3b9afb372176959a8d6eb4c5c5de4382.ppt
- Количество слайдов: 59
Comparative Protein Structure Modeling 7/28/07 Andrej Sali (sali@salilab. org) 10/15/2006
Protein Structure Prediction with Emphasis on Comparative or Homology Modeling 1. 2. 3. 4. 5. 6. 7. 8. Introduction and motivation Types of comparative modeling methods Errors in comparative models Sequence-structure alignment for comparative modeling Modeling of loops in protein structures Prediction of errors in comparative models Structural genomics Applications A. Šali & T. Blundell. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779, 1993. D. Baker & A. Sali. Protein structure prediction and structural genomics. Science 294, 93, 2001. M. A. Marti-Renom et al. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29, 291 -325, 2000. N. Eswar, D. Eramian, B. Webb, M. -Y. Shen, A. Sali. Protein Structure Modeling with MODELLER, in press. Available at: http: //salilab. org/publications/ 10/15/2006

Introduction and motivation 10/15/2006
Sequence, Network versus Structure for understanding, controlling, modifying, designing GDCAGDFKIWYFGRTLLVAGAKDEFGAIDAW… RTLAWYAGHLVAGAKDEFGGDFKIWYFGAID… DFLLVAGAKDEFGKIWYFGGIDAWRTAGDCA… ARTHLVAGFGGGAIDWYFKIWYAKLAFGDED… GCUAGCUUAAGGCCUUCAUGAUCUUCUGAG… AGGGCUCCUUCAUGAUAGCUUAAGGCUUAA… 10/15/2006
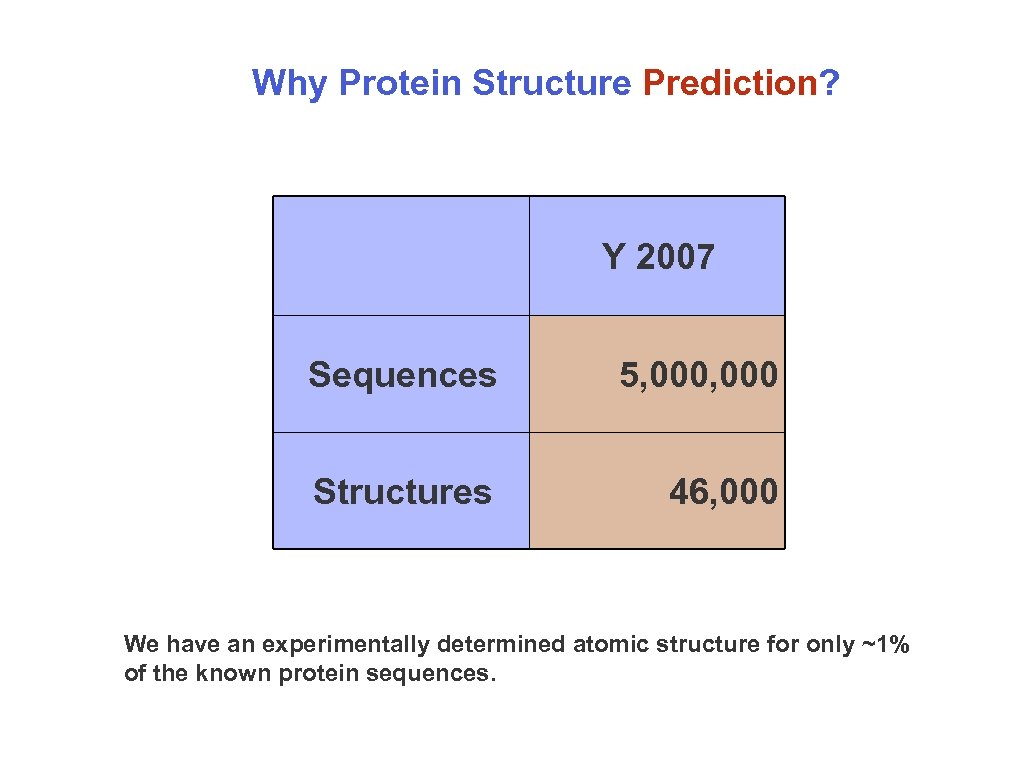
Why Protein Structure Prediction? Y 2007 Sequences 5, 000 Structures 46, 000 We have an experimentally determined atomic structure for only ~1% of the known protein sequences.
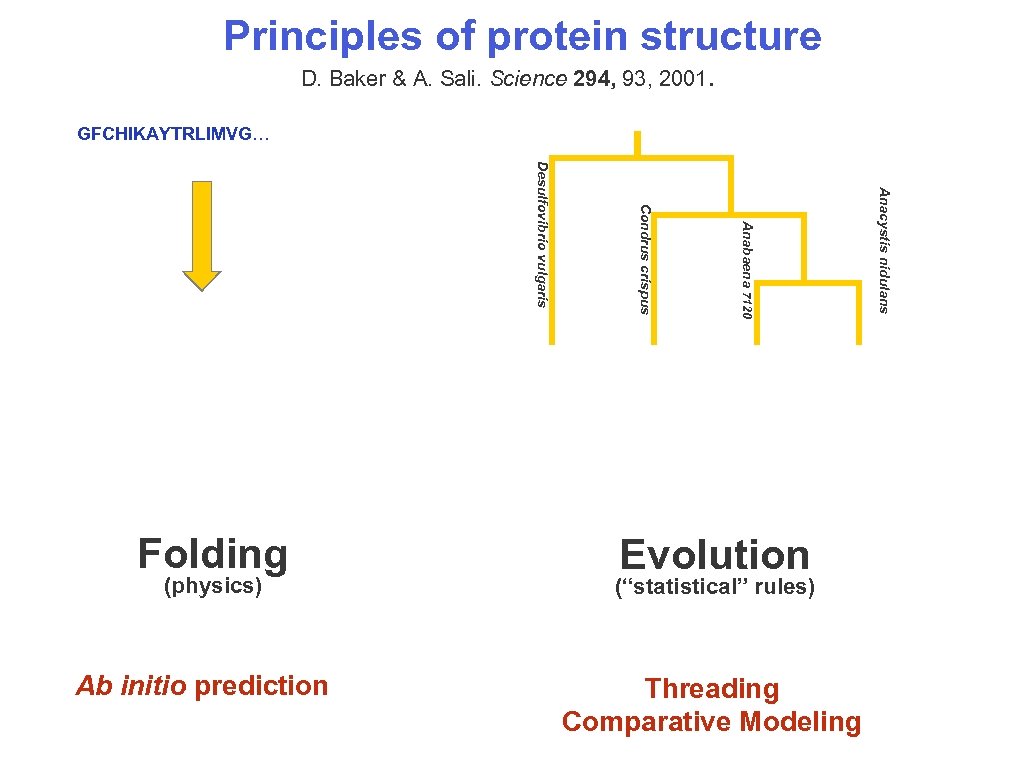
Principles of protein structure D. Baker & A. Sali. Science 294, 93, 2001. GFCHIKAYTRLIMVG… Anacystis nidulans Ab initio prediction Anabaena 7120 (physics) Condrus crispus Desulfovibrio vulgaris Folding Evolution (“statistical” rules) Threading Comparative Modeling 10/15/2006
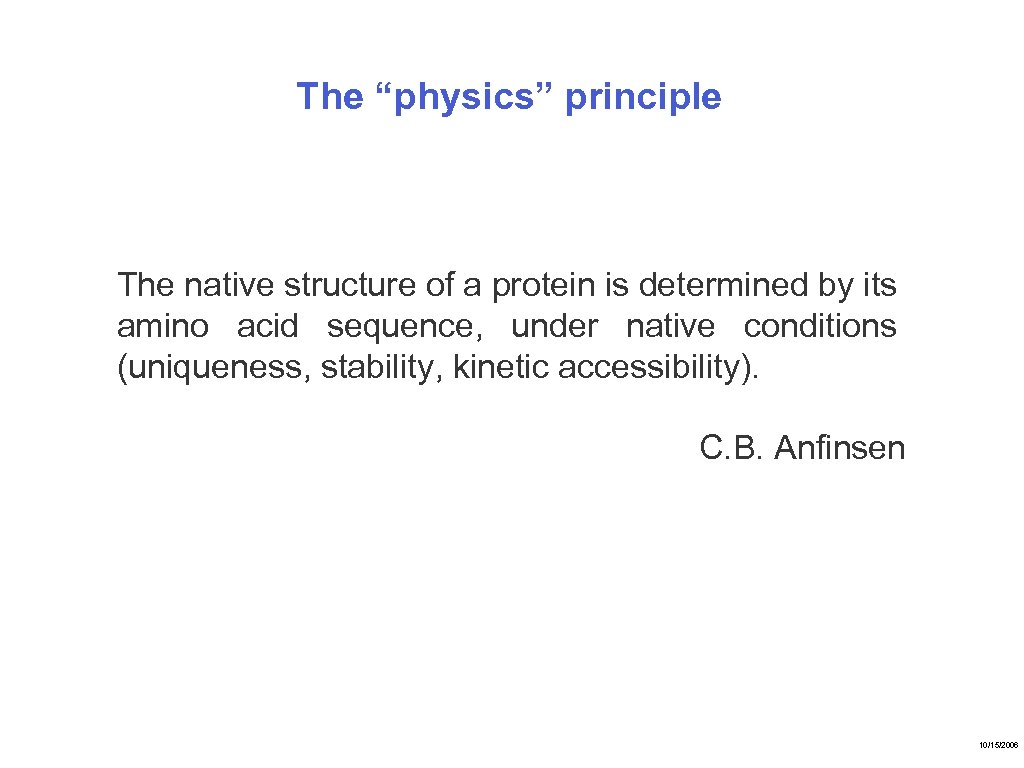
The “physics” principle The native structure of a protein is determined by its amino acid sequence, under native conditions (uniqueness, stability, kinetic accessibility). C. B. Anfinsen 10/15/2006
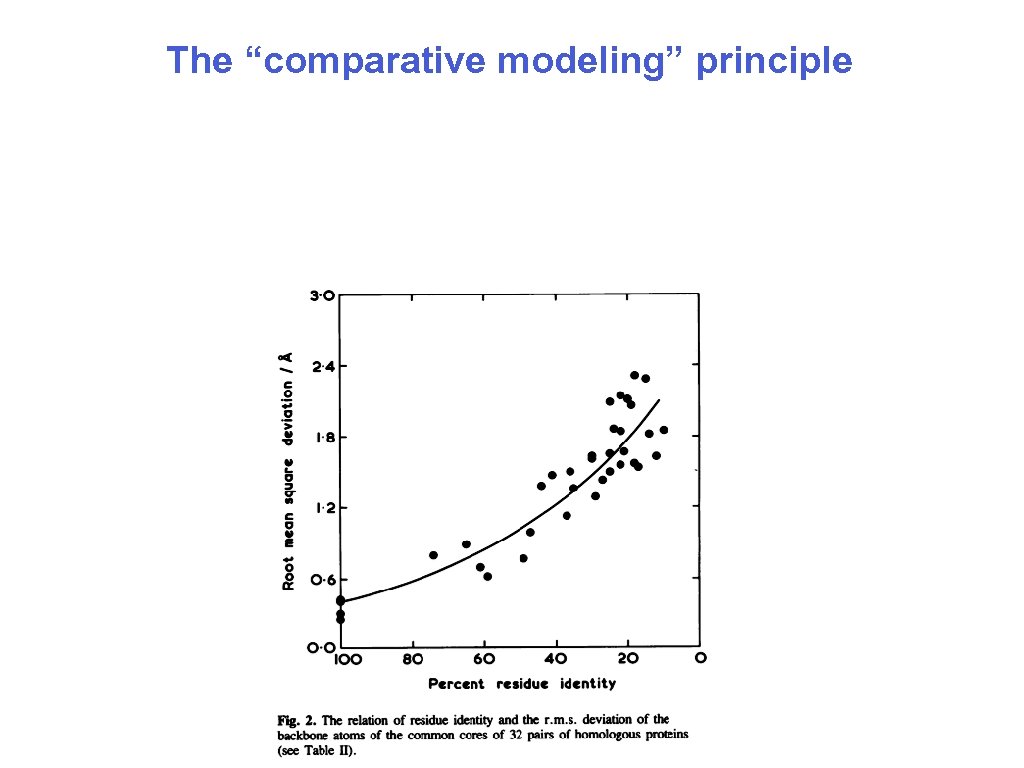
The “comparative modeling” principle 10/15/2006
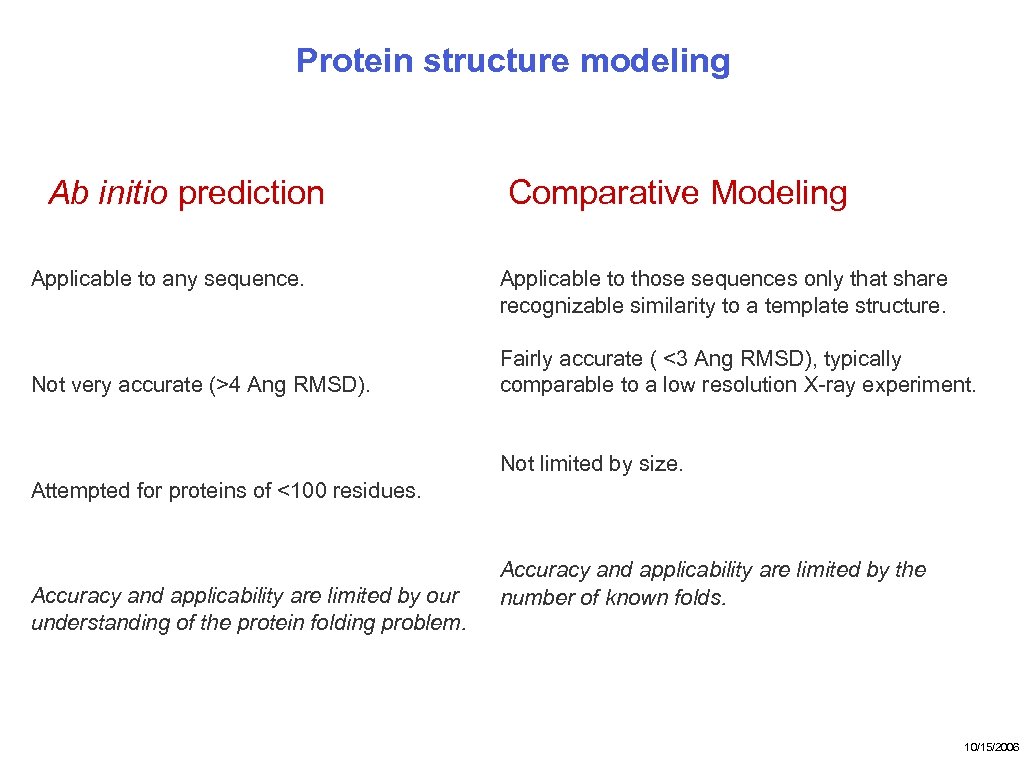
