATCTGTCCTATGTTCAAGCTGTGCCTATCCACAAAGTCCAGGATGACACCAAAACCCTCA TCAAGACC ATTGTCACCAGGATCAATGACATTTCACACACGCAGTCGGTATCCGCCAGGCAGAGGGTCA CCGGTTTGG ACTTCATTCCCGGGCTTCACCCCATTCTGAGTTTGTCCAAGATGGACCAGACCCTGGCAGT CTATCAACA GATCCTCACCAGCTTGCCTTCCCAAAACGTGCTGCAGATAGCTCATGACCTGGAGAACCTG CGAGACCTC CTCCATCTGCTGGCCTTCTCCAAGAGCTGCTCCCTGCCGCAGACCCGTGGCCTGCAGAAG CCAGAGAGCC TGGATGGCGTCCTGGAAGCCTCGCTCTACTCCACAGAGGTGGTGGCTCTGAGCAGGCTGC AGGGCTCTCT GCAGGACATTCTTCAACAGTTGGACCTTAGCCCTGAATGCTGAGGTTTC
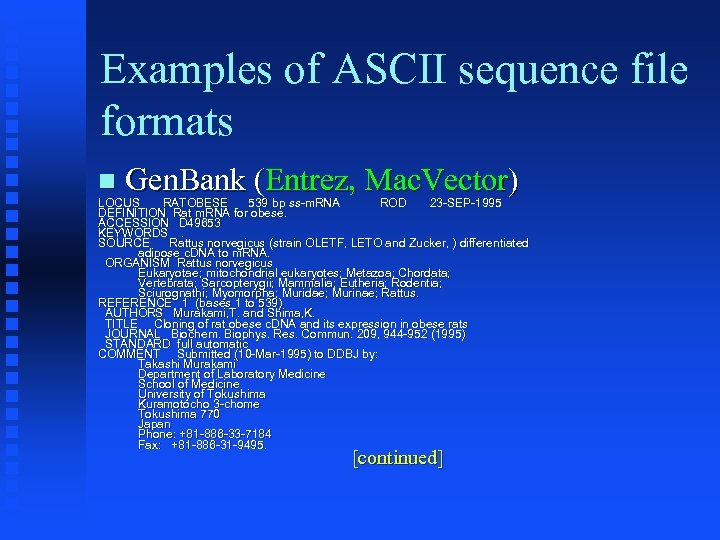
Examples of ASCII sequence file formats Gen. Bank (Entrez, Mac. Vector) LOCUS RATOBESE 539 bp ss-m. RNA ROD 23 -SEP-1995 ss. DEFINITION Rat m. RNA for obese. ACCESSION D 49653 KEYWORDS. SOURCE Rattus norvegicus (strain OLETF, LETO and Zucker, ) differentiated Zucker, adipose c. DNA to m. RNA. ORGANISM Rattus norvegicus Eukaryotae; mitochondrial eukaryotes; Metazoa; Chordata; Eukaryotae; eukaryotes; Metazoa; Chordata; Vertebrata; Sarcopterygii; Mammalia; Eutheria; Rodentia; Sciurognathi; Myomorpha; Muridae; Murinae; Rattus. REFERENCE 1 (bases 1 to 539) AUTHORS Murakami, T. and Shima, K. TITLE Cloning of rat obese c. DNA and its expression in obese rats JOURNAL Biochem. Biophys. Res. Commun. 209, 944 -952 (1995) Biochem. Biophys. Res. Commun. STANDARD full automatic COMMENT Submitted (10 -Mar-1995) to DDBJ by: Takashi Murakami Department of Laboratory Medicine School of Medicine University of Tokushima Kuramotocho 3 -chome Tokushima 770 Japan Phone: +81 -886 -33 -7184 Fax: +81 -886 -31 -9495. [continued]
