61752bf07d54e90d2b5dcecc57eac86f.ppt
- Количество слайдов: 43
 Classifying the protein universe Synapse. Associated Protein 97 Ashwin Sivakumar Wu et al, 2002. EMBO J 19: 5740 -5751
Classifying the protein universe Synapse. Associated Protein 97 Ashwin Sivakumar Wu et al, 2002. EMBO J 19: 5740 -5751
 Domain Analysis and Protein Families § Introduction What are protein families? § Protein families Description & Definition Motifs and Profiles § The modular architecture of proteins § Domain Properties and Classification
Domain Analysis and Protein Families § Introduction What are protein families? § Protein families Description & Definition Motifs and Profiles § The modular architecture of proteins § Domain Properties and Classification
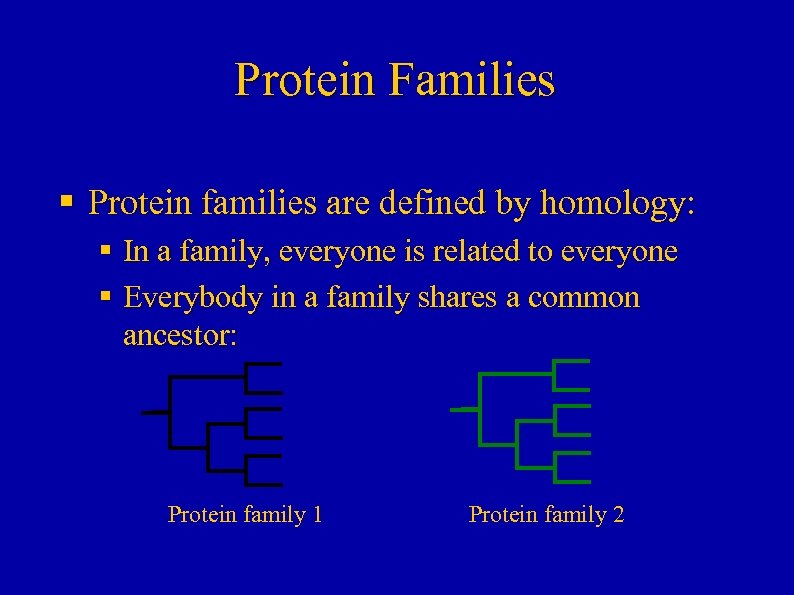 Protein Families § Protein families are defined by homology: § In a family, everyone is related to everyone § Everybody in a family shares a common ancestor: Protein family 1 Protein family 2
Protein Families § Protein families are defined by homology: § In a family, everyone is related to everyone § Everybody in a family shares a common ancestor: Protein family 1 Protein family 2
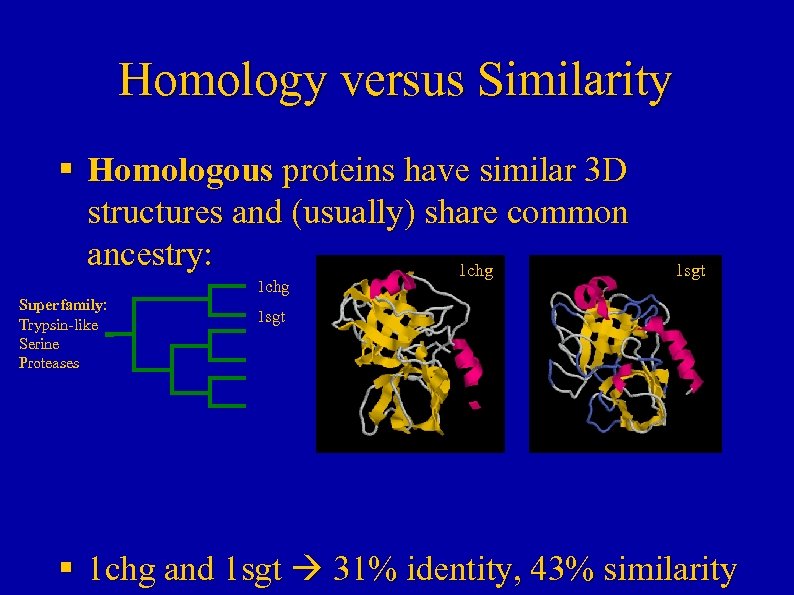 Homology versus Similarity § Homologous proteins have similar 3 D structures and (usually) share common ancestry: 1 chg Superfamily: Trypsin-like Serine Proteases 1 sgt § 1 chg and 1 sgt 31% identity, 43% similarity
Homology versus Similarity § Homologous proteins have similar 3 D structures and (usually) share common ancestry: 1 chg Superfamily: Trypsin-like Serine Proteases 1 sgt § 1 chg and 1 sgt 31% identity, 43% similarity
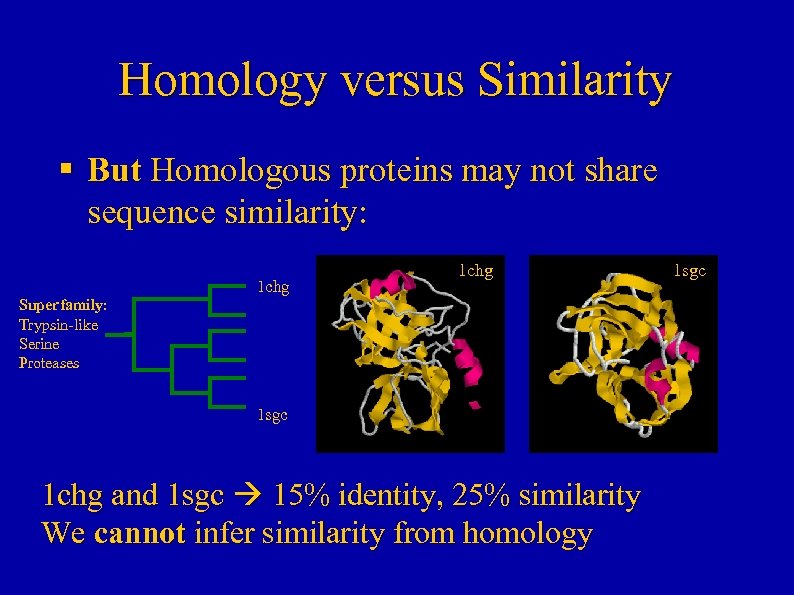 Homology versus Similarity § But Homologous proteins may not share sequence similarity: 1 chg Superfamily: Trypsin-like Serine Proteases 1 sgc 1 chg and 1 sgc 15% identity, 25% similarity We cannot infer similarity from homology 1 sgc
Homology versus Similarity § But Homologous proteins may not share sequence similarity: 1 chg Superfamily: Trypsin-like Serine Proteases 1 sgc 1 chg and 1 sgc 15% identity, 25% similarity We cannot infer similarity from homology 1 sgc
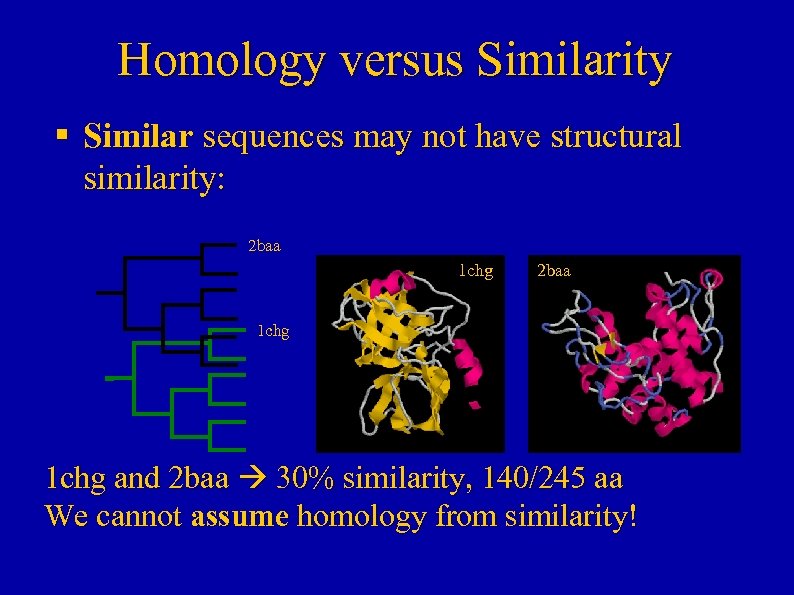 Homology versus Similarity § Similar sequences may not have structural similarity: 2 baa 1 chg and 2 baa 30% similarity, 140/245 aa We cannot assume homology from similarity!
