f325e510c4c98ebd19c17f7ec86741b4.ppt
- Количество слайдов: 11
 CHROMOSOME 7 SEQUENCING T 0848 TM 15 T 0731 T 0966 TG 438 T 1257 TG 216 CT 54 TM 18 T 1347 T 0676 T 1497 T 1962 T 1414 T 1428 T 1355 T 1112 Current status and perspective EU-SOL 2008 November 13 -16, Toulouse, FRANCE
CHROMOSOME 7 SEQUENCING T 0848 TM 15 T 0731 T 0966 TG 438 T 1257 TG 216 CT 54 TM 18 T 1347 T 0676 T 1497 T 1962 T 1414 T 1428 T 1355 T 1112 Current status and perspective EU-SOL 2008 November 13 -16, Toulouse, FRANCE
 INTRODUCTION • Chromosome 7 : – 27 Mbases of gene-dense euchromatin first estimated – 25. 1 Mb according to Chang et al, 2008 – Initially 270 BACs to be sequenced (new estimation 250 BACs) • BAC selection: – Selection and verification of the BACs on chromosome 7 by GBF (INP-T) – Generation of new tools and resources for rapid selection of the BACs to be sequenced • BAC sequencing: – Private Company "Cogenics" – From draft production to BAC finishing (phase 3) EU-SOL 2008 November 13 -16, Toulouse, FRANCE
INTRODUCTION • Chromosome 7 : – 27 Mbases of gene-dense euchromatin first estimated – 25. 1 Mb according to Chang et al, 2008 – Initially 270 BACs to be sequenced (new estimation 250 BACs) • BAC selection: – Selection and verification of the BACs on chromosome 7 by GBF (INP-T) – Generation of new tools and resources for rapid selection of the BACs to be sequenced • BAC sequencing: – Private Company "Cogenics" – From draft production to BAC finishing (phase 3) EU-SOL 2008 November 13 -16, Toulouse, FRANCE
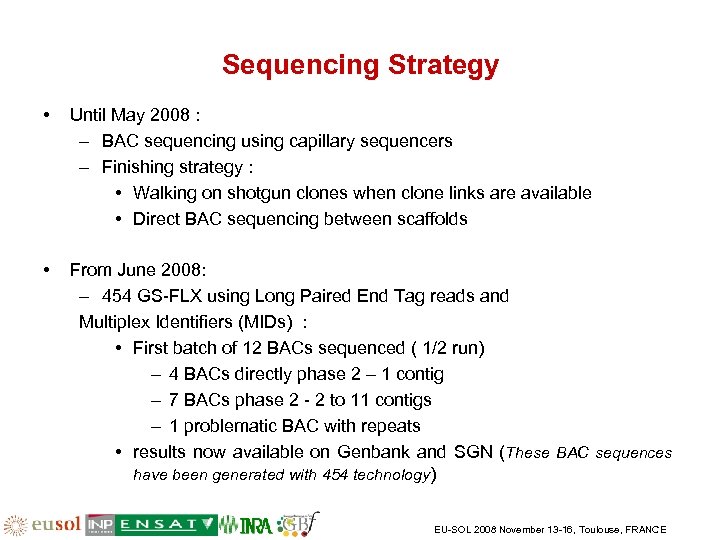 Sequencing Strategy • Until May 2008 : – BAC sequencing using capillary sequencers – Finishing strategy : • Walking on shotgun clones when clone links are available • Direct BAC sequencing between scaffolds • From June 2008: – 454 GS-FLX using Long Paired End Tag reads and Multiplex Identifiers (MIDs) : • First batch of 12 BACs sequenced ( 1/2 run) – 4 BACs directly phase 2 – 1 contig – 7 BACs phase 2 - 2 to 11 contigs – 1 problematic BAC with repeats • results now available on Genbank and SGN (These BAC sequences have been generated with 454 technology) EU-SOL 2008 November 13 -16, Toulouse, FRANCE
