718405a604a6fd6083b5cbd7ee2763eb.ppt
- Количество слайдов: 30
 Classifying Chemistry: Current Efforts in Canada Chemistry, Data & the Semantic Web 251 st ACS Chemistry Meeting, San Diego CA David Wishart University of Alberta March 15, 2016
Classifying Chemistry: Current Efforts in Canada Chemistry, Data & the Semantic Web 251 st ACS Chemistry Meeting, San Diego CA David Wishart University of Alberta March 15, 2016
 Chemical Classification – Why Do It? Zoologists Do It
Chemical Classification – Why Do It? Zoologists Do It
 Chemical Classification – Why Do It? Geologists Do It
Chemical Classification – Why Do It? Geologists Do It
 Chemical Classification – Why Do It? Astronomers Do It
Chemical Classification – Why Do It? Astronomers Do It
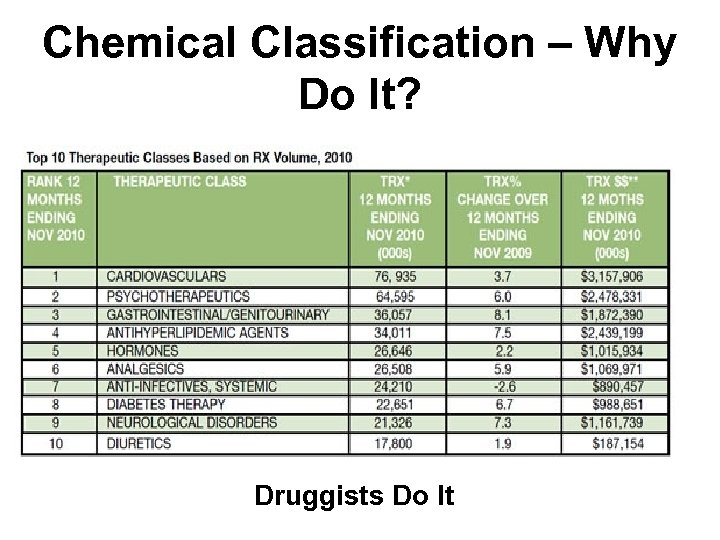 Chemical Classification – Why Do It? Druggists Do It
Chemical Classification – Why Do It? Druggists Do It
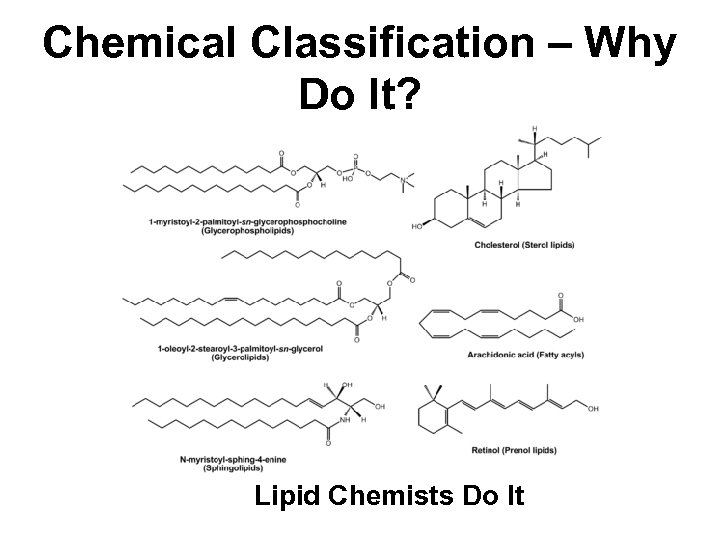 Chemical Classification – Why Do It? Lipid Chemists Do It
Chemical Classification – Why Do It? Lipid Chemists Do It
 Chemical Classification • Chemists started doing it (the periodic table of the elements) • Chemists led the way with standardized naming (IUPAC nomenclature) • Chemists led the way with hash keys and standardized identifiers (In. Ch. I) • But chemists are now far behind other fields with respect to compound classification
Chemical Classification • Chemists started doing it (the periodic table of the elements) • Chemists led the way with standardized naming (IUPAC nomenclature) • Chemists led the way with hash keys and standardized identifiers (In. Ch. I) • But chemists are now far behind other fields with respect to compound classification
 Chemical Classification Benefits • Gives “order” to a complex field • Forces clear and near-universal definitions • Establishes an ontology (a vocabulary of terms, concepts and relationships) • Improves searching and comparisons • Helps identify relationships or origins • Allows for automated annotations
Chemical Classification Benefits • Gives “order” to a complex field • Forces clear and near-universal definitions • Establishes an ontology (a vocabulary of terms, concepts and relationships) • Improves searching and comparisons • Helps identify relationships or origins • Allows for automated annotations