Homology versus Similarity § Similar sequences may not have structural similarity: 2 baa 1 chg and 2 baa 30% similarity, 140/245 aa We cannot assume homology from similarity!
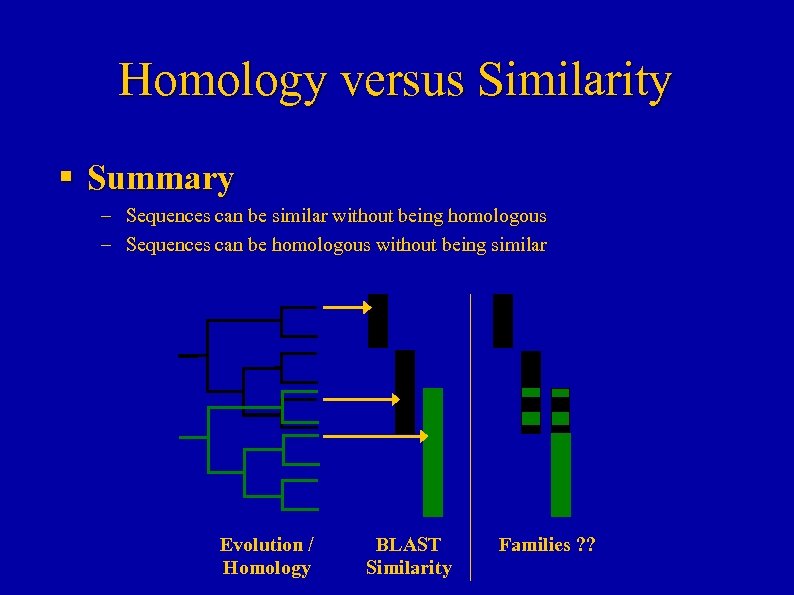 Homology versus Similarity § Summary – Sequences can be similar without being homologous – Sequences can be homologous without being similar Evolution / Homology BLAST Similarity Families ? ?
Homology versus Similarity § Summary – Sequences can be similar without being homologous – Sequences can be homologous without being similar Evolution / Homology BLAST Similarity Families ? ?
 Domain Analysis and Protein Families § Introduction What are protein families? § Protein families Description & Definition Motifs and Profiles § The modular architecture of proteins § Domain Properties and Classification
Domain Analysis and Protein Families § Introduction What are protein families? § Protein families Description & Definition Motifs and Profiles § The modular architecture of proteins § Domain Properties and Classification
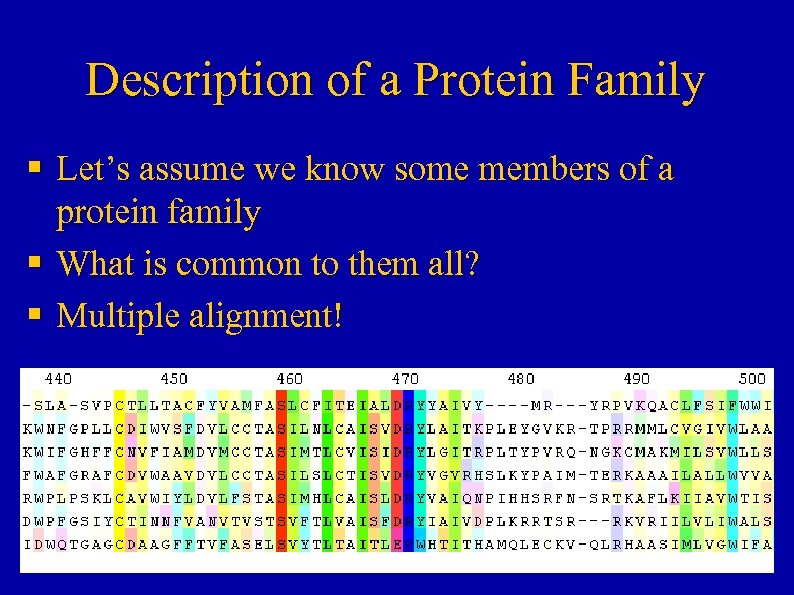 Description of a Protein Family § Let’s assume we know some members of a protein family § What is common to them all? § Multiple alignment!
Description of a Protein Family § Let’s assume we know some members of a protein family § What is common to them all? § Multiple alignment!
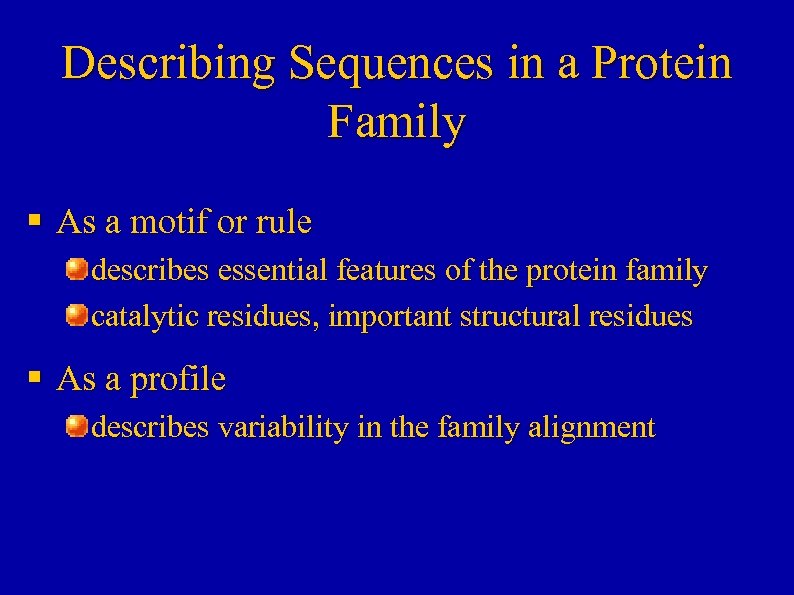 Describing Sequences in a Protein Family § As a motif or rule describes essential features of the protein family catalytic residues, important structural residues § As a profile describes variability in the family alignment
Describing Sequences in a Protein Family § As a motif or rule describes essential features of the protein family catalytic residues, important structural residues § As a profile describes variability in the family alignment
 Techniques for searching sequence databases to Some common strategies to uncover common domains/motifs of biological significance that categorize a protein into a family • Pattern - a deterministic syntax that describes multiple combinations of possible residues within a protein string • Profile - probabilistic generalizations that assign to every segment position, a probability that each of the 20 aa will occur
Techniques for searching sequence databases to Some common strategies to uncover common domains/motifs of biological significance that categorize a protein into a family • Pattern - a deterministic syntax that describes multiple combinations of possible residues within a protein string • Profile - probabilistic generalizations that assign to every segment position, a probability that each of the 20 aa will occur
 Consensus - mathematical probability that a particular amino acid will be located at a given position. • Probabilistic pattern constructed from a MSA. Opportunity to assign penalties for insertions and deletions • PSSM - (Position Specific Scoring Matrix) – Represents the sequence profile in tabular form – Columns of weights for every aa corresponding to each column of a MSA.
Consensus - mathematical probability that a particular amino acid will be located at a given position. • Probabilistic pattern constructed from a MSA. Opportunity to assign penalties for insertions and deletions • PSSM - (Position Specific Scoring Matrix) – Represents the sequence profile in tabular form – Columns of weights for every aa corresponding to each column of a MSA.