Protein structure modeling Ab initio prediction Applicable to any sequence. Not very accurate (>4 Ang RMSD). Comparative Modeling Applicable to those sequences only that share recognizable similarity to a template structure. Fairly accurate ( <3 Ang RMSD), typically comparable to a low resolution X-ray experiment. Not limited by size. Attempted for proteins of <100 residues. Accuracy and applicability are limited by our understanding of the protein folding problem. Accuracy and applicability are limited by the number of known folds. 10/15/2006
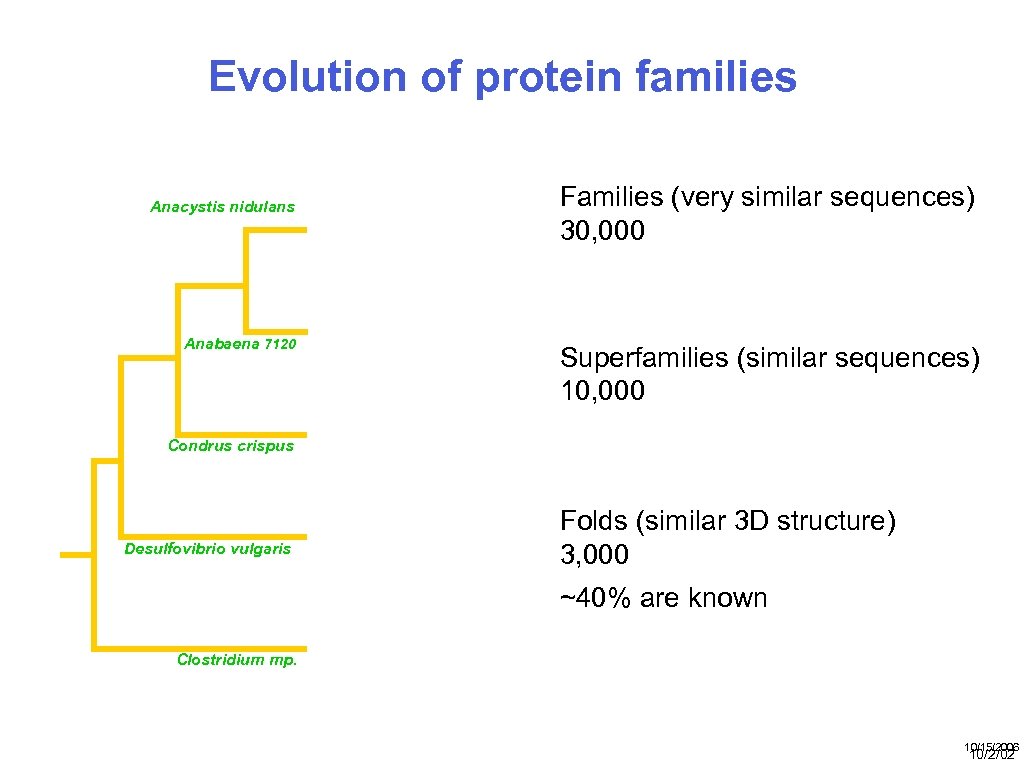
Evolution of protein families Cα RMSD Å (% EQV) 2 (50) 1 (80) 0 (100) Anacystis nidulans Anabaena 7120 Families (very similar sequences) 30, 000 Superfamilies (similar sequences) 10, 000 Condrus crispus Desulfovibrio vulgaris Folds (similar 3 D structure) 3, 000 ~40% are known Clostridium mp. 20 50 100 % SEQUENCE IDENTITY 10/15/2006 10/2/02
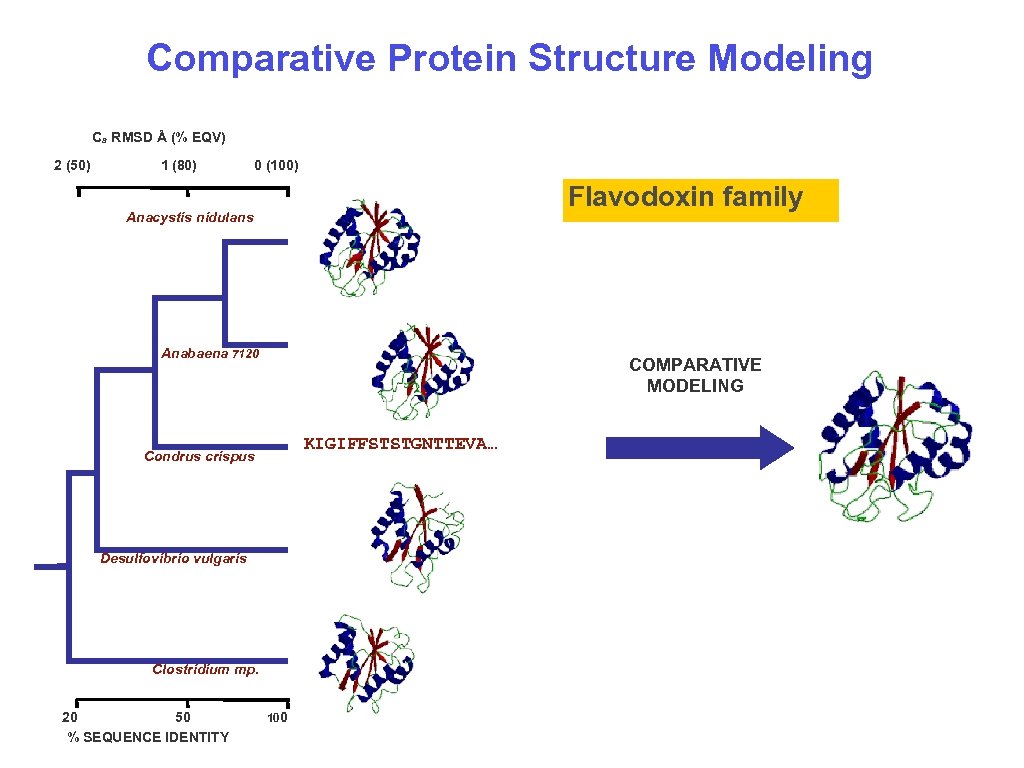
Comparative Protein Structure Modeling Ca RMSD Å (% EQV) 2 (50) 1 (80) 0 (100) Flavodoxin family Anacystis nidulans Anabaena 7120 COMPARATIVE MODELING KIGIFFSTSTGNTTEVA… Condrus crispus Desulfovibrio vulgaris Clostridium mp. 20 50 % SEQUENCE IDENTITY 100
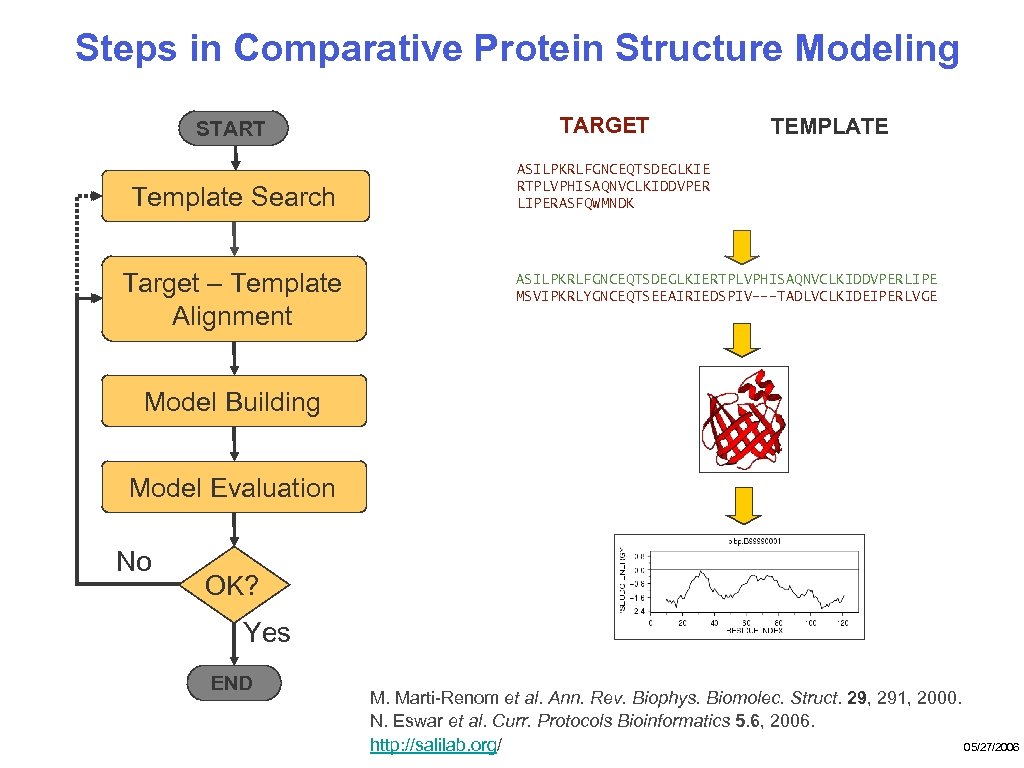
Steps in Comparative Protein Structure Modeling START Template Search Target – Template Alignment TARGET TEMPLATE ASILPKRLFGNCEQTSDEGLKIE RTPLVPHISAQNVCLKIDDVPER LIPERASFQWMNDK ASILPKRLFGNCEQTSDEGLKIERTPLVPHISAQNVCLKIDDVPERLIPE MSVIPKRLYGNCEQTSEEAIRIEDSPIV---TADLVCLKIDEIPERLVGE Model Building Model Evaluation No OK? Yes END M. Marti-Renom et al. Ann. Rev. Biophys. Biomolec. Struct. 29, 291, 2000. N. Eswar et al. Curr. Protocols Bioinformatics 5. 6, 2006. 05/27/2006 http: //salilab. org/
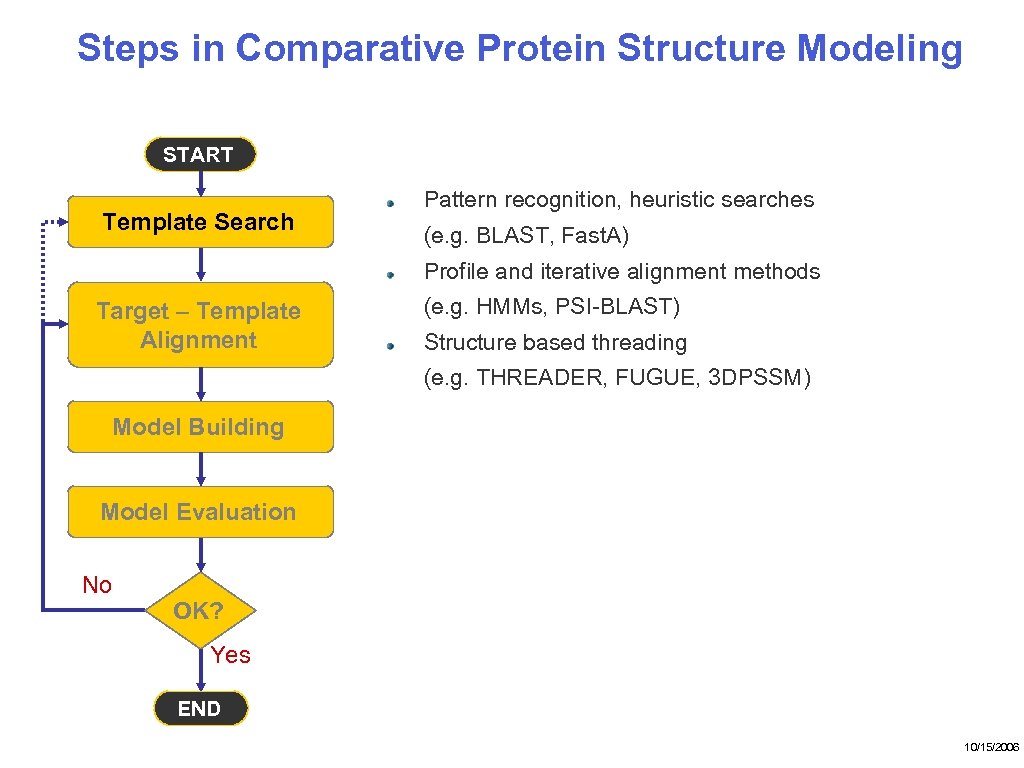
Steps in Comparative Protein Structure Modeling START Template Search Pattern recognition, heuristic searches (e. g. BLAST, Fast. A) Profile and iterative alignment methods Target – Template Alignment (e. g. HMMs, PSI-BLAST) Structure based threading (e. g. THREADER, FUGUE, 3 DPSSM) Model Building Model Evaluation No OK? Yes END 10/15/2006
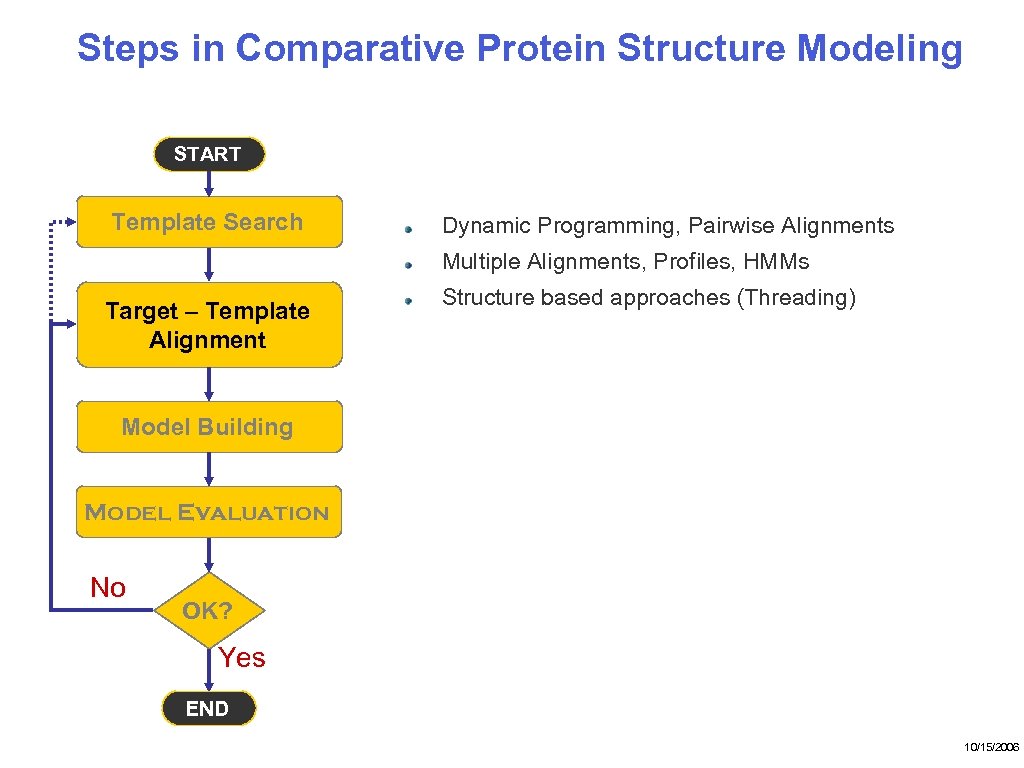
Steps in Comparative Protein Structure Modeling START Template Search Dynamic Programming, Pairwise Alignments Multiple Alignments, Profiles, HMMs Target – Template Alignment Structure based approaches (Threading) Model Building Model Evaluation No OK? Yes END 10/15/2006
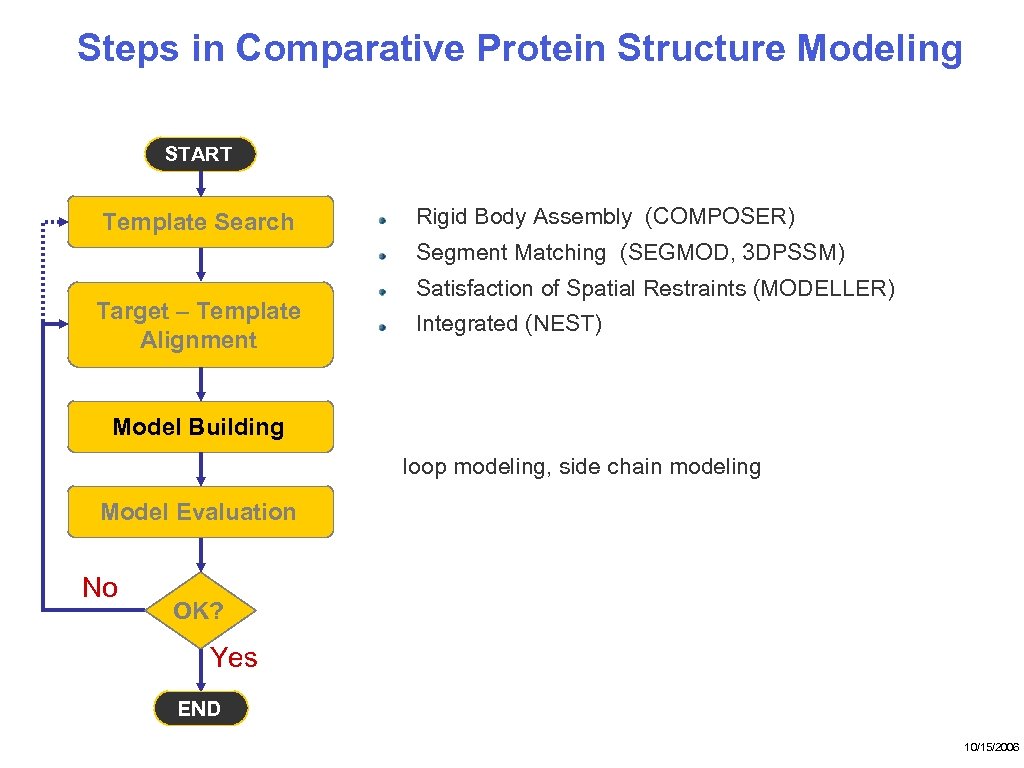