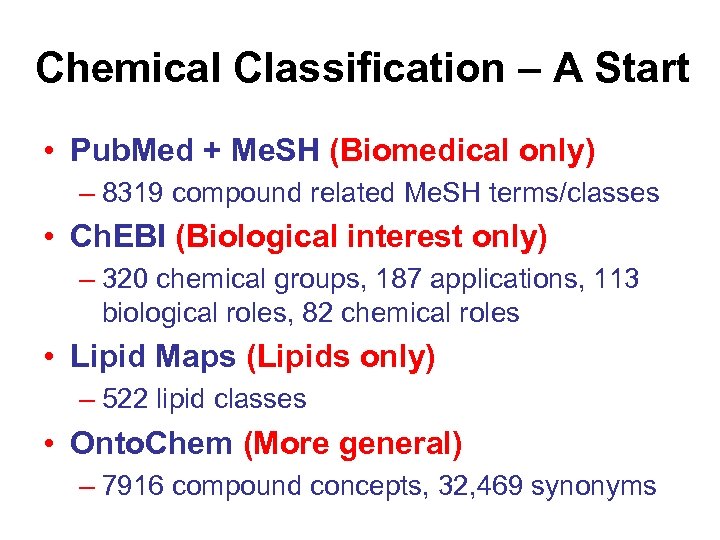 Chemical Classification – A Start • Pub. Med + Me. SH (Biomedical only) – 8319 compound related Me. SH terms/classes • Ch. EBI (Biological interest only) – 320 chemical groups, 187 applications, 113 biological roles, 82 chemical roles • Lipid Maps (Lipids only) – 522 lipid classes • Onto. Chem (More general) – 7916 compound concepts, 32, 469 synonyms
Chemical Classification – A Start • Pub. Med + Me. SH (Biomedical only) – 8319 compound related Me. SH terms/classes • Ch. EBI (Biological interest only) – 320 chemical groups, 187 applications, 113 biological roles, 82 chemical roles • Lipid Maps (Lipids only) – 522 lipid classes • Onto. Chem (More general) – 7916 compound concepts, 32, 469 synonyms
 Why We Got Into It
Why We Got Into It
 Why We Got Into It • Started manual classification for HMDB and Drug. Bank in 2005 (partial) • Growing number of requests from metabolomics researchers wanting compounds clustered by chemical type • Challenges with manually annotating multiple chemical/metabolite databases with large numbers (100, 000+) of different compounds
Why We Got Into It • Started manual classification for HMDB and Drug. Bank in 2005 (partial) • Growing number of requests from metabolomics researchers wanting compounds clustered by chemical type • Challenges with manually annotating multiple chemical/metabolite databases with large numbers (100, 000+) of different compounds
 How To Classify? • By chemical or biological origin? – Not always clear, requires manual annotation, not many classification categories • By function, application, chemical role? – Requires considerable manual effort, always changing, lots of categories, needs ontologies • By biological pathways or biological role? – Only works for biological compounds, not for industrial or synthetic compounds, lots of unknowns • By structure? – Lots of categories, existing classifications to build on, potentially automatable, works for all chemicals
How To Classify? • By chemical or biological origin? – Not always clear, requires manual annotation, not many classification categories • By function, application, chemical role? – Requires considerable manual effort, always changing, lots of categories, needs ontologies • By biological pathways or biological role? – Only works for biological compounds, not for industrial or synthetic compounds, lots of unknowns • By structure? – Lots of categories, existing classifications to build on, potentially automatable, works for all chemicals