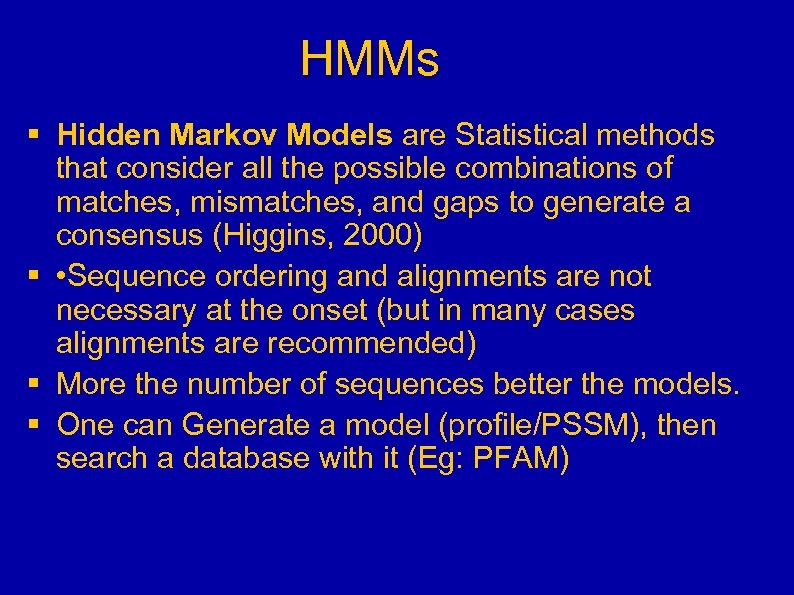 HMMs § Hidden Markov Models are Statistical methods that consider all the possible combinations of matches, mismatches, and gaps to generate a consensus (Higgins, 2000) § • Sequence ordering and alignments are not necessary at the onset (but in many cases alignments are recommended) § More the number of sequences better the models. § One can Generate a model (profile/PSSM), then search a database with it (Eg: PFAM)
HMMs § Hidden Markov Models are Statistical methods that consider all the possible combinations of matches, mismatches, and gaps to generate a consensus (Higgins, 2000) § • Sequence ordering and alignments are not necessary at the onset (but in many cases alignments are recommended) § More the number of sequences better the models. § One can Generate a model (profile/PSSM), then search a database with it (Eg: PFAM)
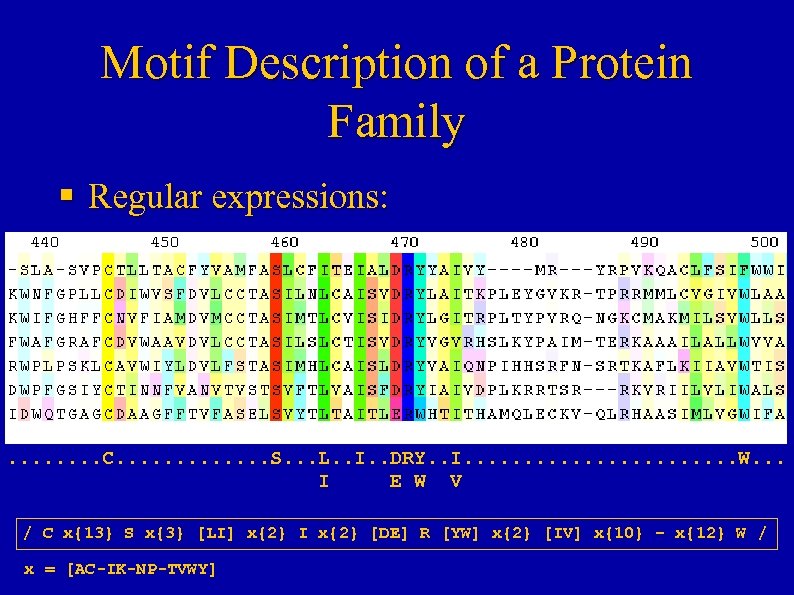 Motif Description of a Protein Family § Regular expressions: . . . . C. . . S. . . L. . I. . DRY. . I. . . W. . . I E W V / C x{13} S x{3} [LI] x{2} I x{2} [DE] R [YW] x{2} [IV] x{10} – x{12} W / x = [AC-IK-NP-TVWY]
Motif Description of a Protein Family § Regular expressions: . . . . C. . . S. . . L. . I. . DRY. . I. . . W. . . I E W V / C x{13} S x{3} [LI] x{2} I x{2} [DE] R [YW] x{2} [IV] x{10} – x{12} W / x = [AC-IK-NP-TVWY]
 Motif Description of a Protein Family §Database: PROSITE “PROSITE is a database of protein families and domains. It is based on the observation that, while there is a huge number of different proteins, most of them can be grouped, on the basis of similarities in their sequences, into a limited number of families. Proteins or protein domains belonging to a particular family generally share functional attributes and are derived from a common ancestor. It is apparent, when studying protein sequence families, that some regions have been better conserved than others during evolution. These regions are generally important for the function of a protein and/or for the maintenance of its three-dimensional structure. By analyzing the constant and variable properties of such groups of similar sequences, it is possible to derive a signature for a protein family or domain, which distinguishes its members from all other unrelated proteins. ” http: //au. expasy. org/prosite_details. html
Motif Description of a Protein Family §Database: PROSITE “PROSITE is a database of protein families and domains. It is based on the observation that, while there is a huge number of different proteins, most of them can be grouped, on the basis of similarities in their sequences, into a limited number of families. Proteins or protein domains belonging to a particular family generally share functional attributes and are derived from a common ancestor. It is apparent, when studying protein sequence families, that some regions have been better conserved than others during evolution. These regions are generally important for the function of a protein and/or for the maintenance of its three-dimensional structure. By analyzing the constant and variable properties of such groups of similar sequences, it is possible to derive a signature for a protein family or domain, which distinguishes its members from all other unrelated proteins. ” http: //au. expasy. org/prosite_details. html