Steps in Comparative Protein Structure Modeling START Template Search Rigid Body Assembly (COMPOSER) Segment Matching (SEGMOD, 3 DPSSM) Target – Template Alignment Satisfaction of Spatial Restraints (MODELLER) Integrated (NEST) Model Building loop modeling, side chain modeling Model Evaluation No OK? Yes END 10/15/2006
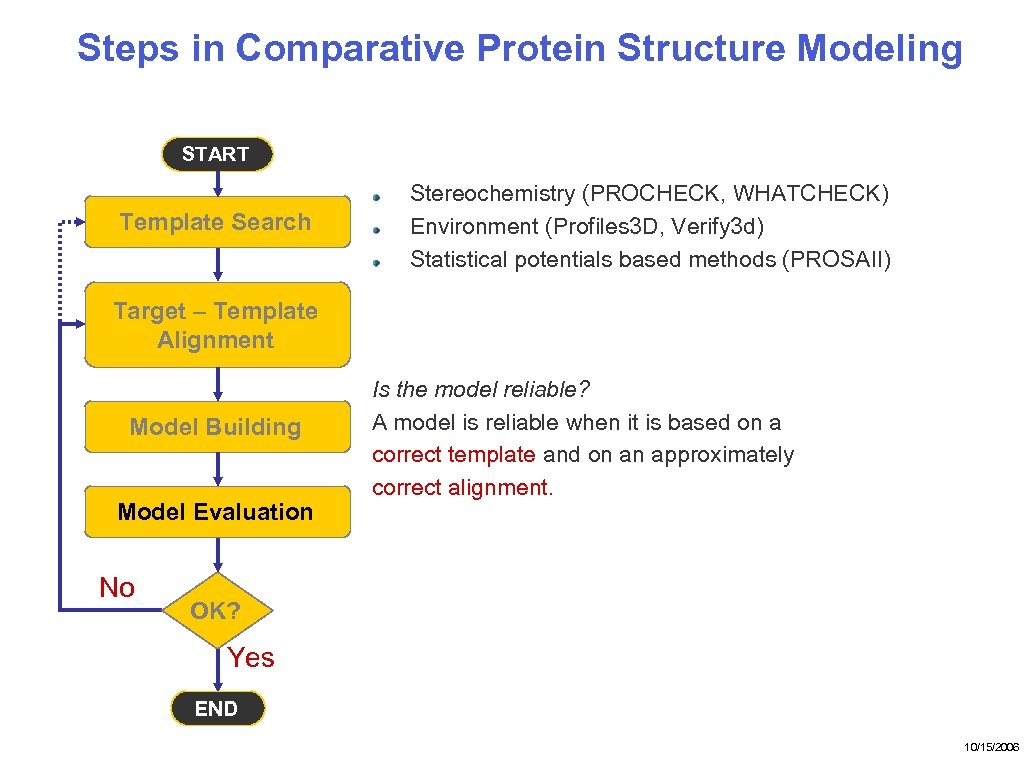
Steps in Comparative Protein Structure Modeling START Template Search Target – Template Alignment Model Building Model Evaluation No Stereochemistry (PROCHECK, WHATCHECK) Environment (Profiles 3 D, Verify 3 d) Statistical potentials based methods (PROSAII) Is the model reliable? A model is reliable when it is based on a correct template and on an approximately correct alignment. OK? Yes END 10/15/2006
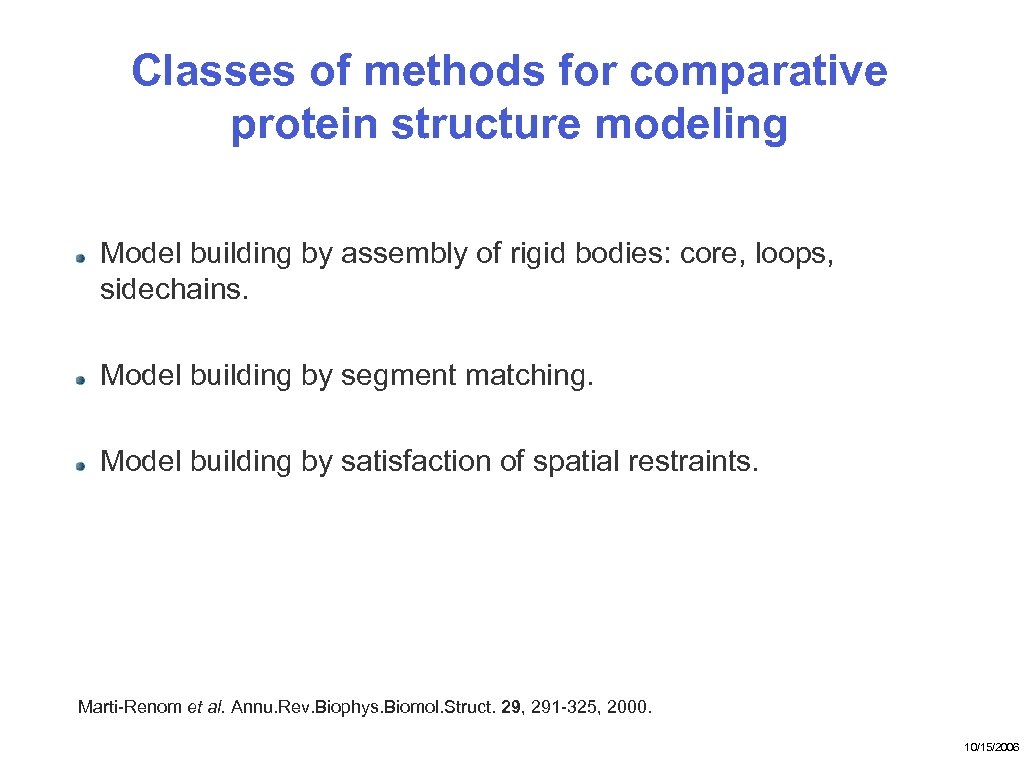
Classes of methods for comparative protein structure modeling Model building by assembly of rigid bodies: core, loops, sidechains. Model building by segment matching. Model building by satisfaction of spatial restraints. Marti-Renom et al. Annu. Rev. Biophys. Biomol. Struct. 29, 291 -325, 2000. 10/15/2006
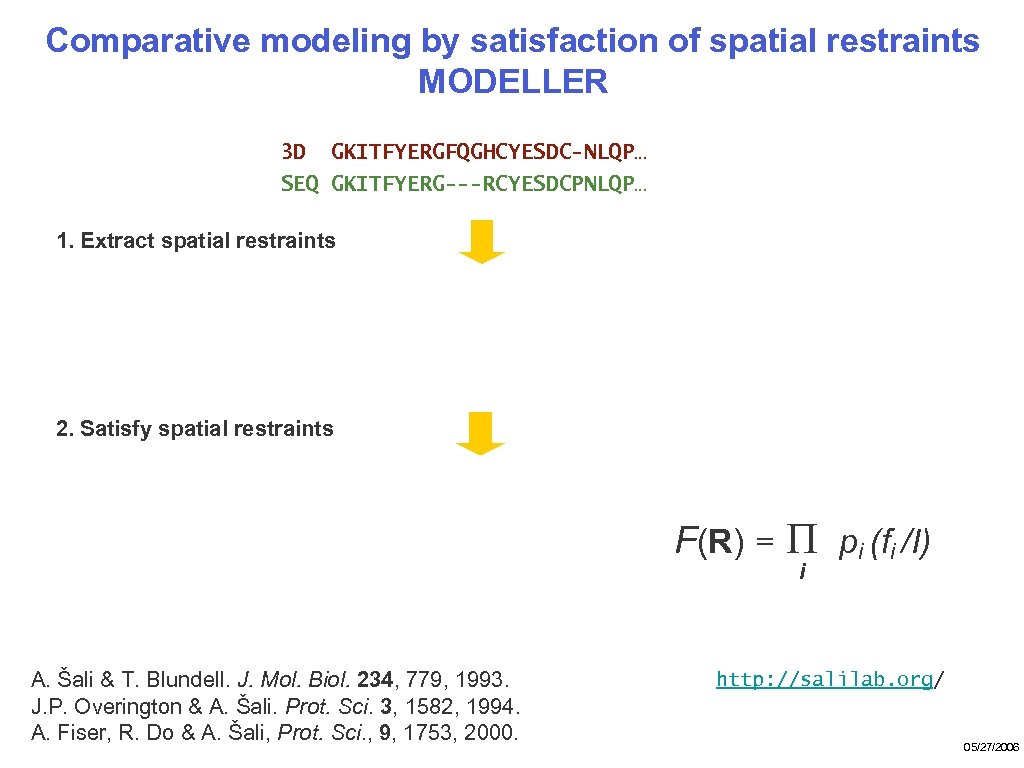
Comparative modeling by satisfaction of spatial restraints MODELLER 3 D GKITFYERGFQGHCYESDC-NLQP… SEQ GKITFYERG---RCYESDCPNLQP… 1. Extract spatial restraints 2. Satisfy spatial restraints F(R) = Π pi (fi /I) i A. Šali & T. Blundell. J. Mol. Biol. 234, 779, 1993. J. P. Overington & A. Šali. Prot. Sci. 3, 1582, 1994. A. Fiser, R. Do & A. Šali, Prot. Sci. , 9, 1753, 2000. http: //salilab. org/ 05/27/2006
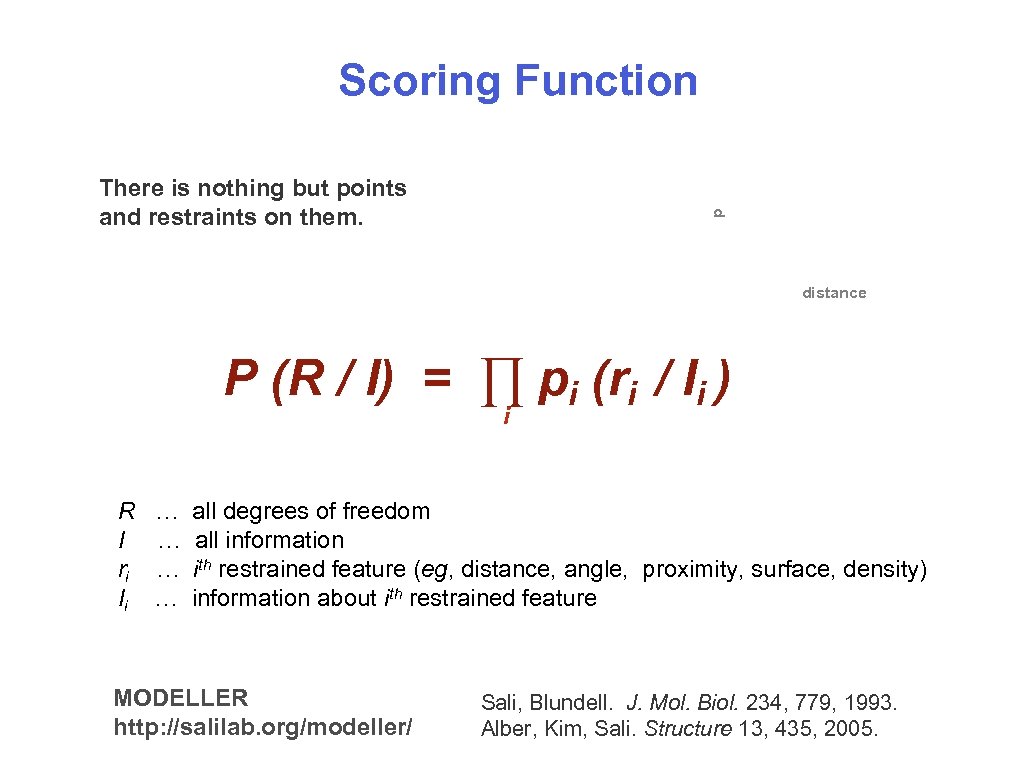
Scoring Function p There is nothing but points and restraints on them. distance P (R / I) = ∏ pi (ri / Ii ) i R … all degrees of freedom I … all information ri … ith restrained feature (eg, distance, angle, proximity, surface, density) Ii … information about ith restrained feature MODELLER http: //salilab. org/modeller/ Sali, Blundell. J. Mol. Biol. 234, 779, 1993. Alber, Kim, Sali. Structure 13, 435, 2005. 06/01/2006
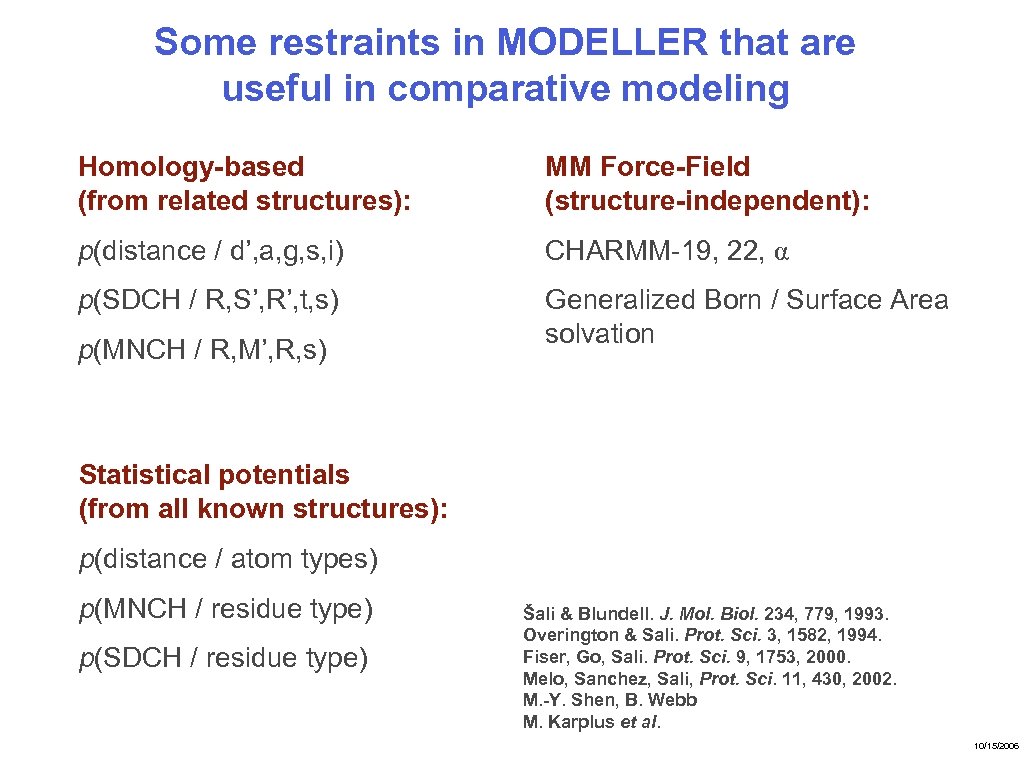
Some restraints in MODELLER that are useful in comparative modeling Homology-based (from related structures): MM Force-Field (structure-independent): p(distance / d’, a, g, s, i) CHARMM-19, 22, α p(SDCH / R, S’, R’, t, s) Generalized Born / Surface Area solvation p(MNCH / R, M’, R, s) Statistical potentials (from all known structures): p(distance / atom types) p(MNCH / residue type) p(SDCH / residue type) Šali & Blundell. J. Mol. Biol. 234, 779, 1993. Overington & Sali. Prot. Sci. 3, 1582, 1994. Fiser, Go, Sali. Prot. Sci. 9, 1753, 2000. Melo, Sanchez, Sali, Prot. Sci. 11, 430, 2002. M. -Y. Shen, B. Webb M. Karplus et al. 10/15/2006