![Examples of ASCII sequence file formats Gen. Bank [continued] NCBI gi: 995614 gi: FEATURES Examples of ASCII sequence file formats Gen. Bank [continued] NCBI gi: 995614 gi: FEATURES](https://present5.com/presentation/2f539f5ecc5deca1ee1594d6ea6b7ca0/image-7.jpg)
Examples of ASCII sequence file formats Gen. Bank [continued] NCBI gi: 995614 gi: FEATURES Location/Qualifiers source 1. . 539 /organism=" Rattus norvegicus" /strain="OLETF, LETO and Zucker" / dev_stage="differentiated" /sequenced_ mol="c. DNA to m. RNA" mol="c. DNA m. RNA" /tissue_type="adipose" CDS 30. . 533 /partial /note="NCBI gi: 995615" gi: / codon_start=1 /product="obese" /translation="MCWRPLCRFLWLWSYLSYVQAVPIHKVQDDTKTLIKTIVTRIND ISHTQSVSARQRVTGLDFIPGLHPILSLSKMDQTLAVYQQILTSLPSQNVLQIAHDLE NLRDLLHLLAFSKSCSLPQTRGLQKPESLDGVLEASLYSTEVVALSRLQGSLQDILQQ LDLSPEC" BASE COUNT 121 a 167 c 133 g 118 t ORIGIN 1 ccaagaagaa gaagacccca gcgaggaaaa tgtgctggag acccctgtgc cggttcctgt 61 ggctttggtc ctatctgtcc tatgttcaag ctgtgcctat ccacaaagtc caggatgaca 121 ccaaaaccct catcaagacc attgtcacca ggatcaatga catttcacac acgcagtcgg 181 tatccgccag gcagagggtc accggtttgg acttcattcc cgggcttcac cccattctga 241 gtttgtccaa gatggaccag accctggcag tctatcaaca gatcctcacc agcttgcctt 301 cccaaaacgt gctgcagata gctcatgacc tggagaacct gcgagacctc ctccatctgc 361 tggccttctc caagagctgc tccctgccgc agacccgtgg cctgcagaag ccagagagcc 421 tggatggcgt cctggaagcc tcgctctact ccacagaggt ggtggctctg agcaggctgc 481 agggctctct gcaggacatt cttcaacagt tggaccttag ccctgaatgc tgaggtttc //
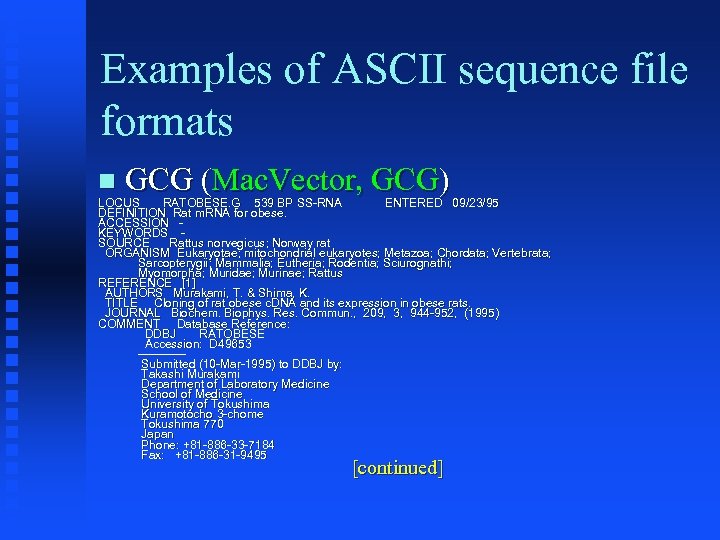