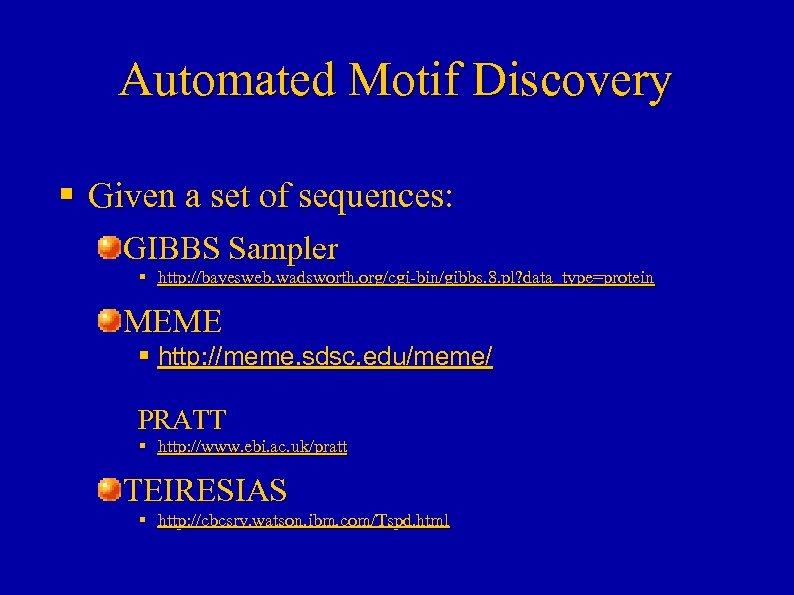 Automated Motif Discovery § Given a set of sequences: GIBBS Sampler § http: //bayesweb. wadsworth. org/cgi-bin/gibbs. 8. pl? data_type=protein MEME § http: //meme. sdsc. edu/meme/ PRATT § http: //www. ebi. ac. uk/pratt TEIRESIAS § http: //cbcsrv. watson. ibm. com/Tspd. html
Automated Motif Discovery § Given a set of sequences: GIBBS Sampler § http: //bayesweb. wadsworth. org/cgi-bin/gibbs. 8. pl? data_type=protein MEME § http: //meme. sdsc. edu/meme/ PRATT § http: //www. ebi. ac. uk/pratt TEIRESIAS § http: //cbcsrv. watson. ibm. com/Tspd. html
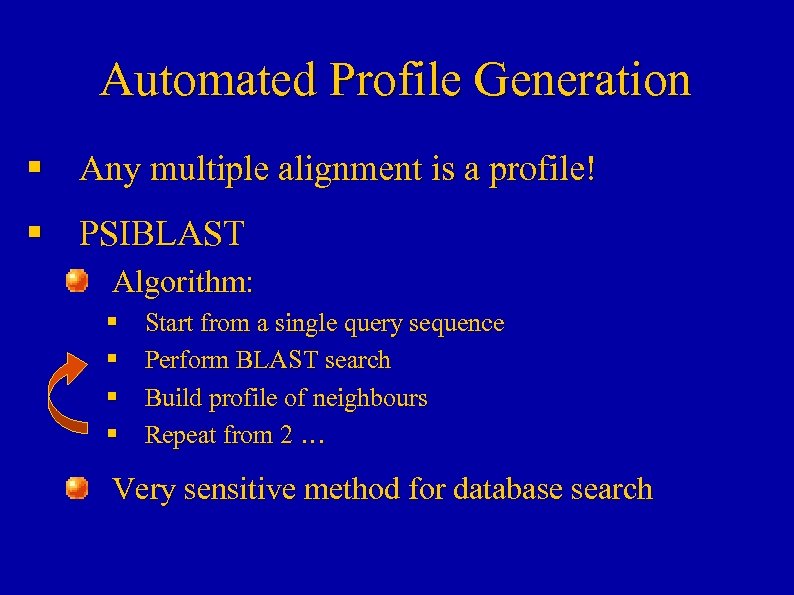 Automated Profile Generation § Any multiple alignment is a profile! § PSIBLAST Algorithm: § § Start from a single query sequence Perform BLAST search Build profile of neighbours Repeat from 2 … Very sensitive method for database search
Automated Profile Generation § Any multiple alignment is a profile! § PSIBLAST Algorithm: § § Start from a single query sequence Perform BLAST search Build profile of neighbours Repeat from 2 … Very sensitive method for database search
 PSI-Blast § Starts with a sequence, BLAST it, § align select results to query sequence, estimate a profile with the MSA, search database with the profile - constructs PSSM § Iterate until process stabilizes § Focus here is on domains, not entire sequences § Greatly improves sensitivity
PSI-Blast § Starts with a sequence, BLAST it, § align select results to query sequence, estimate a profile with the MSA, search database with the profile - constructs PSSM § Iterate until process stabilizes § Focus here is on domains, not entire sequences § Greatly improves sensitivity
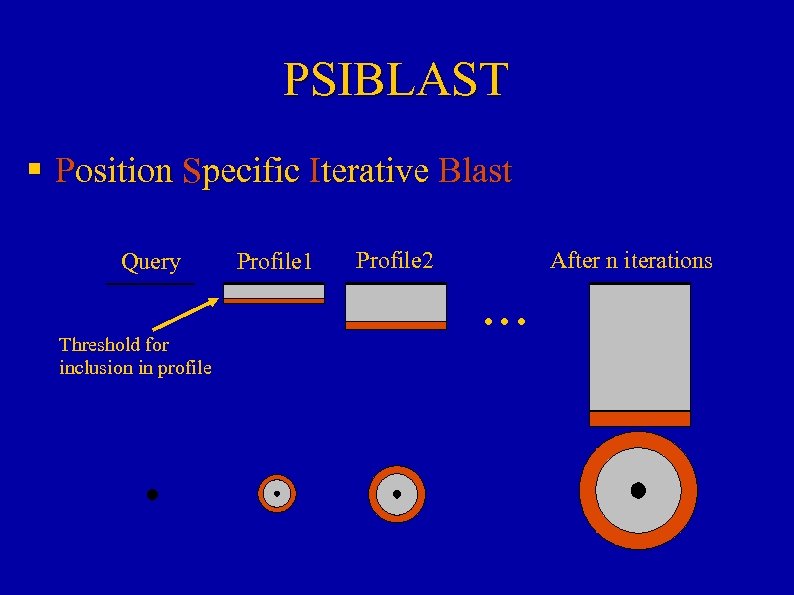 PSIBLAST § Position Specific Iterative Blast Query Threshold for inclusion in profile Profile 1 Profile 2 . . . After n iterations
PSIBLAST § Position Specific Iterative Blast Query Threshold for inclusion in profile Profile 1 Profile 2 . . . After n iterations
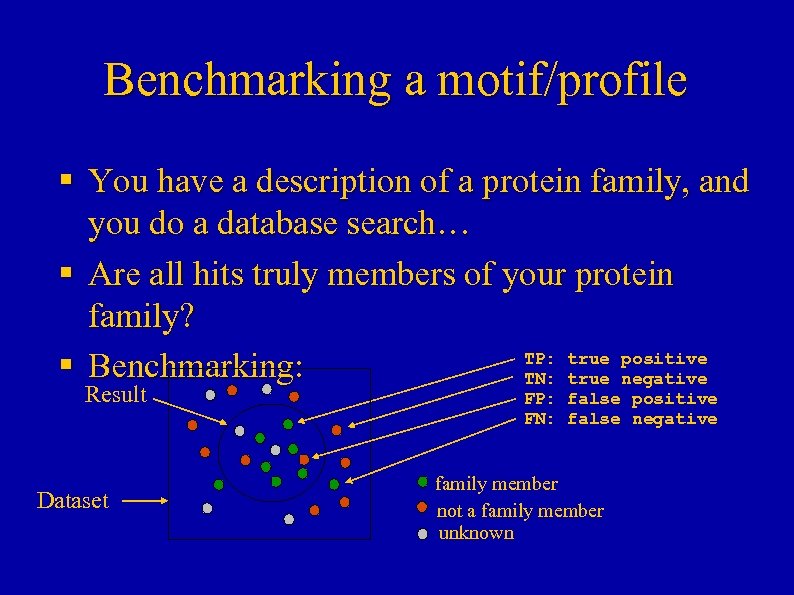 Benchmarking a motif/profile § You have a description of a protein family, and you do a database search… § Are all hits truly members of your protein family? TP: true positive § Benchmarking: TN: true negative Result Dataset FP: false positive FN: false negative family member not a family member unknown
Benchmarking a motif/profile § You have a description of a protein family, and you do a database search… § Are all hits truly members of your protein family? TP: true positive § Benchmarking: TN: true negative Result Dataset FP: false positive FN: false negative family member not a family member unknown
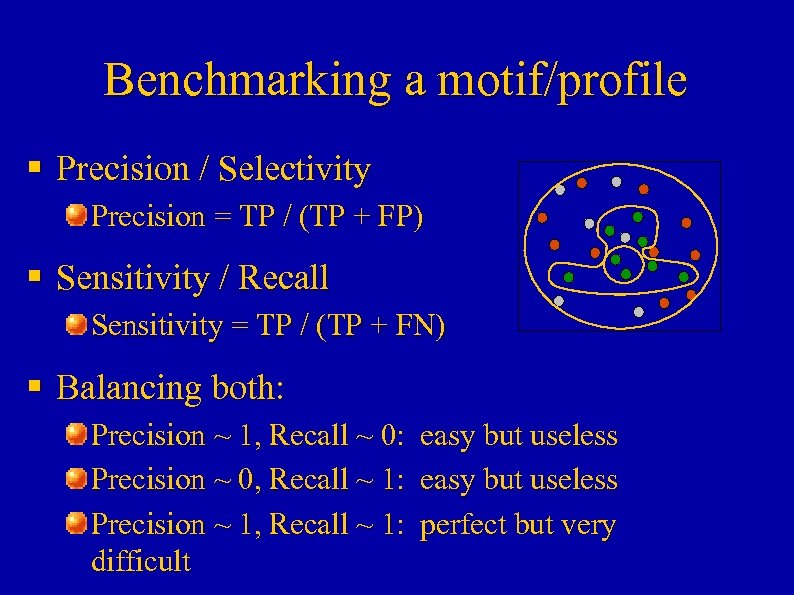 Benchmarking a motif/profile § Precision / Selectivity Precision = TP / (TP + FP) § Sensitivity / Recall Sensitivity = TP / (TP + FN) § Balancing both: Precision ~ 1, Recall ~ 0: easy but useless Precision ~ 0, Recall ~ 1: easy but useless Precision ~ 1, Recall ~ 1: perfect but very difficult
Benchmarking a motif/profile § Precision / Selectivity Precision = TP / (TP + FP) § Sensitivity / Recall Sensitivity = TP / (TP + FN) § Balancing both: Precision ~ 1, Recall ~ 0: easy but useless Precision ~ 0, Recall ~ 1: easy but useless Precision ~ 1, Recall ~ 1: perfect but very difficult