 Classifying by Structure • Need to respect previous classification schemes (amino acids, lipids, etc. ) even if they are not “purely” structural • Need to find a preferred nomenclature (many names can exist for the same compound class) • Need clear definitions and well-defined categories • Need to handle hybrid or chimeric structures consistently and logically
Classifying by Structure • Need to respect previous classification schemes (amino acids, lipids, etc. ) even if they are not “purely” structural • Need to find a preferred nomenclature (many names can exist for the same compound class) • Need clear definitions and well-defined categories • Need to handle hybrid or chimeric structures consistently and logically
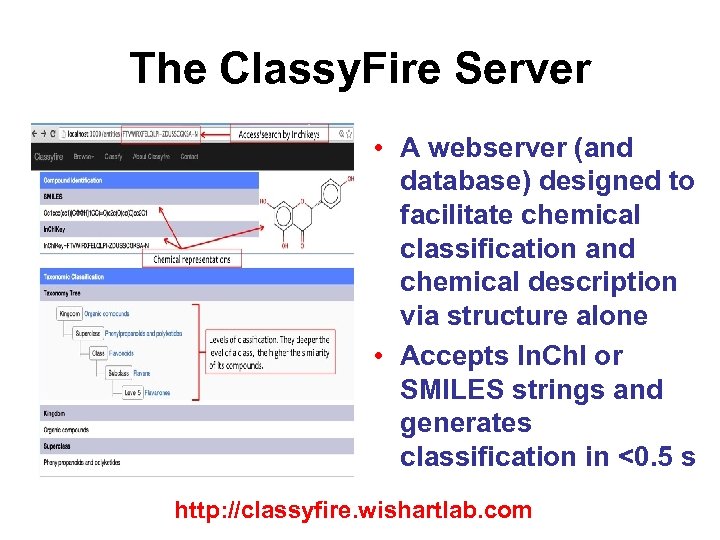 The Classy. Fire Server • A webserver (and database) designed to facilitate chemical classification and chemical description via structure alone • Accepts In. Ch. I or SMILES strings and generates classification in <0. 5 s http: //classyfire. wishartlab. com
The Classy. Fire Server • A webserver (and database) designed to facilitate chemical classification and chemical description via structure alone • Accepts In. Ch. I or SMILES strings and generates classification in <0. 5 s http: //classyfire. wishartlab. com
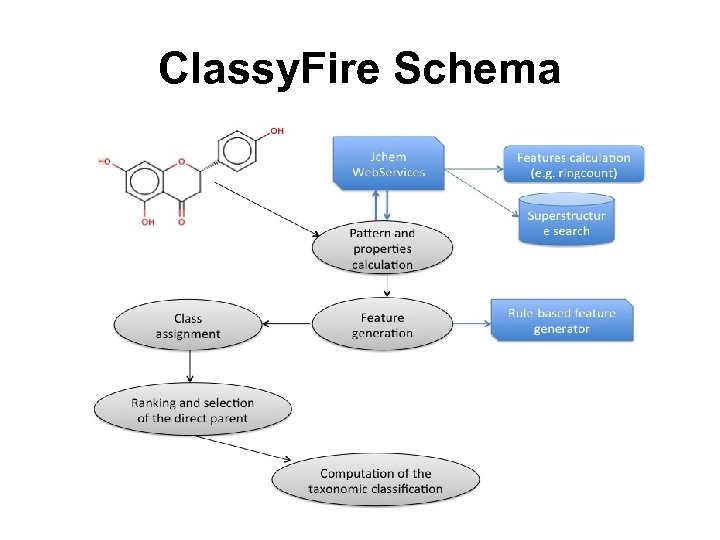 Classy. Fire Schema
Classy. Fire Schema
 Classy. Fire Features • Spans the chemical space from natural products to polymers, biomimmetics, inorganic & organic chemicals • Fully automated • Consistent and aligned with manual classification schemes in Me. SH (Pub. Chem), Ch. EBI and Lipid Maps • Every compound class is fully defined via a text description and a computable structure definition • Provides consistent naming system for chemical classes • Includes 4822 inorganic and organic compound classes • Has been used to classify all chemicals in Pub. Chem, Ch. EBI, KEGG, Lipid Maps, Drug. Bank, HMDB, YMDB, etc. All Public Chemicals Are Now Classified
Classy. Fire Features • Spans the chemical space from natural products to polymers, biomimmetics, inorganic & organic chemicals • Fully automated • Consistent and aligned with manual classification schemes in Me. SH (Pub. Chem), Ch. EBI and Lipid Maps • Every compound class is fully defined via a text description and a computable structure definition • Provides consistent naming system for chemical classes • Includes 4822 inorganic and organic compound classes • Has been used to classify all chemicals in Pub. Chem, Ch. EBI, KEGG, Lipid Maps, Drug. Bank, HMDB, YMDB, etc. All Public Chemicals Are Now Classified