Examples of ASCII sequence file formats GCG (Mac. Vector, GCG) LOCUS RATOBESE. G 539 BP SS-RNA ENTERED 09/23/95 DEFINITION Rat m. RNA for obese. ACCESSION KEYWORDS SOURCE Rattus norvegicus; Norway rat norvegicus; ORGANISM Eukaryotae; mitochondrial eukaryotes; Metazoa; Chordata; Vertebrata; Eukaryotae; eukaryotes; Metazoa; Chordata; Sarcopterygii; Mammalia; Eutheria; Rodentia; Sciurognathi; Myomorpha; Muridae; Murinae; Rattus Myomorpha; Muridae; Murinae; REFERENCE [1] AUTHORS Murakami, T. & Shima, K. Shima, TITLE Cloning of rat obese c. DNA and its expression in obese rats. JOURNAL Biochem. Biophys. Res. Commun. , 209, 3, 944 -952, (1995) Biochem. Biophys. Res. Commun. , COMMENT Database Reference: DDBJ RATOBESE Accession: D 49653 ------Submitted (10 -Mar-1995) to DDBJ by: Takashi Murakami Department of Laboratory Medicine School of Medicine University of Tokushima Kuramotocho 3 -chome Tokushima 770 Japan Phone: +81 -886 -33 -7184 Fax: +81 -886 -31 -9495 [continued]
![Examples of ASCII sequence file formats GCG [continued] FEATURES From To/Span Description pept 30 Examples of ASCII sequence file formats GCG [continued] FEATURES From To/Span Description pept 30](https://present5.com/presentation/2f539f5ecc5deca1ee1594d6ea6b7ca0/image-9.jpg)
Examples of ASCII sequence file formats GCG [continued] FEATURES From To/Span Description pept 30 533 obese ? ? 1 539 source; /organism=Rattus norvegicus; /strain=OLETF, LETO and Zucker; / dev_stage=differentiated; /sequenced_mol=c. DNA dev_stage=differentiated; /sequenced_mol= to m. RNA; /tissue_type=adipose m. RNA; BASE COUNT 121 A 167 C 133 G 118 T 0 OTHER ORIGIN ? RATOBESE. G Length: 539 Jan 30, 1996 - 05: 32 PM Check: 5797. . 