 Domain Analysis and Protein Families § Introduction What are protein families? § Protein families Description & Definition Motifs and Profiles § The modular architecture of proteins § Domain Properties and Classification
Domain Analysis and Protein Families § Introduction What are protein families? § Protein families Description & Definition Motifs and Profiles § The modular architecture of proteins § Domain Properties and Classification
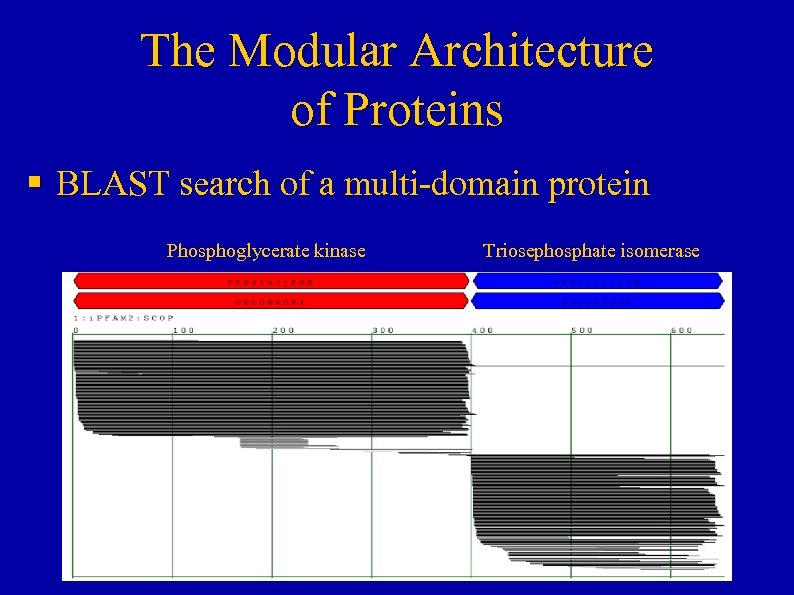 The Modular Architecture of Proteins § BLAST search of a multi-domain protein Phosphoglycerate kinase Triosephosphate isomerase
The Modular Architecture of Proteins § BLAST search of a multi-domain protein Phosphoglycerate kinase Triosephosphate isomerase
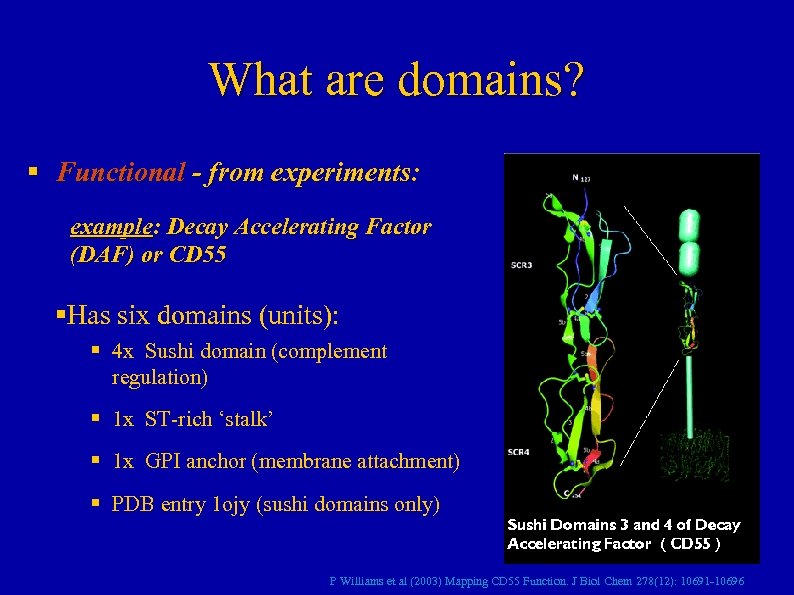 What are domains? § Functional - from experiments: example: Decay Accelerating Factor (DAF) or CD 55 §Has six domains (units): § 4 x Sushi domain (complement regulation) § 1 x ST-rich ‘stalk’ § 1 x GPI anchor (membrane attachment) § PDB entry 1 ojy (sushi domains only) P Williams et al (2003) Mapping CD 55 Function. J Biol Chem 278(12): 10691 -10696
What are domains? § Functional - from experiments: example: Decay Accelerating Factor (DAF) or CD 55 §Has six domains (units): § 4 x Sushi domain (complement regulation) § 1 x ST-rich ‘stalk’ § 1 x GPI anchor (membrane attachment) § PDB entry 1 ojy (sushi domains only) P Williams et al (2003) Mapping CD 55 Function. J Biol Chem 278(12): 10691 -10696
 There is only so much we can conclude… § Classifying domains [To aid structure prediction (predict structural domains, molecular function of the domain)] § Classifying complete sequences (predicting molecular function of proteins, large scale annotation) § Majority of proteins are multi-domain proteins.
There is only so much we can conclude… § Classifying domains [To aid structure prediction (predict structural domains, molecular function of the domain)] § Classifying complete sequences (predicting molecular function of proteins, large scale annotation) § Majority of proteins are multi-domain proteins.
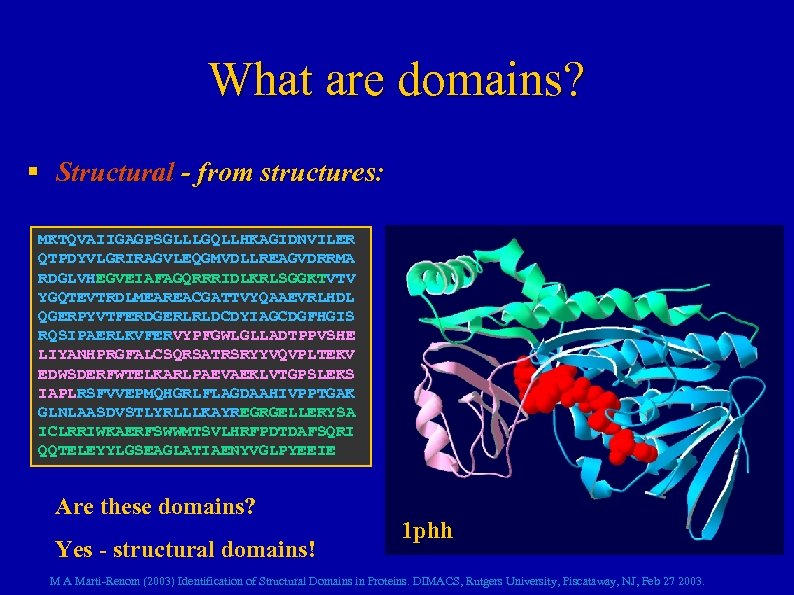 What are domains? § Structural - from structures: MKTQVAIIGAGPSGLLLGQLLHKAGIDNVILER QTPDYVLGRIRAGVLEQGMVDLLREAGVDRRMA RDGLVHEGVEIAFAGQRRRIDLKRLSGGKTVTV YGQTEVTRDLMEAREACGATTVYQAAEVRLHDL QGERPYVTFERDGERLRLDCDYIAGCDGFHGIS RQSIPAERLKVFERVYPFGWLGLLADTPPVSHE LIYANHPRGFALCSQRSATRSRYYVQVPLTEKV EDWSDERFWTELKARLPAEVAEKLVTGPSLEKS IAPLRSFVVEPMQHGRLFLAGDAAHIVPPTGAK GLNLAASDVSTLYRLLLKAYREGRGELLERYSA ICLRRIWKAERFSWWMTSVLHRFPDTDAFSQRI QQTELEYYLGSEAGLATIAENYVGLPYEEIE Are these domains? Yes - structural domains! 1 phh M A Marti-Renom (2003) Identification of Structural Domains in Proteins. DIMACS, Rutgers University, Piscataway, NJ, Feb 27 2003.
What are domains? § Structural - from structures: MKTQVAIIGAGPSGLLLGQLLHKAGIDNVILER QTPDYVLGRIRAGVLEQGMVDLLREAGVDRRMA RDGLVHEGVEIAFAGQRRRIDLKRLSGGKTVTV YGQTEVTRDLMEAREACGATTVYQAAEVRLHDL QGERPYVTFERDGERLRLDCDYIAGCDGFHGIS RQSIPAERLKVFERVYPFGWLGLLADTPPVSHE LIYANHPRGFALCSQRSATRSRYYVQVPLTEKV EDWSDERFWTELKARLPAEVAEKLVTGPSLEKS IAPLRSFVVEPMQHGRLFLAGDAAHIVPPTGAK GLNLAASDVSTLYRLLLKAYREGRGELLERYSA ICLRRIWKAERFSWWMTSVLHRFPDTDAFSQRI QQTELEYYLGSEAGLATIAENYVGLPYEEIE Are these domains? Yes - structural domains! 1 phh M A Marti-Renom (2003) Identification of Structural Domains in Proteins. DIMACS, Rutgers University, Piscataway, NJ, Feb 27 2003.