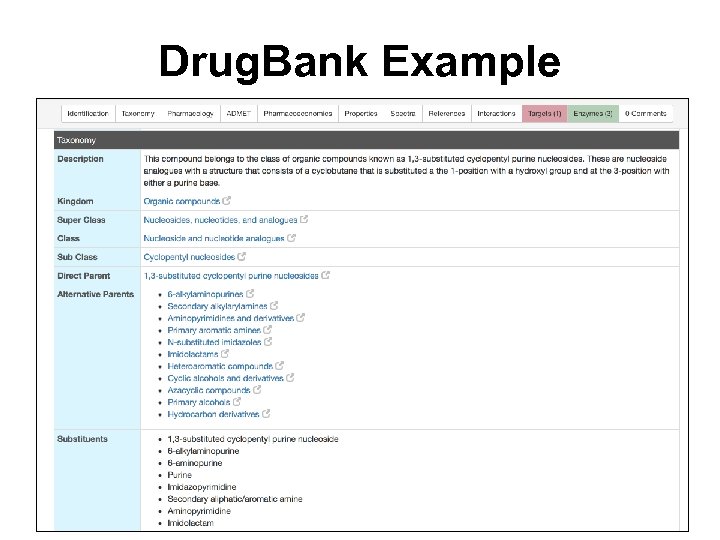 Drug. Bank Example
Drug. Bank Example
 Classification - What’s Next? • Feature Attributes – – Name/In. Ch. I 2 D/3 D structure Structural taxonomy Similarity • Physical Attributes – QSAR descriptors – Phys. Chem properties – Organoleptic qualities The 4 “F’s” • Functional Attributes – – – Hazard properties Health effects Disease associations Biological role Industrial role Membership list • Fate Attributes – Biological location – Origin – Pollutant status/fate
Classification - What’s Next? • Feature Attributes – – Name/In. Ch. I 2 D/3 D structure Structural taxonomy Similarity • Physical Attributes – QSAR descriptors – Phys. Chem properties – Organoleptic qualities The 4 “F’s” • Functional Attributes – – – Hazard properties Health effects Disease associations Biological role Industrial role Membership list • Fate Attributes – Biological location – Origin – Pollutant status/fate
 How To Get These Data? • Calculate chemical properties using open access or commercial tools • Automatically extract information or labels from existing public databases • Use software to cross-check data consistencies • Perform text mining to extract detailed relationships and/or knowledge • Do it manually
How To Get These Data? • Calculate chemical properties using open access or commercial tools • Automatically extract information or labels from existing public databases • Use software to cross-check data consistencies • Perform text mining to extract detailed relationships and/or knowledge • Do it manually
 The Tools
The Tools
 Data. Wrangler • Automated tool to calculate, extract and verify structure, In. Ch. I, names, synonyms, formula, MW, chemical properties, descriptions (local or Wikipedia), chemical classification (Classy. Fire), pathways, targets, reactions and “some” ontological terms • Accepts CAS, In. Ch. I, SMILES or name
Data. Wrangler • Automated tool to calculate, extract and verify structure, In. Ch. I, names, synonyms, formula, MW, chemical properties, descriptions (local or Wikipedia), chemical classification (Classy. Fire), pathways, targets, reactions and “some” ontological terms • Accepts CAS, In. Ch. I, SMILES or name
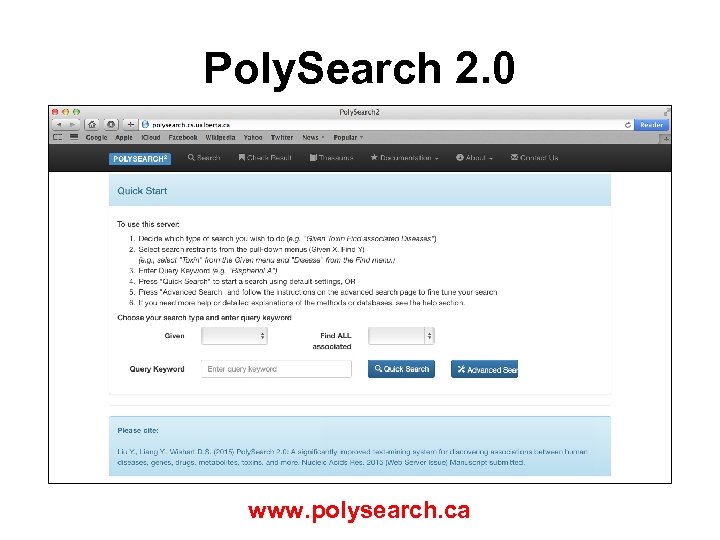