1 CCAAGAAGAA GAAGACCCCA GCGAGGAAAA TGTGCTGGAG ACCCCTGTGC CGGTTCCTGT 61 GGCTTTGGTC CTATCTGTCC TATGTTCAAG CTGTGCCTAT CCACAAAGTC CAGGATGACA 121 CCAAAACCCT CATCAAGACC ATTGTCACCA GGATCAATGA CATTTCACAC ACGCAGTCGG 181 TATCCGCCAG GCAGAGGGTC ACCGGTTTGG ACTTCATTCC CGGGCTTCAC CCCATTCTGA 241 GTTTGTCCAA GATGGACCAG ACCCTGGCAG TCTATCAACA GATCCTCACC AGCTTGCCTT 301 CCCAAAACGT GCTGCAGATA GCTCATGACC TGGAGAACCT GCGAGACCTC CTCCATCTGC 361 TGGCCTTCTC CAAGAGCTGC TCCCTGCCGC AGACCCGTGG CCTGCAGAAG CCAGAGAGCC 421 TGGATGGCGT CCTGGAAGCC TCGCTCTACT CCACAGAGGT GGTGGCTCTG AGCAGGCTGC 481 AGGGCTCTCT GCAGGACATT CTTCAACAGT TGGACCTTAG CCCTGAATGC TGAGGTTTC //
Sequence file format tips When saving a sequence for use in an email message or pasting into a web page, use an unannotated text format such as FASTA When retrieving from a database or exchanging between programs, use an annotated text format such as GCG or Genbank When using sequence again with the same program, use that program’s annotated binary format (or annotated text if binary not available)
Sequence assembly Goal: Assemble pieces of sequence into single, continuous sequence Early commercial system to do sequence assembly was the GCG Gel. Overlap/Gel. Assemble suite (VMS, Unix) We will use Assembly. LIGN (Macintosh), a companion to Mac. Vector