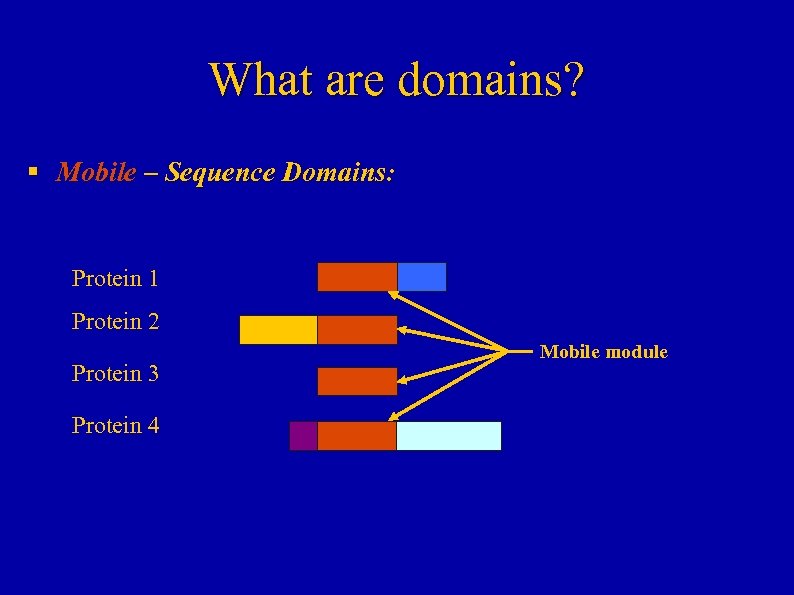 What are domains? § Mobile – Sequence Domains: Protein 1 Protein 2 Protein 3 Protein 4 Mobile module
What are domains? § Mobile – Sequence Domains: Protein 1 Protein 2 Protein 3 Protein 4 Mobile module
 Domains are. . . §. . . evolutionary building blocks: Families of evolutionarily-related sequence segments Domain assignment often coupled with classification § With one or more of the following properties: Globular Independently foldable Recurrence in different contexts § To be precise, we say: “protein family” we mean: “protein domain family”
Domains are. . . §. . . evolutionary building blocks: Families of evolutionarily-related sequence segments Domain assignment often coupled with classification § With one or more of the following properties: Globular Independently foldable Recurrence in different contexts § To be precise, we say: “protein family” we mean: “protein domain family”
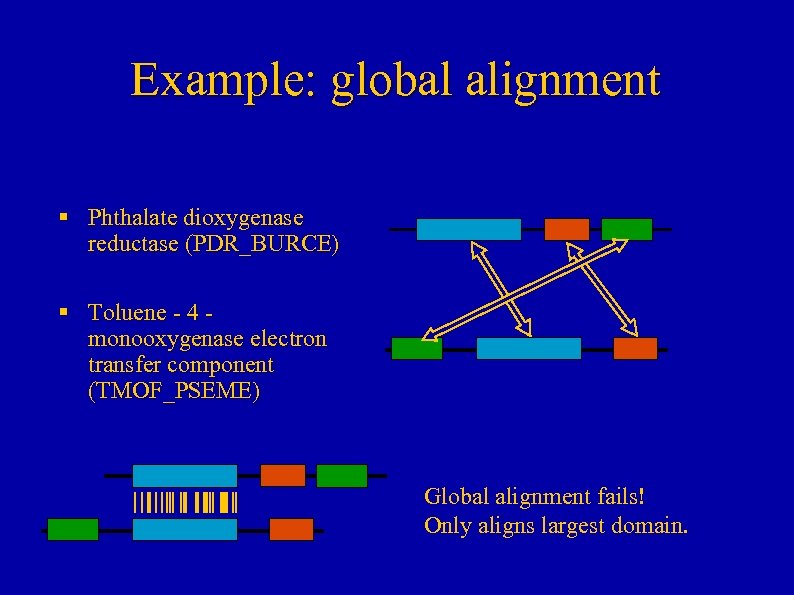 Example: global alignment § Phthalate dioxygenase reductase (PDR_BURCE) § Toluene - 4 monooxygenase electron transfer component (TMOF_PSEME) Global alignment fails! Only aligns largest domain.
Example: global alignment § Phthalate dioxygenase reductase (PDR_BURCE) § Toluene - 4 monooxygenase electron transfer component (TMOF_PSEME) Global alignment fails! Only aligns largest domain.
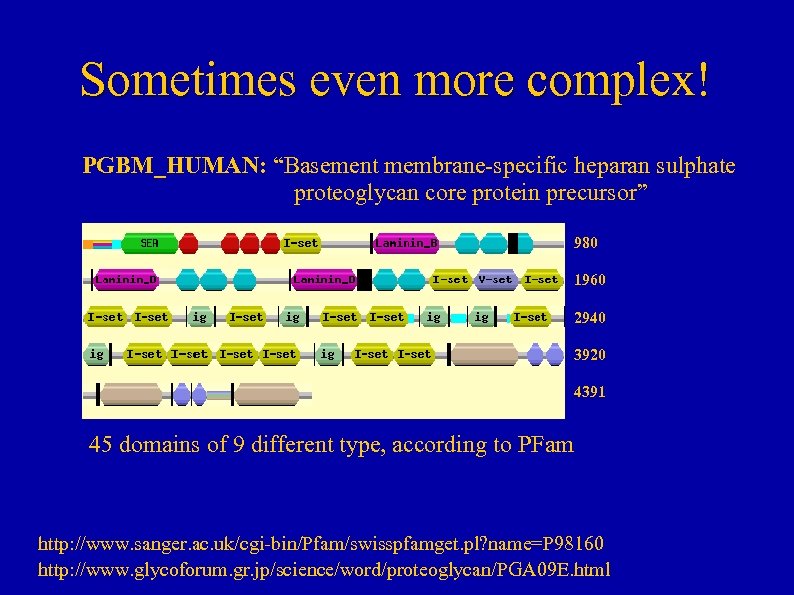 Sometimes even more complex! PGBM_HUMAN: “Basement membrane-specific heparan sulphate proteoglycan core protein precursor” 980 1960 2940 3920 4391 45 domains of 9 different type, according to PFam http: //www. sanger. ac. uk/cgi-bin/Pfam/swisspfamget. pl? name=P 98160 http: //www. glycoforum. gr. jp/science/word/proteoglycan/PGA 09 E. html
Sometimes even more complex! PGBM_HUMAN: “Basement membrane-specific heparan sulphate proteoglycan core protein precursor” 980 1960 2940 3920 4391 45 domains of 9 different type, according to PFam http: //www. sanger. ac. uk/cgi-bin/Pfam/swisspfamget. pl? name=P 98160 http: //www. glycoforum. gr. jp/science/word/proteoglycan/PGA 09 E. html
 Domain Analysis and Protein Families § Introduction What are protein families? § Protein families Description & Definition Motifs and Profiles § The modular architecture of proteins § Domain Properties and Classification
Domain Analysis and Protein Families § Introduction What are protein families? § Protein families Description & Definition Motifs and Profiles § The modular architecture of proteins § Domain Properties and Classification
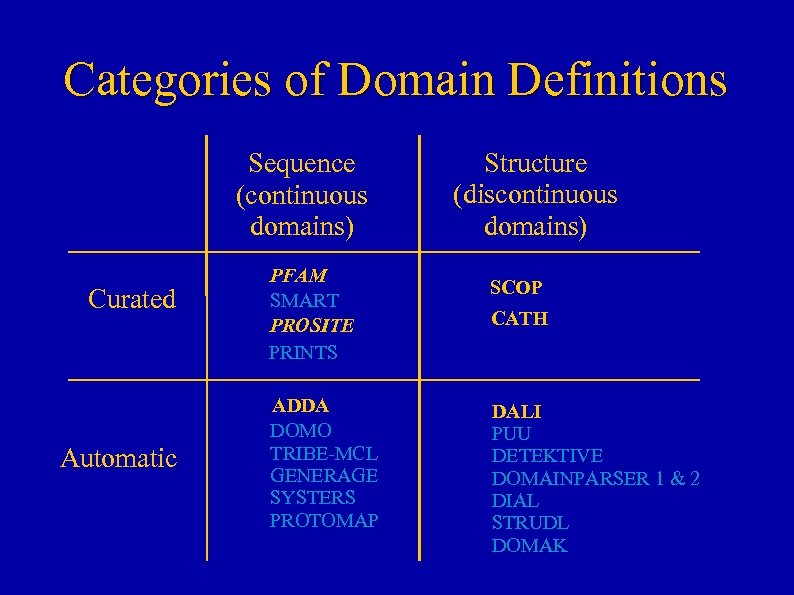 Categories of Domain Definitions Sequence (continuous domains) Curated Automatic PFAM SMART PROSITE PRINTS ADDA DOMO TRIBE-MCL GENERAGE SYSTERS PROTOMAP Structure (discontinuous domains) SCOP CATH DALI PUU DETEKTIVE DOMAINPARSER 1 & 2 DIAL STRUDL DOMAK
Categories of Domain Definitions Sequence (continuous domains) Curated Automatic PFAM SMART PROSITE PRINTS ADDA DOMO TRIBE-MCL GENERAGE SYSTERS PROTOMAP Structure (discontinuous domains) SCOP CATH DALI PUU DETEKTIVE DOMAINPARSER 1 & 2 DIAL STRUDL DOMAK