Errors in comparative models 10/15/2006

Assessing errors is big industry Manual: Critical Assessment of Techniques for Protein Structure Prediction (CASP) (http: //predictioncenter. llnl. gov/) Automated: CAFASP EVA (http: //salilab. org/~eva/) Live. Bench (http: //bioinfo. pl/) 10/15/2006
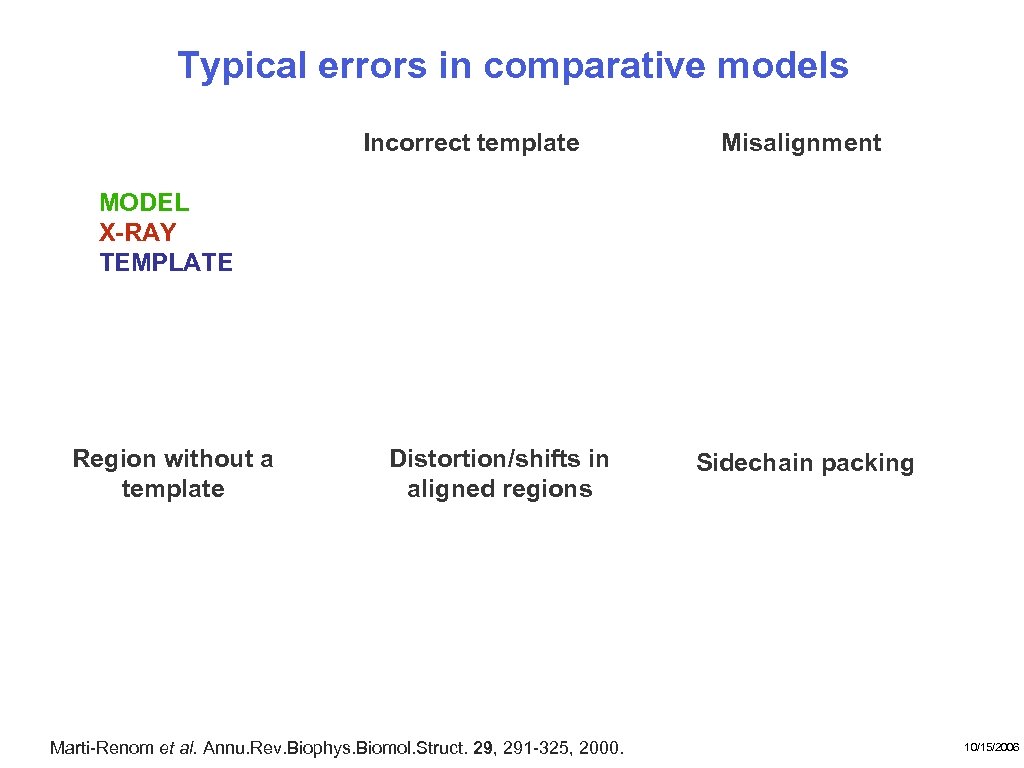
Typical errors in comparative models Incorrect template Misalignment MODEL X-RAY TEMPLATE Region without a template Distortion/shifts in aligned regions Marti-Renom et al. Annu. Rev. Biophys. Biomol. Struct. 29, 291 -325, 2000. Sidechain packing 10/15/2006
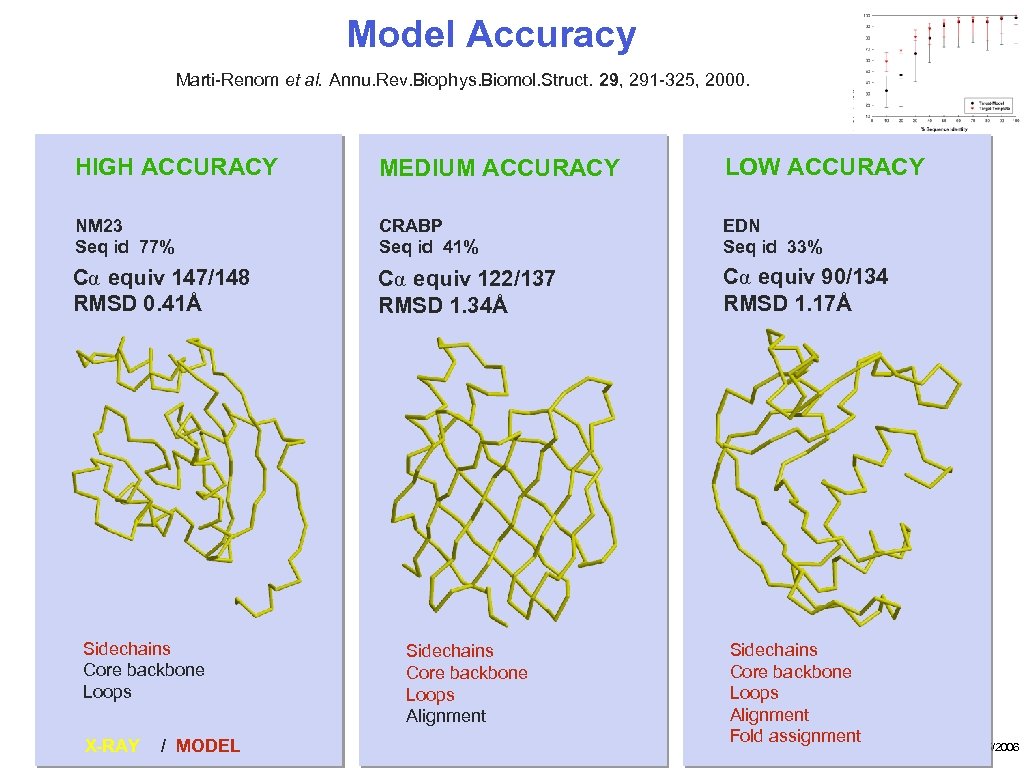
Model Accuracy Marti-Renom et al. Annu. Rev. Biophys. Biomol. Struct. 29, 291 -325, 2000. HIGH ACCURACY MEDIUM ACCURACY LOW ACCURACY NM 23 Seq id 77% CRABP Seq id 41% EDN Seq id 33% Cα equiv 147/148 RMSD 0. 41Å Cα equiv 122/137 RMSD 1. 34Å Cα equiv 90/134 RMSD 1. 17Å Sidechains Core backbone Loops X-RAY / MODEL Sidechains Core backbone Loops Alignment Fold assignment 10/15/2006
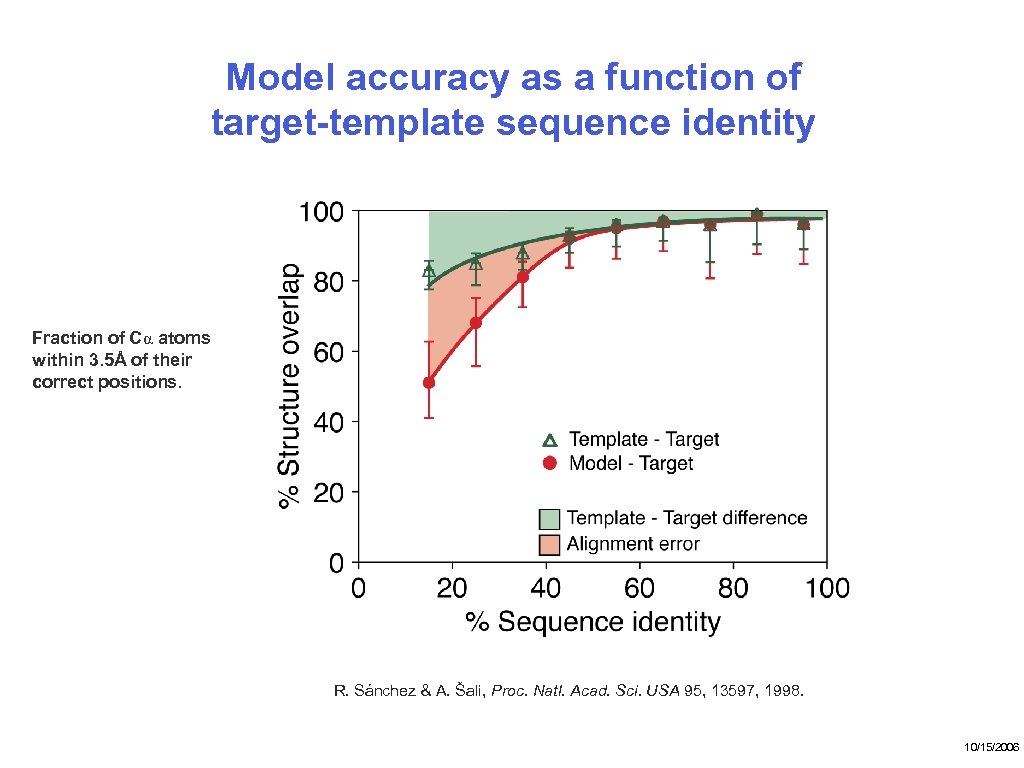
Model accuracy as a function of target-template sequence identity Fraction of Cα atoms within 3. 5Å of their correct positions. R. Sánchez & A. Šali, Proc. Natl. Acad. Sci. USA 95, 13597, 1998. 10/15/2006
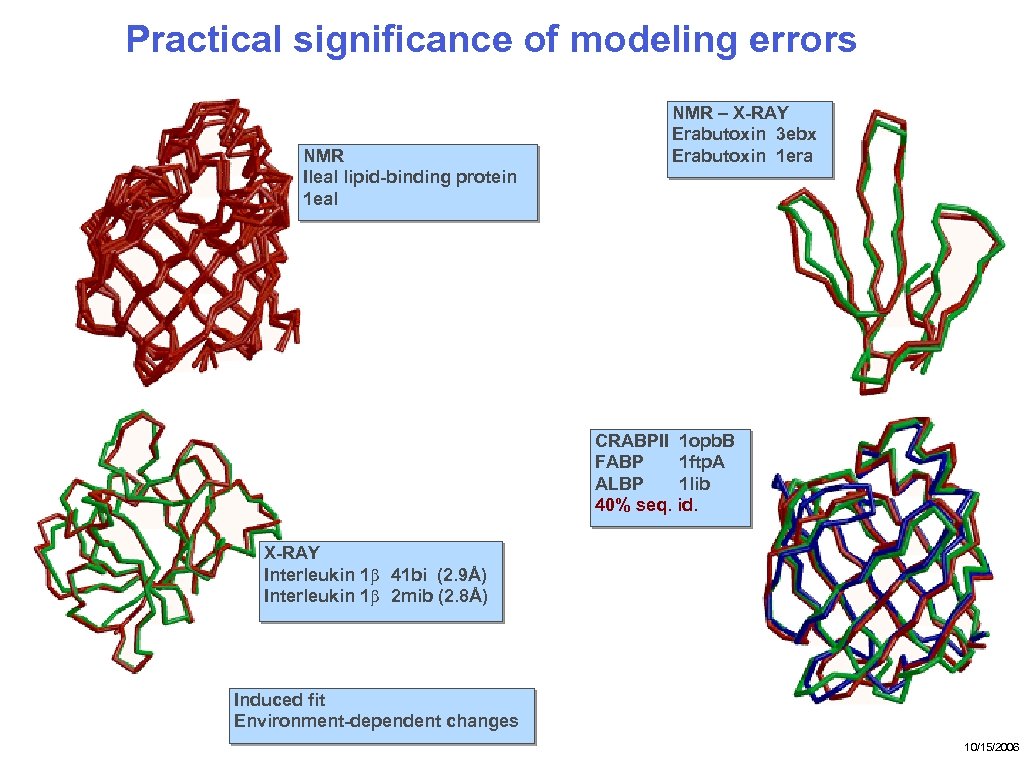
Practical significance of modeling errors NMR Ileal lipid-binding protein 1 eal NMR – X-RAY Erabutoxin 3 ebx Erabutoxin 1 era CRABPII 1 opb. B FABP 1 ftp. A ALBP 1 lib 40% seq. id. X-RAY Interleukin 1β 41 bi (2. 9Å) Interleukin 1β 2 mib (2. 8Å) Induced fit Environment-dependent changes 10/15/2006

Some Models Can Be Surprisingly Accurate (in Some Regions) 24% sequence identity 25% sequence identity YGL 203 C YJL 001 W 1 ac 5 1 ryp. H His 488 Ser 176 Asp 383
Sequence-structure alignment 10/15/2006
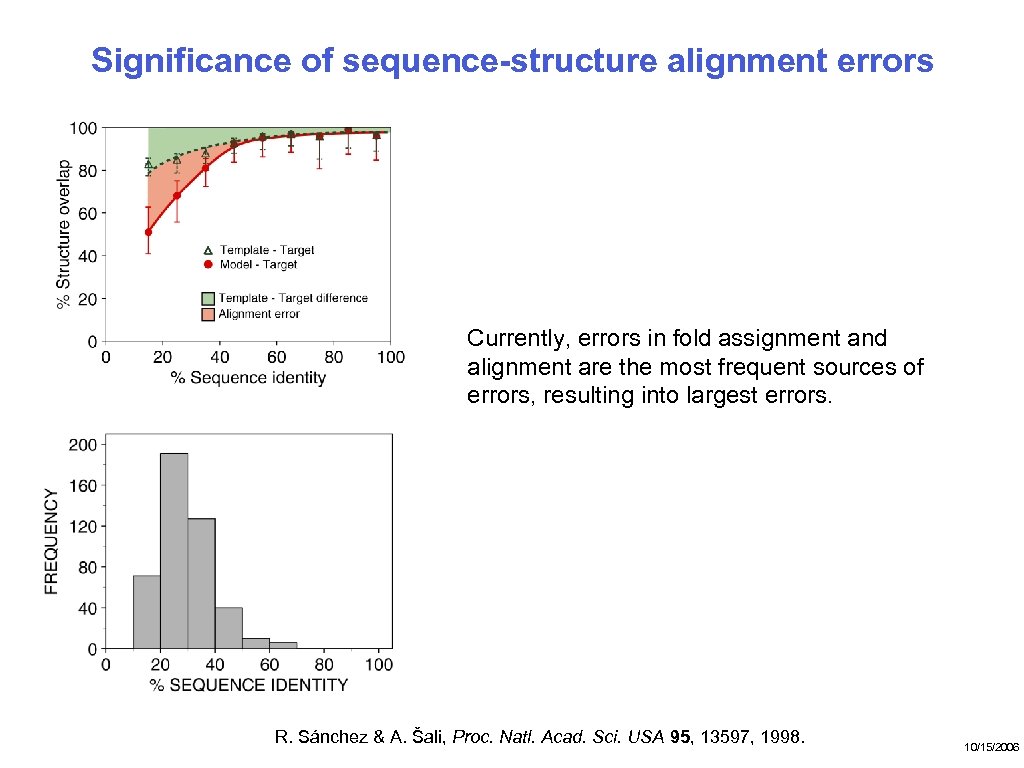
Significance of sequence-structure alignment errors Currently, errors in fold assignment and alignment are the most frequent sources of errors, resulting into largest errors. R. Sánchez & A. Šali, Proc. Natl. Acad. Sci. USA 95, 13597, 1998. 10/15/2006

Minimizing errors in sequence-structure alignment • Complex gap penalty functions. • Multiple sequence profiles. • Hidden Markov Models. • Threading. 10/15/2006