Sequence assembly: Terms project collection of fragments, templates and contigs fragments pieces of sequence entered by user or read from files contigs lists of aligned fragments generated (normally) by program
Sequence assembly: Terms templates any sequence to be searched for can be entered by user can be read from system files most common example is sequence of vector sequences in templates are NOT included in assembled sequences unless they are ALSO present in a fragment (and have not been removed)

Sequence assembly: File organization Assembly. LIGN keeps all information (including sequences) in a single project document GCG keeps all information in a directory (and its subdirectories), with each fragment in a separate file

Sequence assembly: Steps Data entry/import (fragments, templates) Removal of unwanted sequence Automated creation of contigs Manual editing/confirmation Export

Automated creation of contigs Steps 1. Finding pairwise overlaps 2. Resolving overlaps 3. Improving alignment 4. Final assembly and consensus generation

1. Finding pairwise overlaps Compare each fragment (and its complement) with each other fragment Generate list of regions of similarity that meet criteria below Parameters minimum overlap length (default 20) stringency (% of bases that must match, default 70) minimum repeat length (default 30)

1. Finding pairwise overlaps Each fragment may appear in more than one overlap 1 3 6 8 5 7 8 5 3 2 4 9

2. Resolving overlaps Build larger pieces by combining overlaps 1 6 5 8 3 4 3 5 8 2 7 9

2. Resolving overlaps Build larger pieces by combining overlaps 1 8 6 3 5 4 1 3 5 8 2 7 9 8 2