 Poly. Search 2. 0 • Online text-mining system for identifying relationships between human diseases, genes, proteins, drugs, metabolites, toxins, metabolic pathways, organs, tissues, subcellular organelles, positive health effects, negative health effects, drug actions, Gene Ontology terms, Me. SH terms, ICD-10 medical codes, biological taxonomies and chemical taxonomies • Supports generalized 'Given X, find all associated Ys' query, where X and Y can be selected from the above biomedical entities
Poly. Search 2. 0 • Online text-mining system for identifying relationships between human diseases, genes, proteins, drugs, metabolites, toxins, metabolic pathways, organs, tissues, subcellular organelles, positive health effects, negative health effects, drug actions, Gene Ontology terms, Me. SH terms, ICD-10 medical codes, biological taxonomies and chemical taxonomies • Supports generalized 'Given X, find all associated Ys' query, where X and Y can be selected from the above biomedical entities
 Poly. Search 2. 0 www. polysearch. ca
Poly. Search 2. 0 www. polysearch. ca
 Poly. Search 2. 0 • Maintain a local API for “heavy” queries • Searches local versions of Wikipedia, PMC-Central, Pub. Med, NCBI On-line textbooks, US Patent abstracts, Uni. Prot, Drug. Bank, HMDB, T 3 DB, ECMDB, YMDB, Daily. Med, KEGG, OMIM, HPRD, Meta. Cyc, NCBI taxonomy • 165 Gbytes of data
Poly. Search 2. 0 • Maintain a local API for “heavy” queries • Searches local versions of Wikipedia, PMC-Central, Pub. Med, NCBI On-line textbooks, US Patent abstracts, Uni. Prot, Drug. Bank, HMDB, T 3 DB, ECMDB, YMDB, Daily. Med, KEGG, OMIM, HPRD, Meta. Cyc, NCBI taxonomy • 165 Gbytes of data
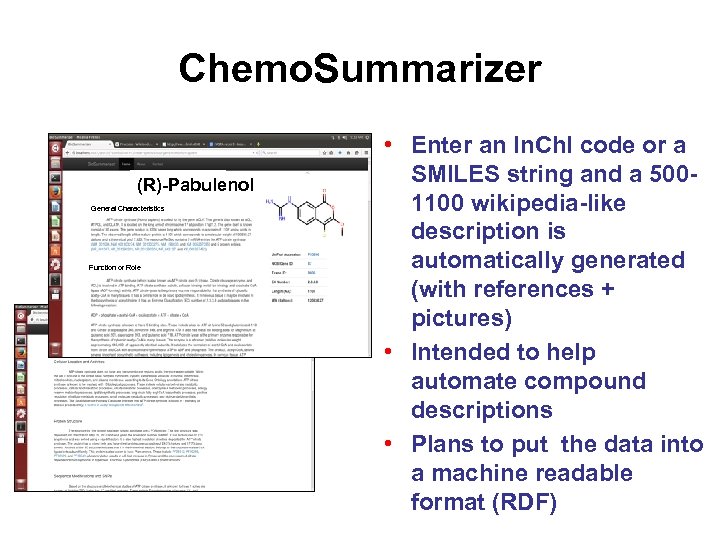 Chemo. Summarizer (R)-Pabulenol General Characteristics Function or Role • Enter an In. Ch. I code or a SMILES string and a 5001100 wikipedia-like description is automatically generated (with references + pictures) • Intended to help automate compound descriptions • Plans to put the data into a machine readable format (RDF)
Chemo. Summarizer (R)-Pabulenol General Characteristics Function or Role • Enter an In. Ch. I code or a SMILES string and a 5001100 wikipedia-like description is automatically generated (with references + pictures) • Intended to help automate compound descriptions • Plans to put the data into a machine readable format (RDF)
 Looking Forward • Consistent and complete mapping between Me. SH, Lipid Maps, Ch. EBI, Onto. Chem and Classyfire – a common chemistry taxonomy • A common, comprehensive chemistry ontology (the 4 “F’s”) • Common data formats and exchange protocols between major chemistry databases (RDF)