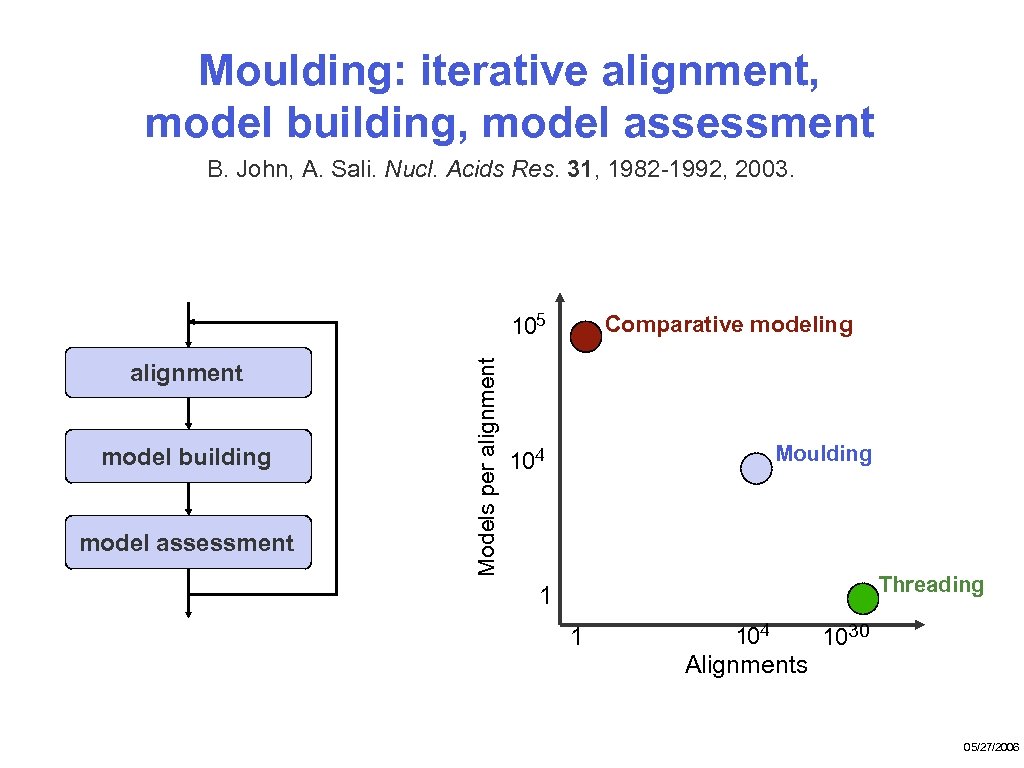
Moulding: iterative alignment, model building, model assessment B. John, A. Sali. Nucl. Acids Res. 31, 1982 -1992, 2003. alignment model building model assessment Models per alignment 105 Comparative modeling Moulding 104 Threading 1 1 104 1030 Alignments 05/27/2006
Modeling of loops in protein structures (modeling of insertions) 10/15/2006
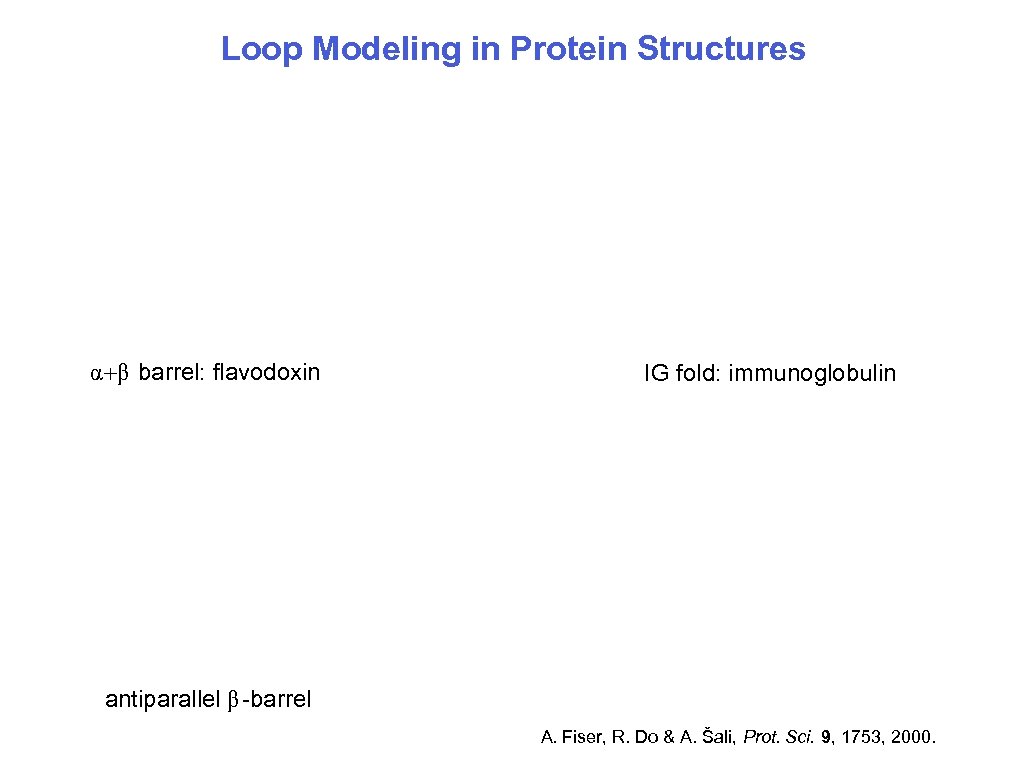
Loop Modeling in Protein Structures α+β barrel: flavodoxin IG fold: immunoglobulin antiparallel β-barrel A. Fiser, R. Do & A. Šali, Prot. Sci. 9, 1753, 2000. 10/15/2006
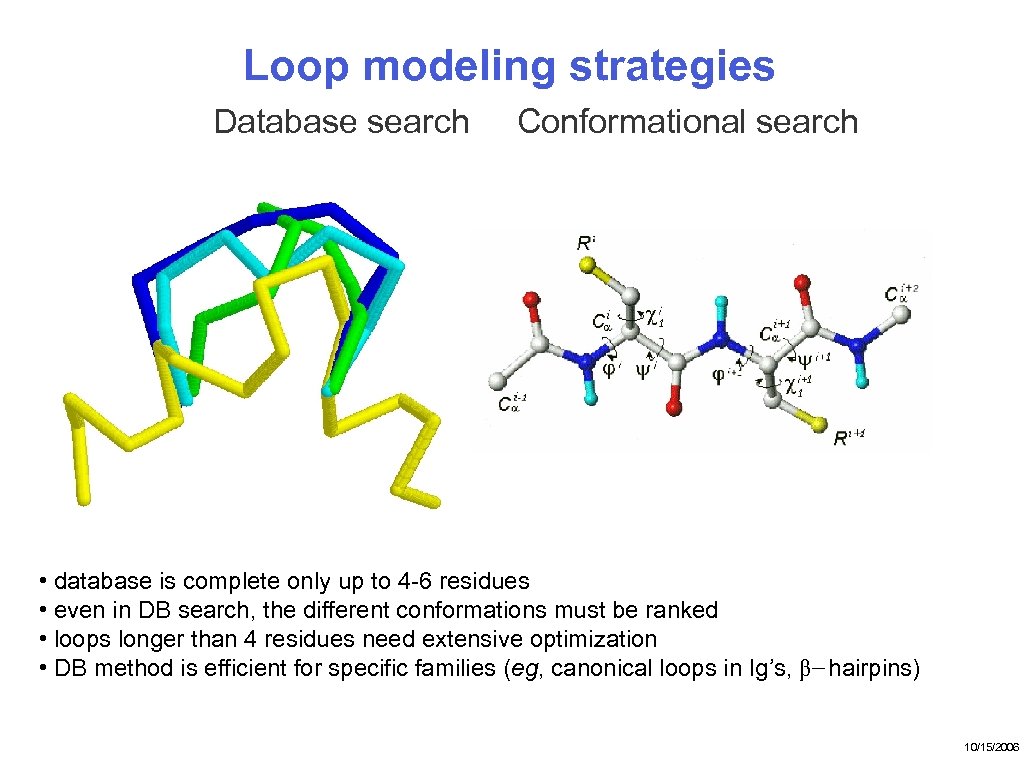
Loop modeling strategies Database search Conformational search • database is complete only up to 4 -6 residues • even in DB search, the different conformations must be ranked • loops longer than 4 residues need extensive optimization • DB method is efficient for specific families (eg, canonical loops in Ig’s, β− hairpins) 10/15/2006
Scoring Function for Loop Modeling The energy function is a sum of many terms: • Stereochemistry (CHARMM). 1. Mainchain conformation (stat. pot. for Φ, Ψ). 2. Non-bonded contacts (stat. pot. for d). 10/15/2006

Mainchain Terms for Loop Modeling 10/15/2006
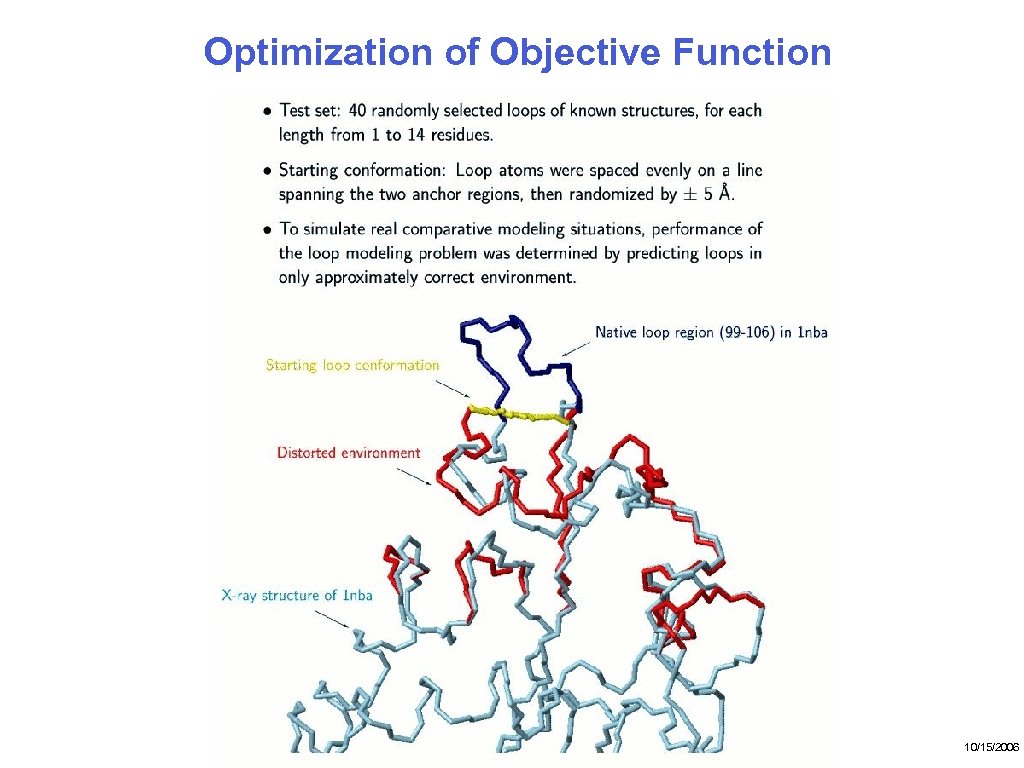
Optimization of Objective Function 10/15/2006
Calculating an Ensemble of Loop Models 10/15/2006
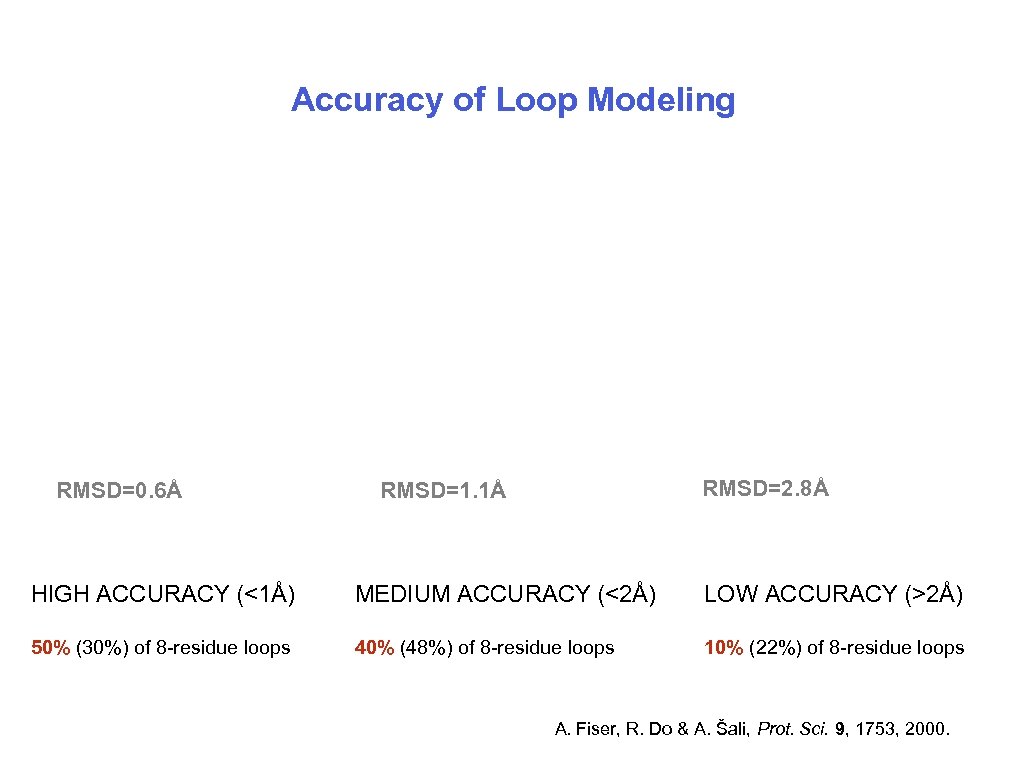
Accuracy of Loop Modeling RMSD=0. 6Å RMSD=2. 8Å RMSD=1. 1Å HIGH ACCURACY (<1Å) MEDIUM ACCURACY (<2Å) LOW ACCURACY (>2Å) 50% (30%) of 8 -residue loops 40% (48%) of 8 -residue loops 10% (22%) of 8 -residue loops A. Fiser, R. Do & A. Šali, Prot. Sci. 9, 1753, 2000. 10/15/2006
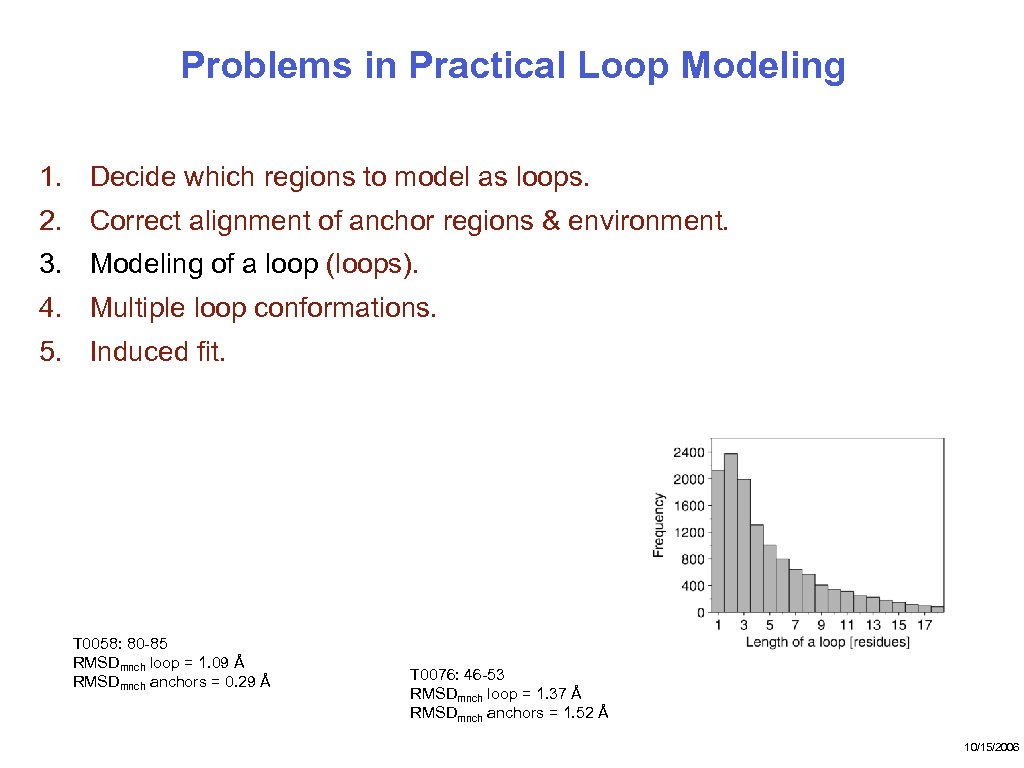
Problems in Practical Loop Modeling 1. Decide which regions to model as loops. 2. Correct alignment of anchor regions & environment. 3. Modeling of a loop (loops). 4. Multiple loop conformations. 5. Induced fit. T 0058: 80 -85 RMSDmnch loop = 1. 09 Å RMSDmnch anchors = 0. 29 Å T 0076: 46 -53 RMSDmnch loop = 1. 37 Å RMSDmnch anchors = 1. 52 Å 10/15/2006

Prediction of errors in comparative models 10/15/2006