2. Resolving overlaps Build larger pieces by combining overlaps 3 6 5 3 5 4 1 7 9 8 2
2. Resolving overlaps Build larger pieces by combining overlaps 3 6 5 3 5 7 4 1 6 9 8 3 2 5
2. Resolving overlaps Build larger pieces by combining overlaps 5 7 4 1 6 9 8 3 2 5
2. Resolving overlaps Build larger pieces by combining overlaps 5 7 4 1 6 9 8 3 2 5 4

2. Resolving overlaps Build larger pieces by combining overlaps 7 1 6 9 8 3 2 5 4

3. Improve alignment Introduce gaps in sequences if they will improve overlaps alignment parameters gap creation penalty (default 2. 0) gap extension penalty (default (0. 1)

4. Final assembly and consensus generation Mark fragments that are now part of a contig (no longer appear by themselves) Form consensus for each contig by “reading” along aligned sequences and converting to IUB codes by consensus rules consensus parameter base designation threshold (% of all bases at a given position that must be the same for that base to be assigned to the consensus; otherwise, less specific IUB code used; default 80%)

Manual consensus editing Crucial to verify alignment and resolve ambiguities (e. g. , sequencing errors) Program keeps an “edit history” that tracks all changes made to the original sequences: essential to be able to “retrace your steps” from original sequencing gels (e. g. , in case of conflicts with sequences in database)
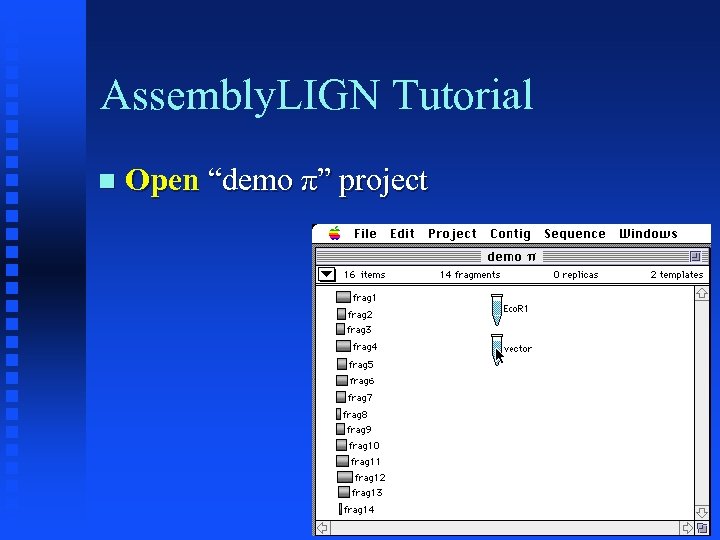
Assembly. LIGN Tutorial Open “demo π” project
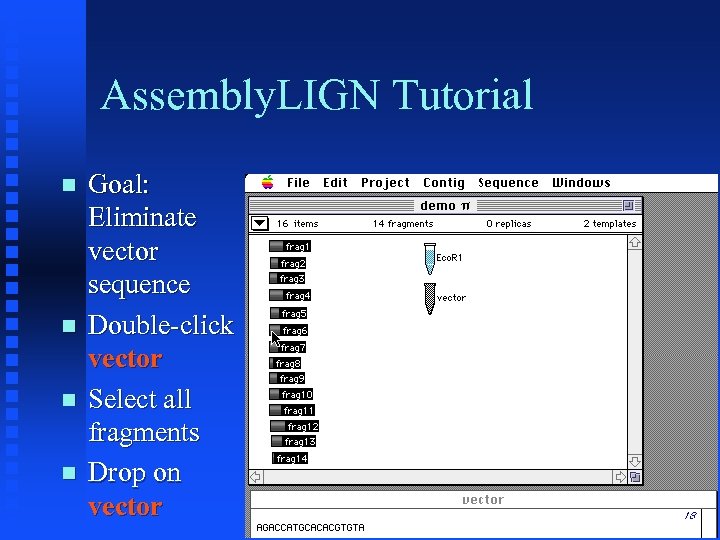