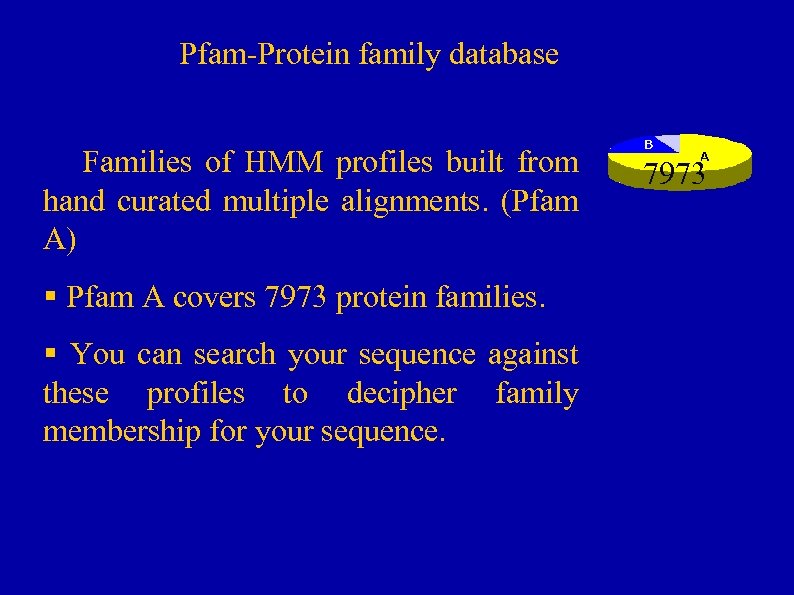 Pfam-Protein family database § Families of HMM profiles built from hand curated multiple alignments. (Pfam A) § Pfam A covers 7973 protein families. § You can search your sequence against these profiles to decipher family membership for your sequence. 7973
Pfam-Protein family database § Families of HMM profiles built from hand curated multiple alignments. (Pfam A) § Pfam A covers 7973 protein families. § You can search your sequence against these profiles to decipher family membership for your sequence. 7973
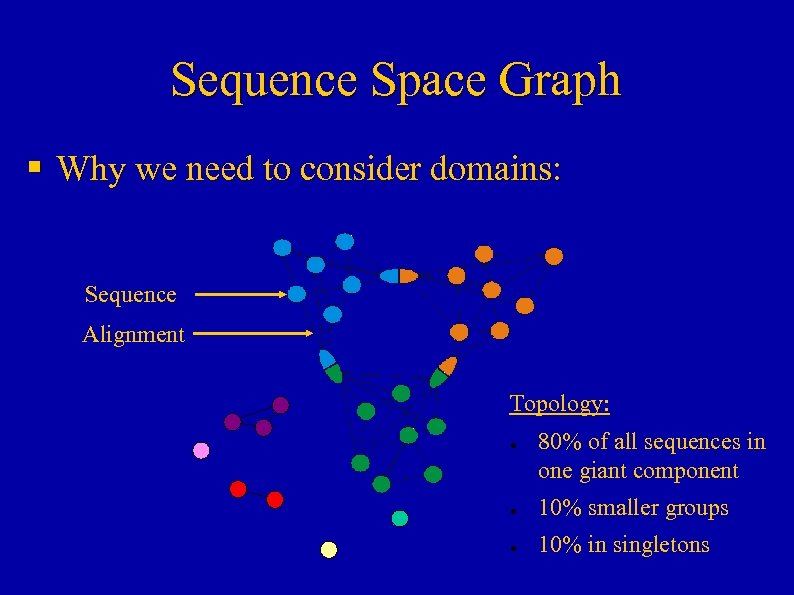 Sequence Space Graph § Why we need to consider domains: Sequence Alignment Topology: ● 80% of all sequences in one giant component ● 10% smaller groups ● 10% in singletons
Sequence Space Graph § Why we need to consider domains: Sequence Alignment Topology: ● 80% of all sequences in one giant component ● 10% smaller groups ● 10% in singletons
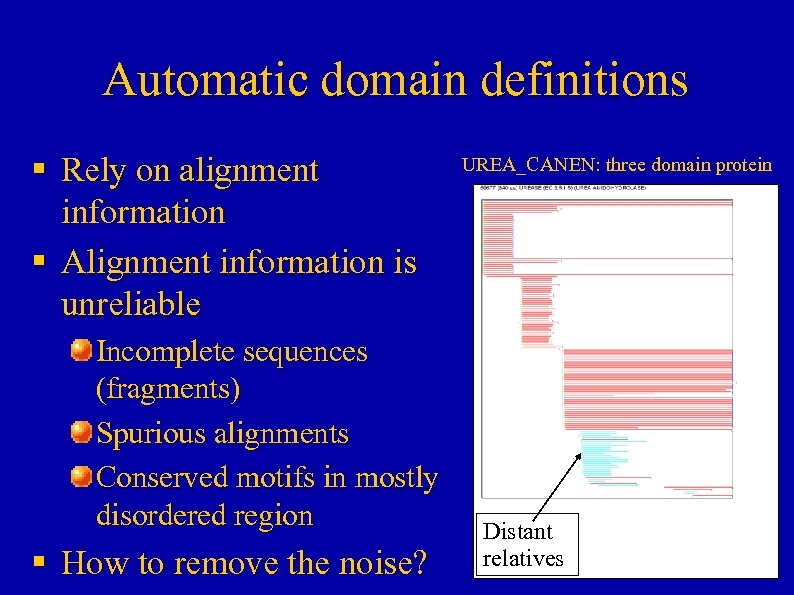 Automatic domain definitions § Rely on alignment information § Alignment information is unreliable Incomplete sequences (fragments) Spurious alignments Conserved motifs in mostly disordered region § How to remove the noise? UREA_CANEN: three domain protein Distant relatives
Automatic domain definitions § Rely on alignment information § Alignment information is unreliable Incomplete sequences (fragments) Spurious alignments Conserved motifs in mostly disordered region § How to remove the noise? UREA_CANEN: three domain protein Distant relatives
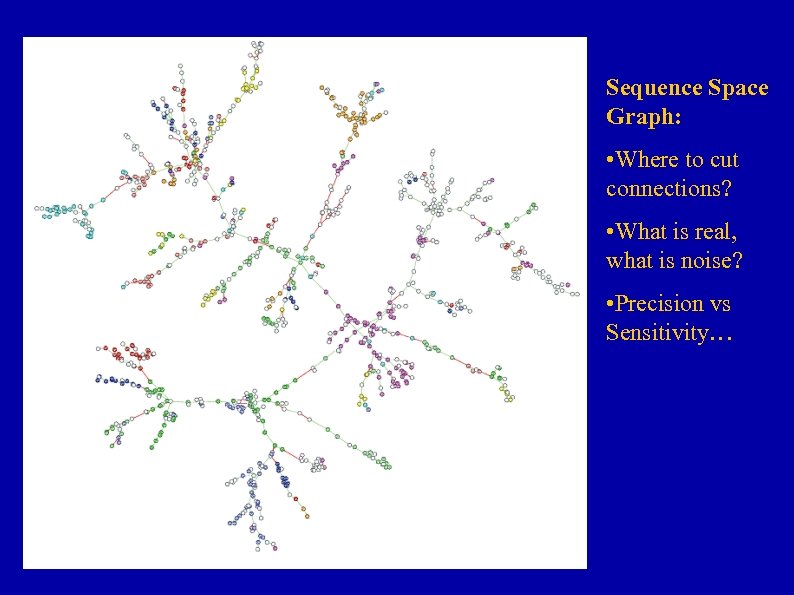 Sequence Space Graph: • Where to cut connections? • What is real, what is noise? • Precision vs Sensitivity…
Sequence Space Graph: • Where to cut connections? • What is real, what is noise? • Precision vs Sensitivity…
 ADDA § Holm. Group in-house database! http: //ekhidna. biocenter. helsinki. fi: 9801/sqgraph/pairsdb § Classification of non-redundant sequences 100% level: 1562243 sequences, 2697368 domains 40% level: 479740 sequences, 827925 domains § PFAM-A benchmark Sensitivity: 87% (average unification in single cluster) Selectivity: 98% (average purity of cluster) Coverage: 100% (all known proteins) [ Pfam ~50% ]
ADDA § Holm. Group in-house database! http: //ekhidna. biocenter. helsinki. fi: 9801/sqgraph/pairsdb § Classification of non-redundant sequences 100% level: 1562243 sequences, 2697368 domains 40% level: 479740 sequences, 827925 domains § PFAM-A benchmark Sensitivity: 87% (average unification in single cluster) Selectivity: 98% (average purity of cluster) Coverage: 100% (all known proteins) [ Pfam ~50% ]
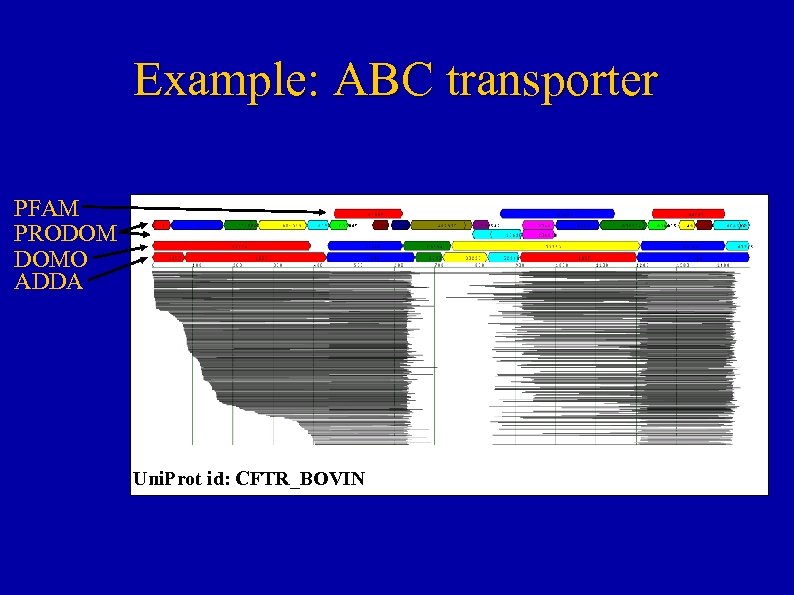 Example: ABC transporter PFAM PRODOM DOMO ADDA Uni. Prot id: CFTR_BOVIN
Example: ABC transporter PFAM PRODOM DOMO ADDA Uni. Prot id: CFTR_BOVIN
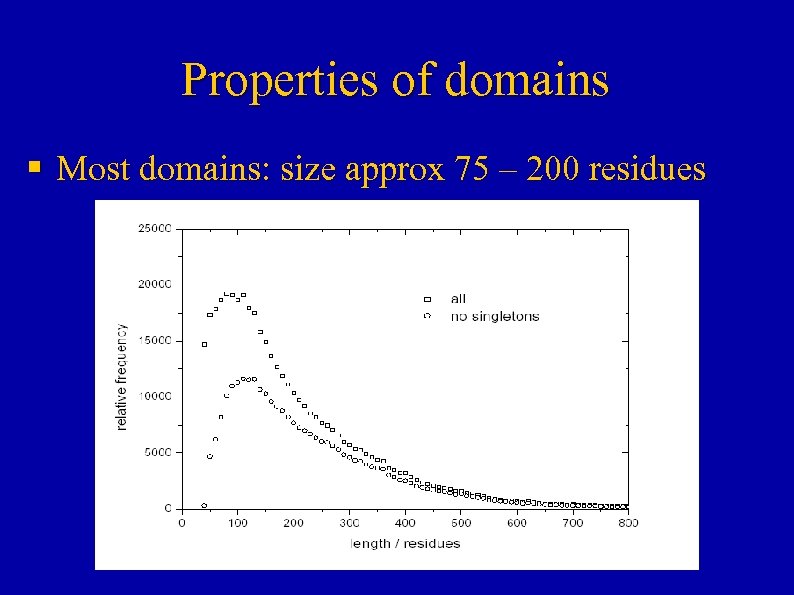 Properties of domains § Most domains: size approx 75 – 200 residues
Properties of domains § Most domains: size approx 75 – 200 residues