Model Evaluation Methods Is the fold correct? How correct is the overall structure? What regions are modeled incorrectly? What is the best model in the set of alternative models? Does the model satisfy the restraints used to calculate it? What regions of the fold are variable? Stereochemistry test (PROCHECK) Residue environment test (Profiles 3 D) Statistical potential tests (PROSAII) Other statistical tests, including tests with multiple criteria (GA 341). Molecular mechanics force field tests. 10/15/2006
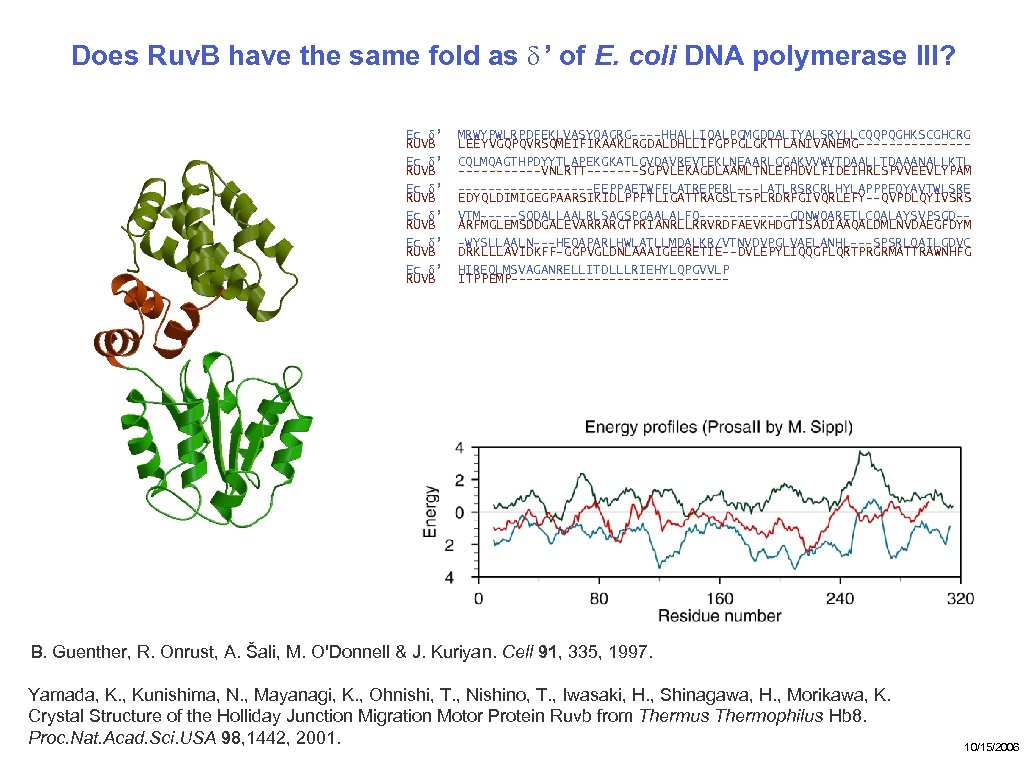
Does Ruv. B have the same fold as δ ’ of E. coli DNA polymerase III? Ec δ’ RUVB Ec δ’ RUVB MRWYPWLRPDFEKLVASYQAGRG----HHALLIQALPGMGDDALIYALSRYLLCQQPQGHKSCGHCRG LEEYVGQPQVRSQMEIFIKAAKLRGDALDHLLIFGPPGLGKTTLANIVANEMG-------CQLMQAGTHPDYYTLAPEKGKATLGVDAVREVTEKLNEAARLGGAKVVWVTDAALLTDAAANALLKTL ------VNLRTT-------SGPVLEKAGDLAAMLTNLEPHDVLFIDEIHRLSPVVEEVLYPAM ---------EEPPAETWFFLATREPERL---LATLRSRCRLHYLAPPPEQYAVTWLSRE EDYQLDIMIGEGPAARSIKIDLPPFTLIGATTRAGSLTSPLRDRFGIVQRLEFY--QVPDLQYIVSRS VTM-----SQDALLAALRLSAGSPGAALALFQ------GDNWQARETLCQALAYSVPSGD-ARFMGLEMSDDGALEVARRARGTPRIANRLLRRVRDFAEVKHDGTISADIAAQALDMLNVDAEGFDYM -WYSLLAALN---HEQAPARLHWLATLLMDALKR/VTNVDVPGLVAELANHL---SPSRLQAILGDVC DRKLLLAVIDKFF-GGPVGLDNLAAAIGEERETIE--DVLEPYLIQQGFLQRTPRGRMATTRAWNHFG HIREQLMSVAGANRELLITDLLLRIEHYLQPGVVLP ITPPEMP--------------- B. Guenther, R. Onrust, A. Šali, M. O'Donnell & J. Kuriyan. Cell 91, 335, 1997. Yamada, K. , Kunishima, N. , Mayanagi, K. , Ohnishi, T. , Nishino, T. , Iwasaki, H. , Shinagawa, H. , Morikawa, K. Crystal Structure of the Holliday Junction Migration Motor Protein Ruvb from Thermus Thermophilus Hb 8. Proc. Nat. Acad. Sci. USA 98, 1442, 2001. 10/15/2006
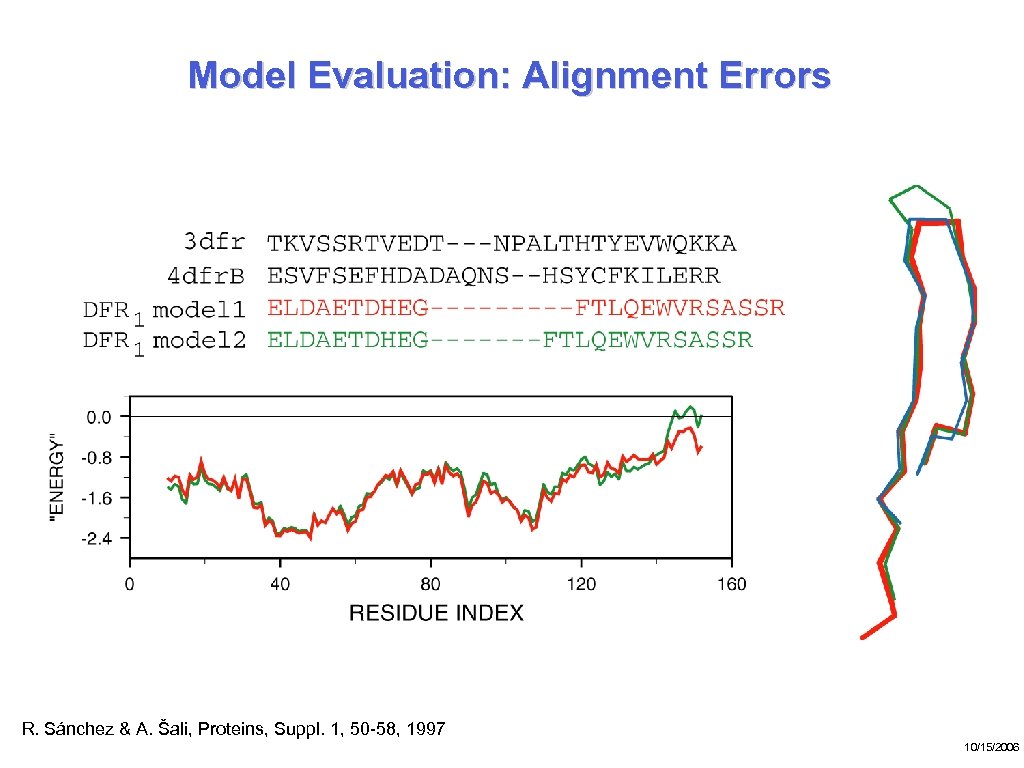
Model Evaluation: Alignment Errors R. Sánchez & A. Šali, Proteins, Suppl. 1, 50 -58, 1997 10/15/2006

Structural Genomics 10/15/2006
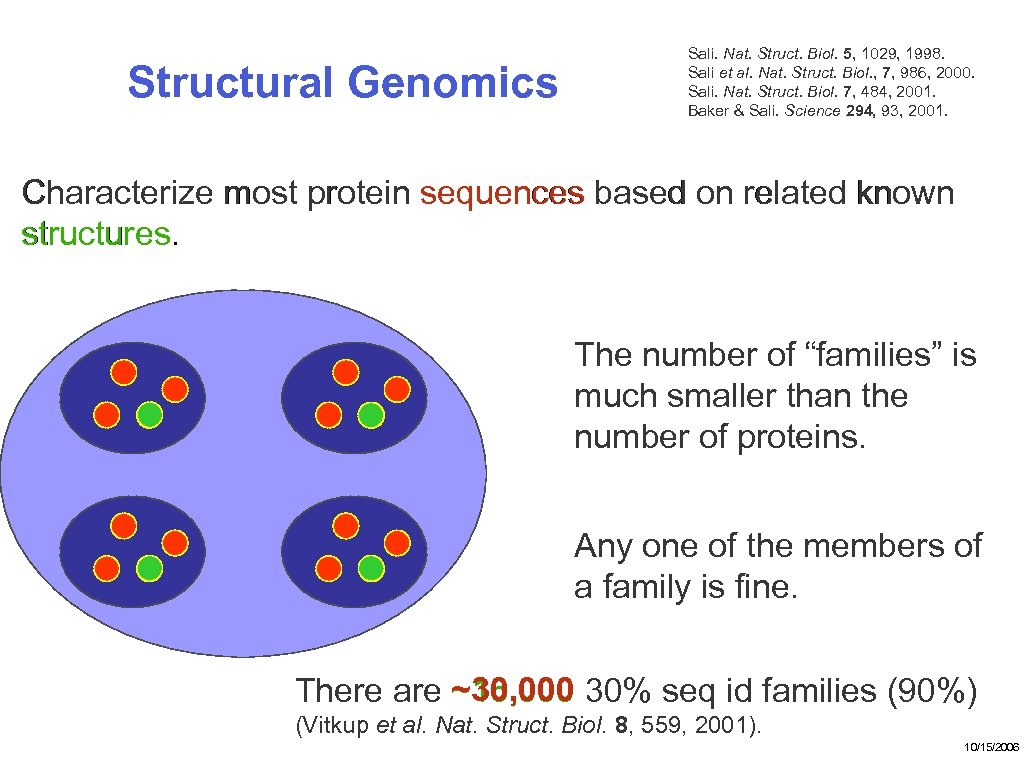
Structural Genomics Sali. Nat. Struct. Biol. 5, 1029, 1998. Sali et al. Nat. Struct. Biol. , 7, 986, 2000. Sali. Nat. Struct. Biol. 7, 484, 2001. Baker & Sali. Science 294, 93, 2001. Characterize most protein sequences based on related known structures. The number of “families” is much smaller than the number of proteins. Any one of the members of a family is fine. ~30, 000 There are ~16, 000 30% seq id families (90%) (Vitkup et al. Nat. Struct. Biol. 8, 559, 2001). 10/15/2006
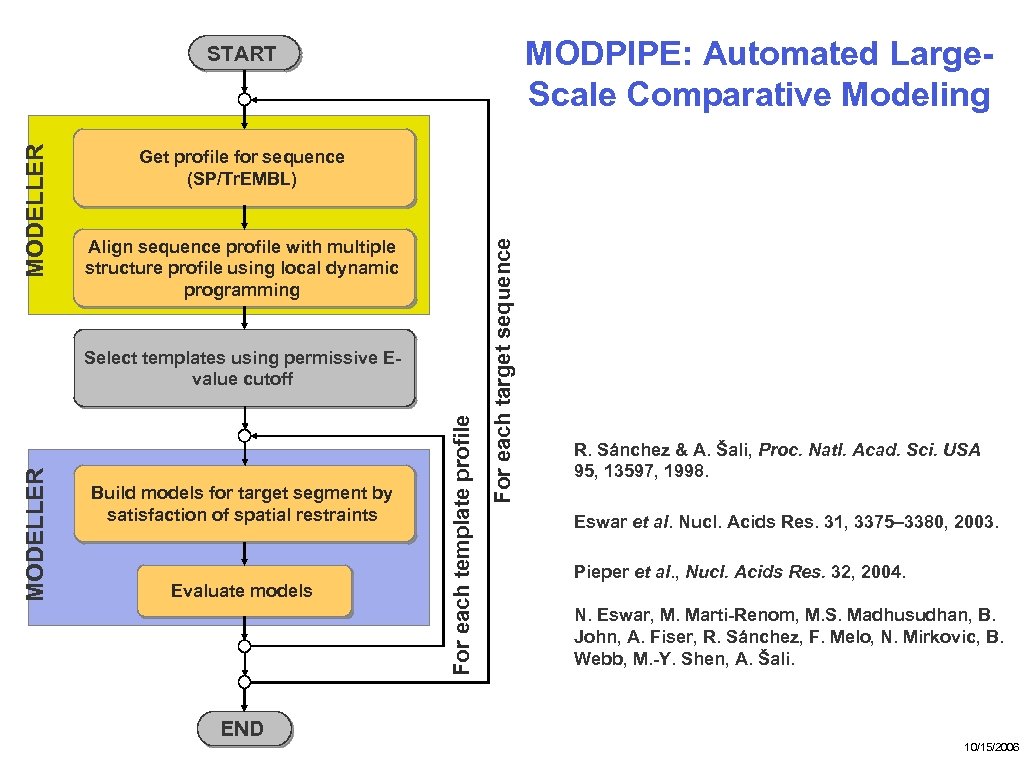