Assembly. LIGN Tutorial Goal: Eliminate vector sequence Double-click vector Select all fragments Drop on vector
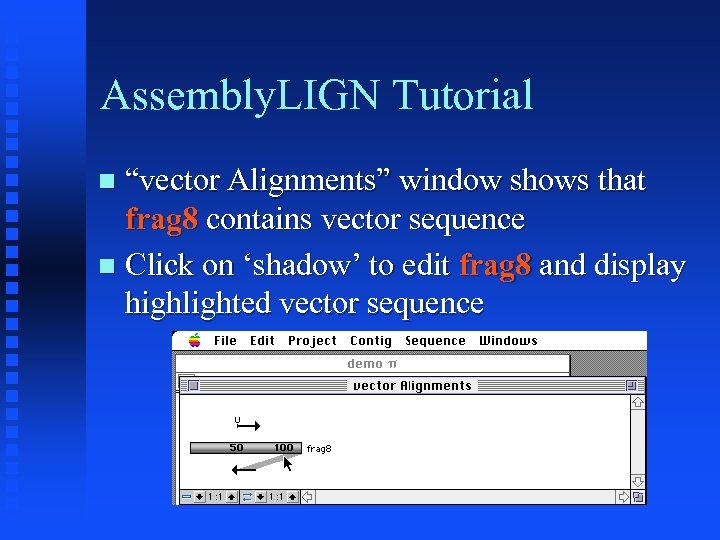
Assembly. LIGN Tutorial “vector Alignments” window shows that frag 8 contains vector sequence Click on ‘shadow’ to edit frag 8 and display highlighted vector sequence
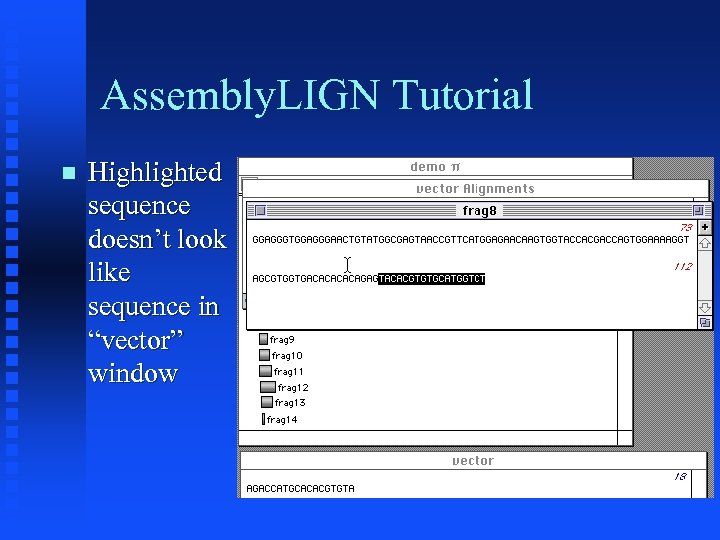
Assembly. LIGN Tutorial Highlighted sequence doesn’t look like sequence in “vector” window
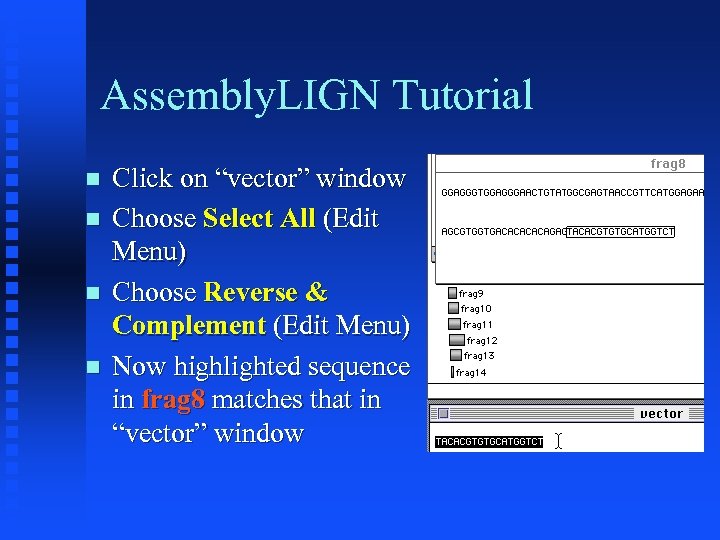
Assembly. LIGN Tutorial Click on “vector” window Choose Select All (Edit Menu) Choose Reverse & Complement (Edit Menu) Now highlighted sequence in frag 8 matches that in “vector” window
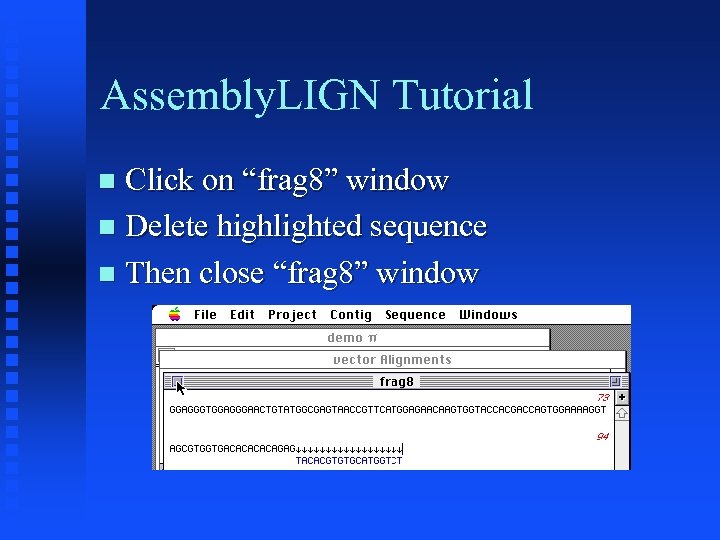
Assembly. LIGN Tutorial Click on “frag 8” window Delete highlighted sequence Then close “frag 8” window
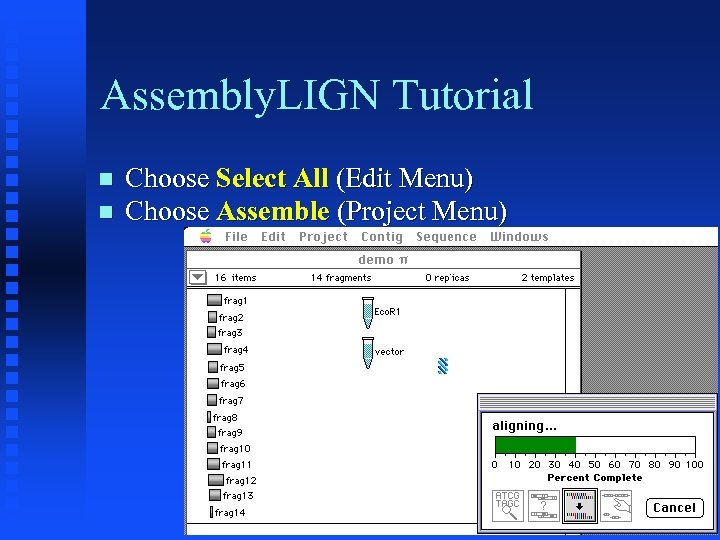
Assembly. LIGN Tutorial Choose Select All (Edit Menu) Choose Assemble (Project Menu)
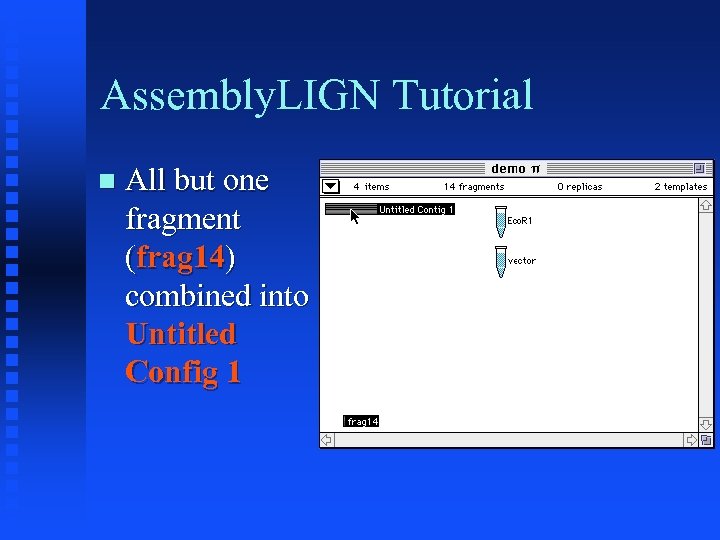
Assembly. LIGN Tutorial All but one fragment (frag 14) combined into Untitled Config 1
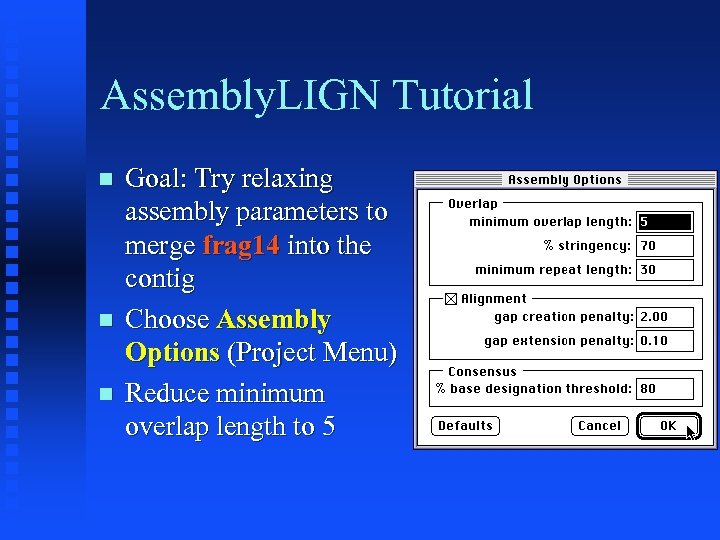
Assembly. LIGN Tutorial Goal: Try relaxing assembly parameters to merge frag 14 into the contig Choose Assembly Options (Project Menu) Reduce minimum overlap length to 5
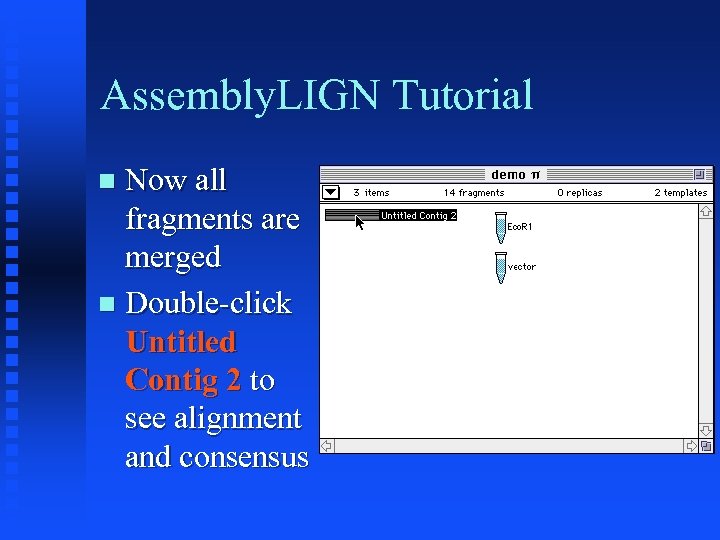
Assembly. LIGN Tutorial Now all fragments are merged Double-click Untitled Contig 2 to see alignment and consensus
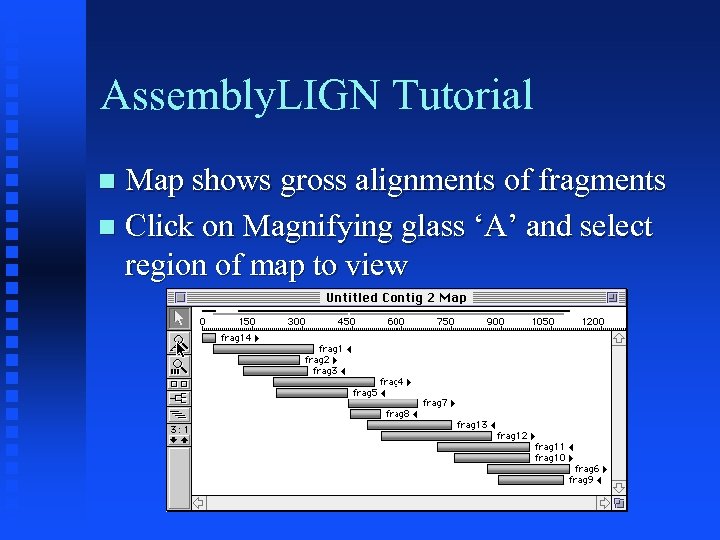
Assembly. LIGN Tutorial Map shows gross alignments of fragments Click on Magnifying glass ‘A’ and select region of map to view