Looking Forward • Consistent and complete mapping between Me. SH, Lipid Maps, Ch. EBI, Onto. Chem and Classyfire – a common chemistry taxonomy • A common, comprehensive chemistry ontology (the 4 “F’s”) • Common data formats and exchange protocols between major chemistry databases (RDF)
 Looking Forward • New tools that use ontologies to generate novel information and novel hypotheses • Tools that extract previously unknown or little-known information from the literature • Creation of automated, open access chem-ontology servers like AMIGO (for gene ontology) – call it AMICO?
Looking Forward • New tools that use ontologies to generate novel information and novel hypotheses • Tools that extract previously unknown or little-known information from the literature • Creation of automated, open access chem-ontology servers like AMIGO (for gene ontology) – call it AMICO?
 Looking Forward • • Filling in missing data on known cmpds Predict Likely Biological Function Predict Likely Industrial Role Predict Likely Source Organisms Predict Likely Toxicity or Hazard Predict Likely Health Effects Predict Likely Pathways/Targets Predict Likely Organoleptic Properties
Looking Forward • • Filling in missing data on known cmpds Predict Likely Biological Function Predict Likely Industrial Role Predict Likely Source Organisms Predict Likely Toxicity or Hazard Predict Likely Health Effects Predict Likely Pathways/Targets Predict Likely Organoleptic Properties
 Looking Forward • Analyzing Newly Synthesized or Newly Discovered Compounds • Predict Likely Biological Function • Predict Likely Industrial Role • Predict Likely Source Organisms • Predict Likely Toxicity or Hazard • Predict Likely Health Effects • Predict Likely Pathways/Targets • Predict Likely Organoleptic Properties
Looking Forward • Analyzing Newly Synthesized or Newly Discovered Compounds • Predict Likely Biological Function • Predict Likely Industrial Role • Predict Likely Source Organisms • Predict Likely Toxicity or Hazard • Predict Likely Health Effects • Predict Likely Pathways/Targets • Predict Likely Organoleptic Properties
 Acknowledgements • • Yannick Djoumbou Tanvir Sajed Yifeng Lu Zachery Budinsky David Arndt Craig Knox Michael Wilson
Acknowledgements • • Yannick Djoumbou Tanvir Sajed Yifeng Lu Zachery Budinsky David Arndt Craig Knox Michael Wilson