MODPIPE: Automated Large. Scale Comparative Modeling Get profile for sequence (SP/Tr. EMBL) Align sequence profile with multiple structure profile using local dynamic programming Build models for target segment by satisfaction of spatial restraints Evaluate models For each template profile MODELLER Select templates using permissive Evalue cutoff For each target sequence MODELLER START R. Sánchez & A. Šali, Proc. Natl. Acad. Sci. USA 95, 13597, 1998. Eswar et al. Nucl. Acids Res. 31, 3375– 3380, 2003. Pieper et al. , Nucl. Acids Res. 32, 2004. N. Eswar, M. Marti-Renom, M. S. Madhusudhan, B. John, A. Fiser, R. Sánchez, F. Melo, N. Mirkovic, B. Webb, M. -Y. Shen, A. Šali. END 10/15/2006
Eswar et al. Nucl. Acids Res. 31, 3375– 3380, 2003. 10/15/2006
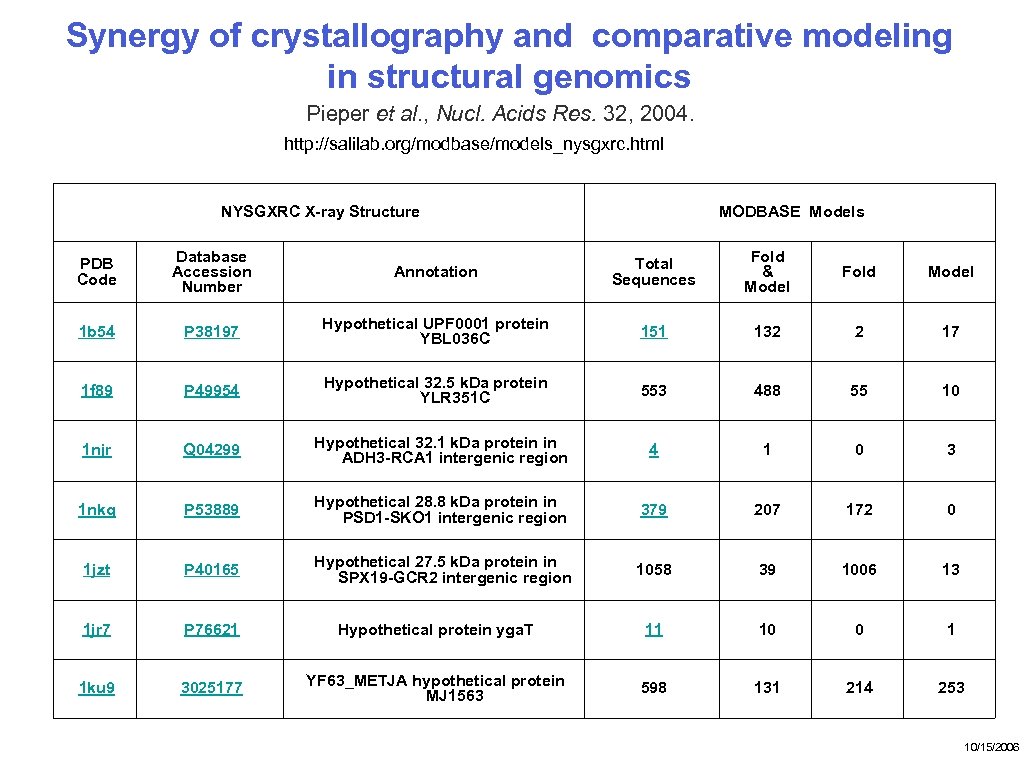
Synergy of crystallography and comparative modeling in structural genomics Pieper et al. , Nucl. Acids Res. 32, 2004. http: //salilab. org/modbase/models_nysgxrc. html NYSGXRC X-ray Structure PDB Code Database Accession Number 1 b 54 MODBASE Models Annotation Total Sequences Fold & Model Fold Model P 38197 Hypothetical UPF 0001 protein YBL 036 C 151 132 2 17 1 f 89 P 49954 Hypothetical 32. 5 k. Da protein YLR 351 C 553 488 55 10 1 njr Q 04299 Hypothetical 32. 1 k. Da protein in ADH 3 -RCA 1 intergenic region 4 1 0 3 1 nkq P 53889 Hypothetical 28. 8 k. Da protein in PSD 1 -SKO 1 intergenic region 379 207 172 0 1 jzt P 40165 Hypothetical 27. 5 k. Da protein in SPX 19 -GCR 2 intergenic region 1058 39 1006 13 1 jr 7 P 76621 Hypothetical protein yga. T 11 10 0 1 1 ku 9 3025177 YF 63_METJA hypothetical protein MJ 1563 598 131 214 253 10/15/2006
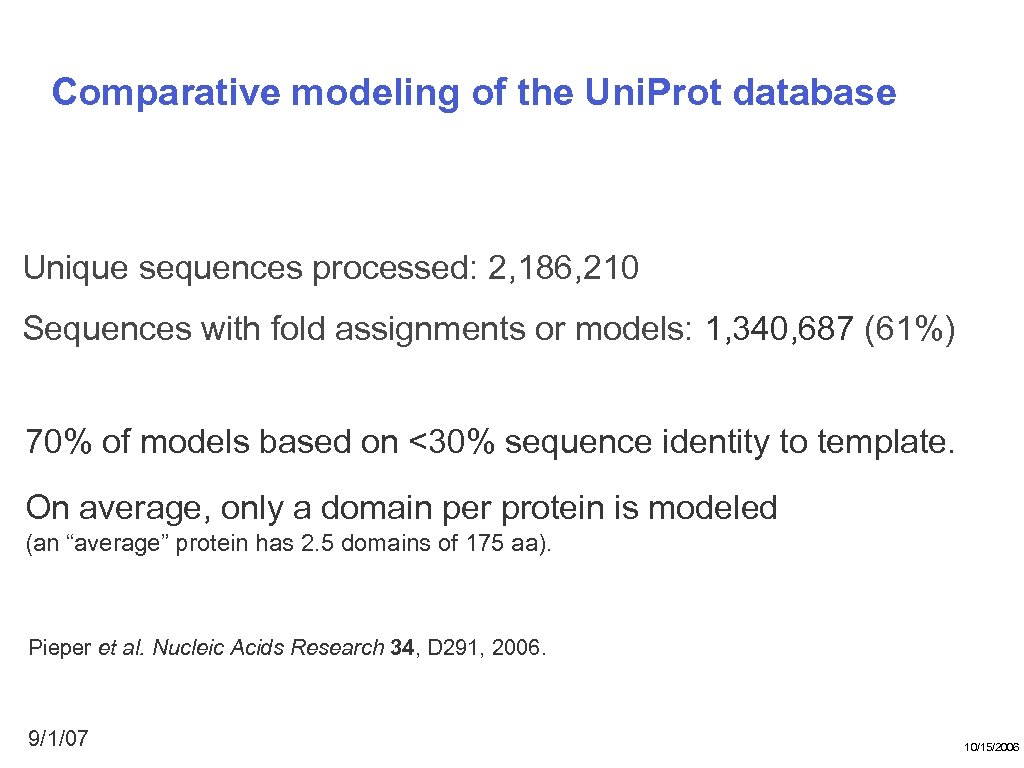
Comparative modeling of the Uni. Prot database Unique sequences processed: 2, 186, 210 Sequences with fold assignments or models: 1, 340, 687 (61%) 70% of models based on <30% sequence identity to template. On average, only a domain per protein is modeled (an “average” protein has 2. 5 domains of 175 aa). Pieper et al. Nucleic Acids Research 34, D 291, 2006. 9/1/07 10/15/2006
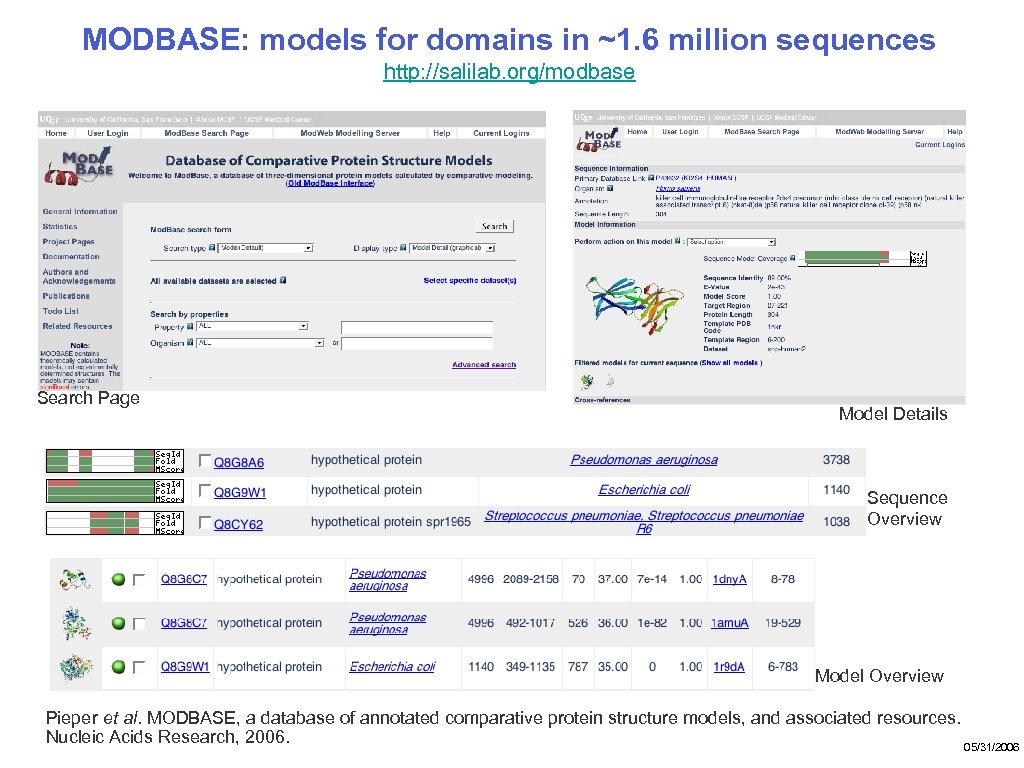
MODBASE: models for domains in ~1. 6 million sequences http: //salilab. org/modbase Search Page Model Details Sequence Overview Model Overview Pieper et al. MODBASE, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Research, 2006. 05/31/2006
Tools for comparative modeling (http: //salilab. org/bioinformatics_resources. shtml) 10/15/2006

Applications of comparative models 10/15/2006
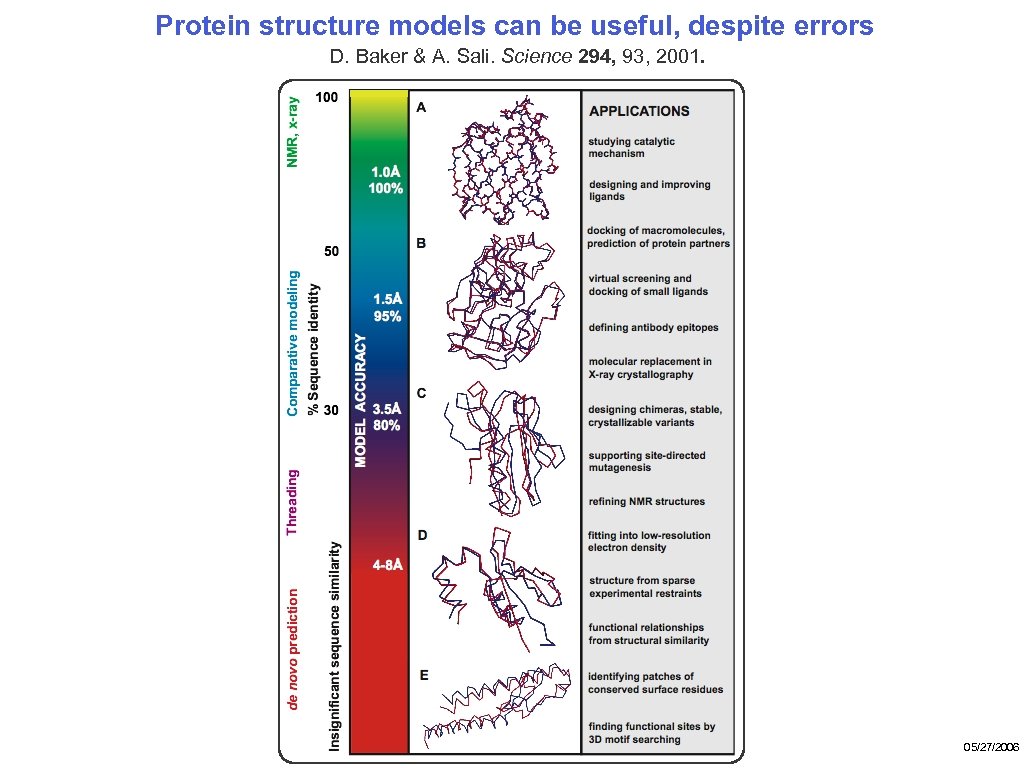