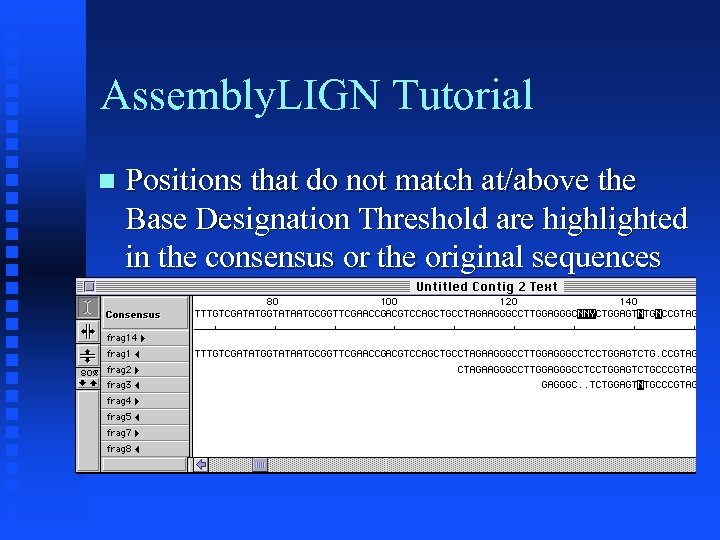
Assembly. LIGN Tutorial Positions that do not match at/above the Base Designation Threshold are highlighted in the consensus or the original sequences
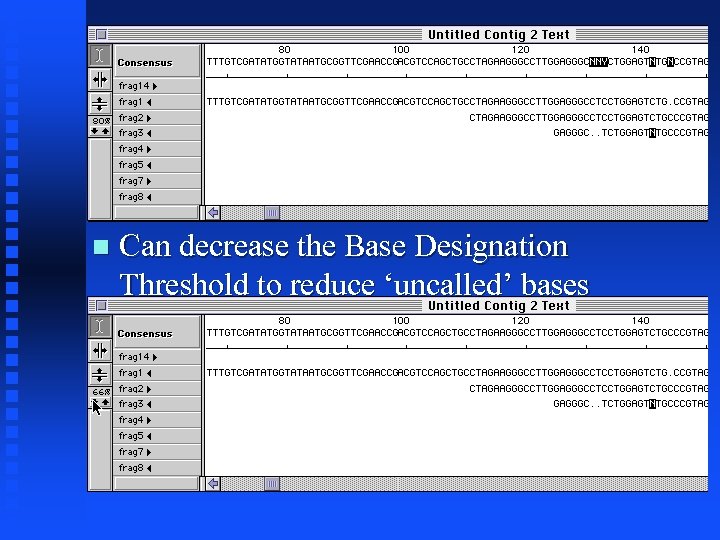
Can decrease the Base Designation Threshold to reduce ‘uncalled’ bases

Reading for Next Class Baxellanis & Ouellette, Chapter 7 Sequence Analysis Primer, pp. 90 -124 “Similarity versus Homology” and “Dot Matrix Methods” (on web page) (03 -510) Durbin et al, pp. 12 -17

Summary, Part 6 A variety of sequence file formats are currently in use. Files can be either text or binary, and can consist only of sequence or also include annotation information. The choice of file format is dictated by the requirements of the analysis desired and the subset of formats compatible between the “writing” and “reading” program.
Summary, Part 6 Sequence assembly requires the ability to compare sequences to find regions of high homology. Pieces of sequence are assembled by “connecting” them via regions of overlap. A consensus sequence can be generated from the connected pieces (using userspecified rules to resolve ambiguity).
Sequence comparisons using BLAST server Web Page Main BLAST web page URL http: //www. ncbi. nlm. nih. gov/BLAST/ Links to Basic and Advanced Search Pages Two main BLAST programs blastn - compares user nucleotide sequence to nucleotide sequences in database blastp - compares user peptide sequence to peptide sequences in database
2f539f5ecc5deca1ee1594d6ea6b7ca0.ppt