 So, you have a sequence. . . §. . . look it up in existing database – – SRS: http: //srs. ebi. ac. uk INTERPRO: http: //www. ebi. ac. uk/interpro §. . . search against existing family descriptions – – PFAM: http: //www. sanger. ac. uk/Software/Pfam SMART: http: //smart. embl-heidelberg. de PRINTS: http: //bioinf. man. ac. uk/dbbrowser/PRINTS PROSITE: http: //us. expasy. org/prosite §. . . look it up in ADDA
So, you have a sequence. . . §. . . look it up in existing database – – SRS: http: //srs. ebi. ac. uk INTERPRO: http: //www. ebi. ac. uk/interpro §. . . search against existing family descriptions – – PFAM: http: //www. sanger. ac. uk/Software/Pfam SMART: http: //smart. embl-heidelberg. de PRINTS: http: //bioinf. man. ac. uk/dbbrowser/PRINTS PROSITE: http: //us. expasy. org/prosite §. . . look it up in ADDA
 Manually Curated Protein Family Databases § PFAM (Hidden Markov Models) – http: //www. sanger. ac. uk/Software/Pfam § SMART (Hidden Markov Models) – http: //smart. embl-heidelberg. de § PROSITE (Regular Expressions, Profiles) – http: //au. expasy. org/prosite § PRINTS (combination of Profiles) – http: //bioinf. man. ac. uk/dbbrowser/PRINTS
Manually Curated Protein Family Databases § PFAM (Hidden Markov Models) – http: //www. sanger. ac. uk/Software/Pfam § SMART (Hidden Markov Models) – http: //smart. embl-heidelberg. de § PROSITE (Regular Expressions, Profiles) – http: //au. expasy. org/prosite § PRINTS (combination of Profiles) – http: //bioinf. man. ac. uk/dbbrowser/PRINTS
 Why a multiple alignment? § With a multiple alignment, we can guess which residues are “important” § secondary structure prediction § transmembrane segments prediction § homology modelling § guide to wet-lab EXPERIMENTATION! build a motif/profile and find more family members build phylogenetic trees Multiple Alignments are THE central object in protein sequence analysis!
Why a multiple alignment? § With a multiple alignment, we can guess which residues are “important” § secondary structure prediction § transmembrane segments prediction § homology modelling § guide to wet-lab EXPERIMENTATION! build a motif/profile and find more family members build phylogenetic trees Multiple Alignments are THE central object in protein sequence analysis!
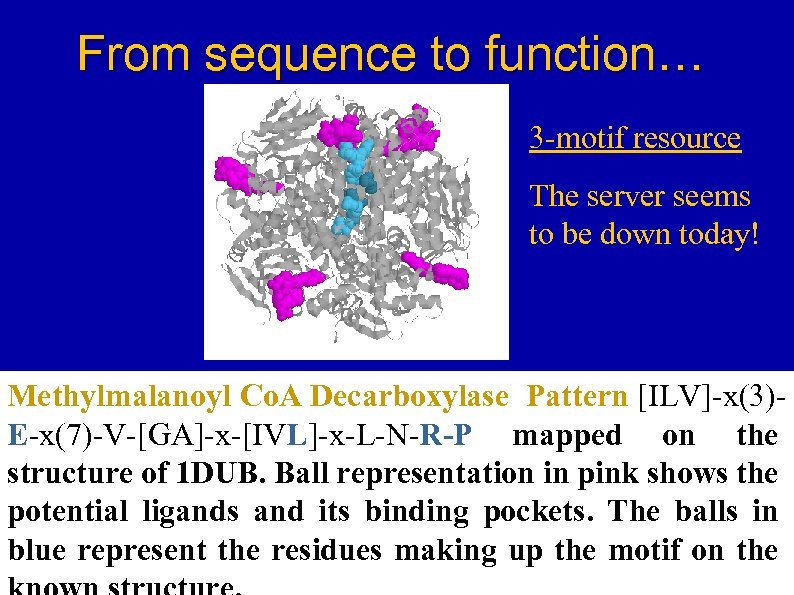 From sequence to function… 3 -motif resource The server seems to be down today! Methylmalanoyl Co. A Decarboxylase Pattern [ILV]-x(3)E-x(7)-V-[GA]-x-[IVL]-x-L-N-R-P mapped on the structure of 1 DUB. Ball representation in pink shows the potential ligands and its binding pockets. The balls in blue represent the residues making up the motif on the
From sequence to function… 3 -motif resource The server seems to be down today! Methylmalanoyl Co. A Decarboxylase Pattern [ILV]-x(3)E-x(7)-V-[GA]-x-[IVL]-x-L-N-R-P mapped on the structure of 1 DUB. Ball representation in pink shows the potential ligands and its binding pockets. The balls in blue represent the residues making up the motif on the