Protein structure models can be useful, despite errors D. Baker & A. Sali. Science 294, 93, 2001. 05/27/2006
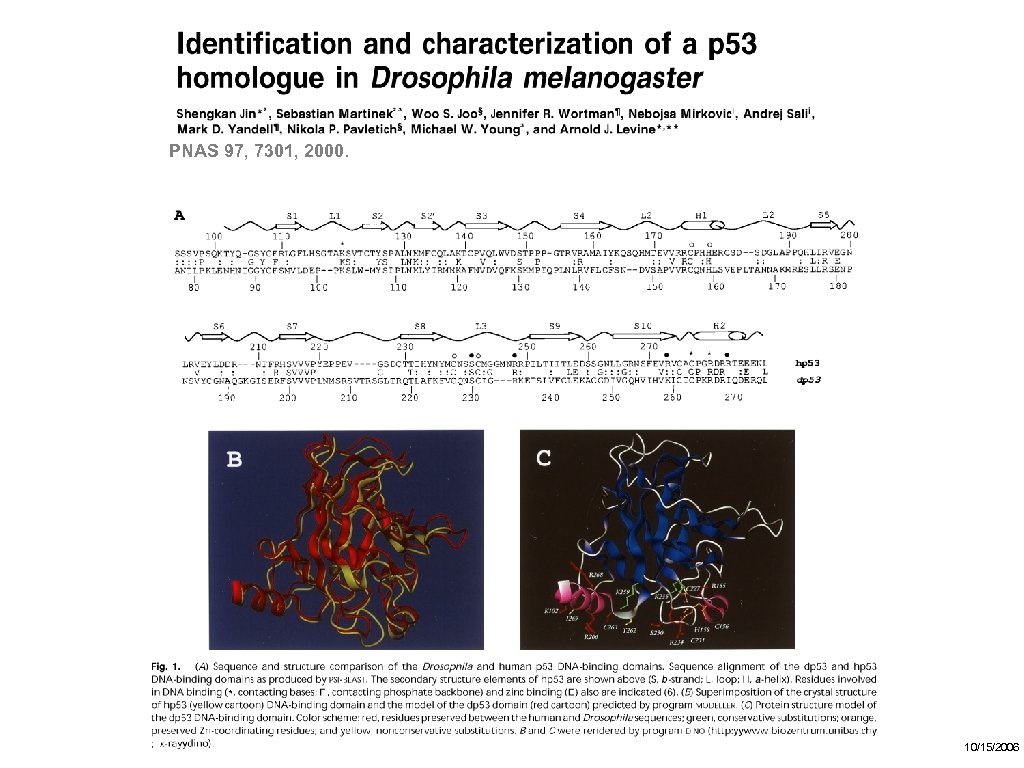
PNAS 97, 7301, 2000. Fly has a p 53 -like protein 10/15/2006
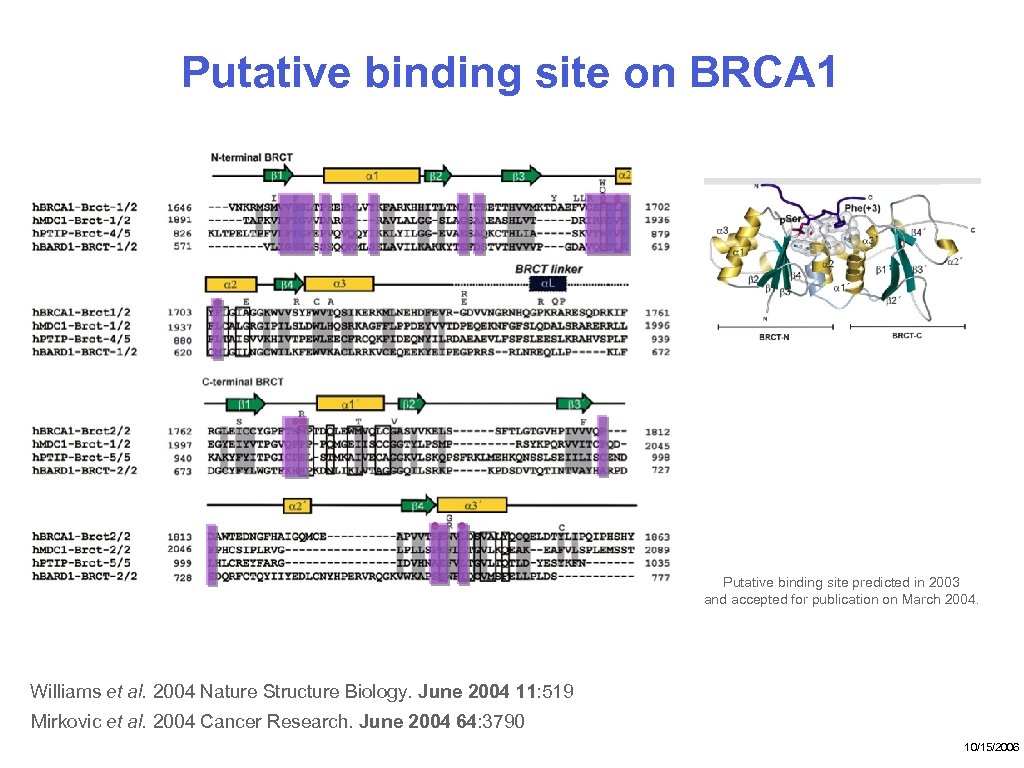
Putative binding site on BRCA 1 Putative binding site predicted in 2003 and accepted for publication on March 2004. Williams et al. 2004 Nature Structure Biology. June 2004 11: 519 Mirkovic et al. 2004 Cancer Research. June 2004 64: 3790 10/15/2006
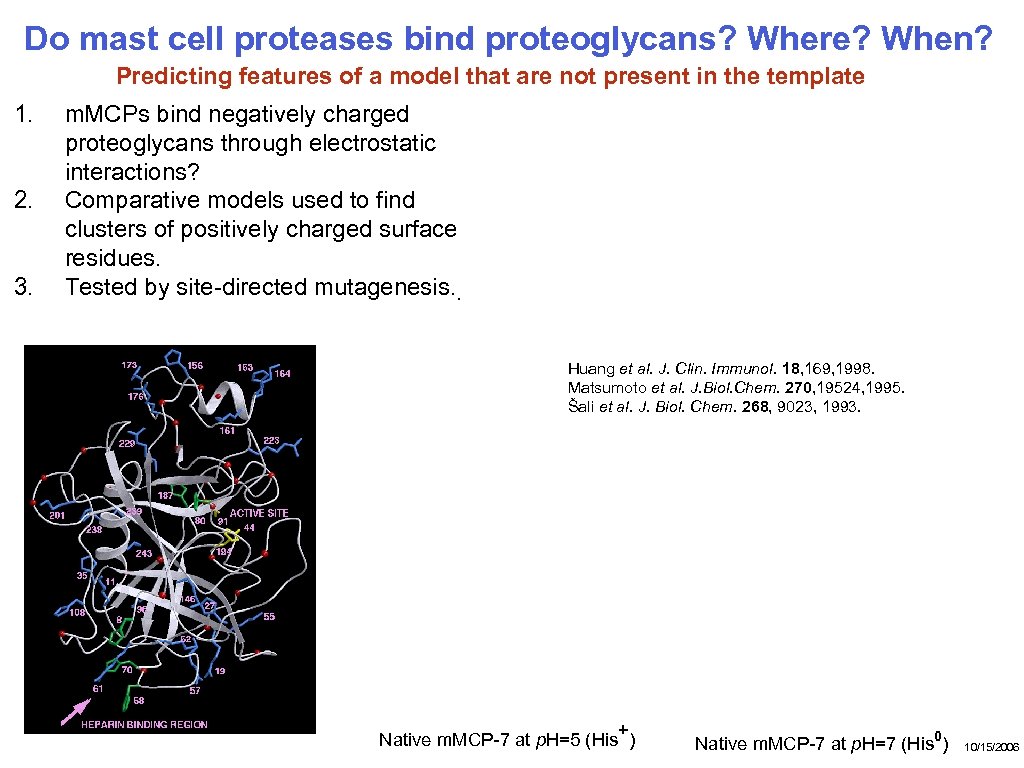
Do mast cell proteases bind proteoglycans? Where? When? Predicting features of a model that are not present in the template 1. 2. 3. m. MCPs bind negatively charged proteoglycans through electrostatic interactions? Comparative models used to find clusters of positively charged surface residues. Tested by site-directed mutagenesis. . Huang et al. J. Clin. Immunol. 18, 169, 1998. Matsumoto et al. J. Biol. Chem. 270, 19524, 1995. Šali et al. J. Biol. Chem. 268, 9023, 1993. + Native m. MCP-7 at p. H=5 (His ) Native m. MCP-7 at p. H=7 (His 0) 10/15/2006
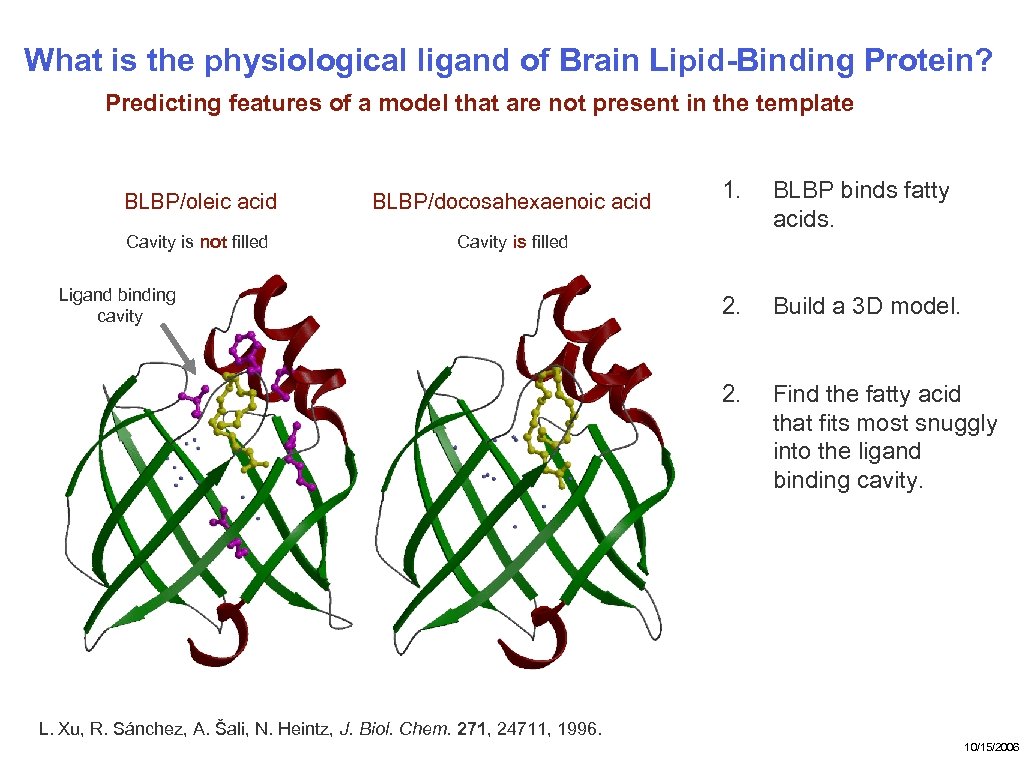
What is the physiological ligand of Brain Lipid-Binding Protein? Predicting features of a model that are not present in the template BLBP/docosahexaenoic acid Cavity is not filled 1. BLBP binds fatty acids. 2. Build a 3 D model. 2. BLBP/oleic acid Find the fatty acid that fits most snuggly into the ligand binding cavity. Cavity is filled Ligand binding cavity L. Xu, R. Sánchez, A. Šali, N. Heintz, J. Biol. Chem. 271, 24711, 1996. 10/15/2006
Protein Structure Prediction with Emphasis on Comparative or Homology Modeling 1. 2. 3. 4. 5. 6. 7. 8. Introduction and motivation Types of comparative modeling methods Errors in comparative models Sequence-structure alignment for comparative modeling Modeling of loops in protein structures Prediction of errors in comparative models Structural genomics Applications A. Šali & T. Blundell. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779, 1993. D. Baker & A. Sali. Protein structure prediction and structural genomics. Science 294, 93, 2001. M. A. Marti-Renom et al. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29, 291 -325, 2000. N. Eswar, D. Eramian, B. Webb, M. -Y. Shen, A. Sali. Protein Structure Modeling with MODELLER, in press. Available at: http: //salilab. org/publications/ 10/15/2006
3b9afb372176959a8d6eb4c5c5de4382.ppt