Sequencing Strategy • Until May 2008 : – BAC sequencing using capillary sequencers – Finishing strategy : • Walking on shotgun clones when clone links are available • Direct BAC sequencing between scaffolds • From June 2008: – 454 GS-FLX using Long Paired End Tag reads and Multiplex Identifiers (MIDs) : • First batch of 12 BACs sequenced ( 1/2 run) – 4 BACs directly phase 2 – 1 contig – 7 BACs phase 2 - 2 to 11 contigs – 1 problematic BAC with repeats • results now available on Genbank and SGN (These BAC sequences have been generated with 454 technology) EU-SOL 2008 November 13 -16, Toulouse, FRANCE
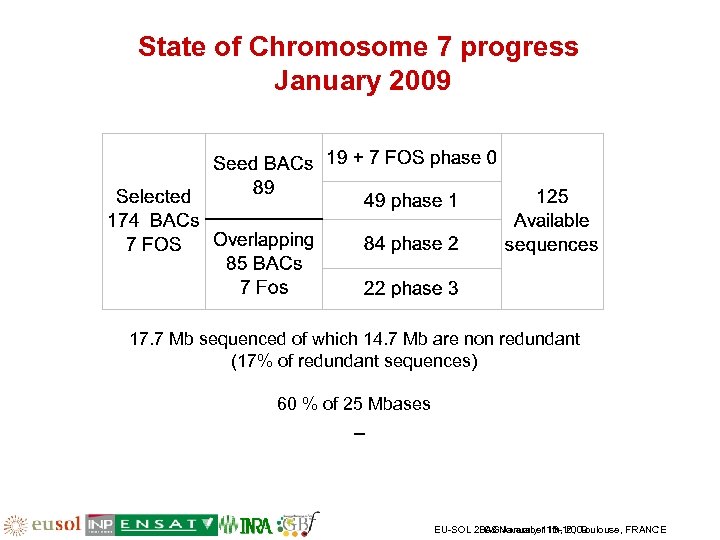 State of Chromosome 7 progress January 2009 17. 7 Mb sequenced of which 14. 7 Mb are non redundant (17% of redundant sequences) 60 % of 25 Mbases EU-SOL 2008 November 13 -16, Toulouse, FRANCE PAG January 11 th, 2009
State of Chromosome 7 progress January 2009 17. 7 Mb sequenced of which 14. 7 Mb are non redundant (17% of redundant sequences) 60 % of 25 Mbases EU-SOL 2008 November 13 -16, Toulouse, FRANCE PAG January 11 th, 2009
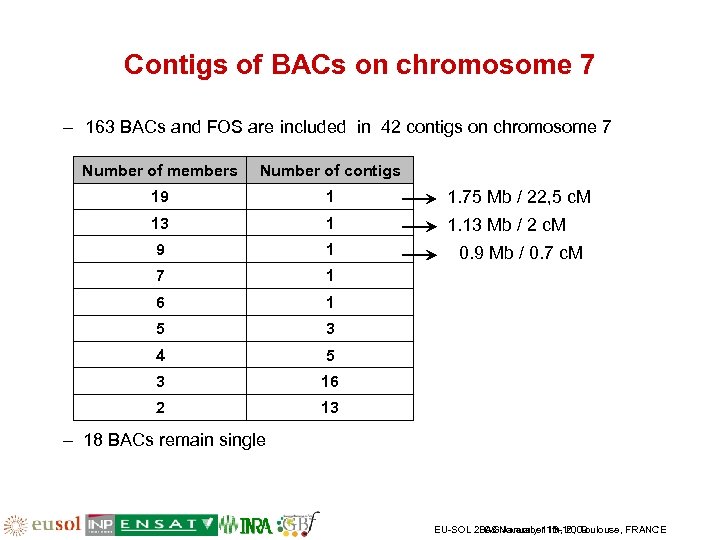 Contigs of BACs on chromosome 7 – 163 BACs and FOS are included in 42 contigs on chromosome 7 Number of members Number of contigs 19 1 1. 75 Mb / 22, 5 c. M 13 1 1. 13 Mb / 2 c. M 9 1 7 1 6 1 5 3 4 5 3 16 2 13 0. 9 Mb / 0. 7 c. M – 18 BACs remain single EU-SOL 2008 November 13 -16, Toulouse, FRANCE PAG January 11 th, 2009
Contigs of BACs on chromosome 7 – 163 BACs and FOS are included in 42 contigs on chromosome 7 Number of members Number of contigs 19 1 1. 75 Mb / 22, 5 c. M 13 1 1. 13 Mb / 2 c. M 9 1 7 1 6 1 5 3 4 5 3 16 2 13 0. 9 Mb / 0. 7 c. M – 18 BACs remain single EU-SOL 2008 November 13 -16, Toulouse, FRANCE PAG January 11 th, 2009
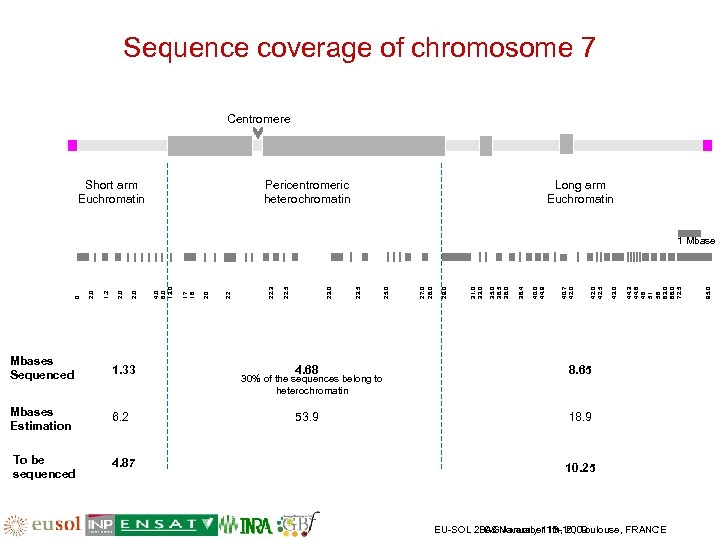 Sequence coverage of chromosome 7 Centromere Short arm Euchromatin Pericentromeric heterochromatin Long arm Euchromatin Mbases Sequenced 1. 33 Mbases Estimation 6. 2 To be sequenced 4. 87 4. 68 18. 9 30% of the sequences belong to heterochromatin 10. 25 EU-SOL 2008 November 13 -16, Toulouse, FRANCE PAG January 11 th, 2009 95. 0 44. 3 44. 6 46 51 56 63. 0 68. 0 72. 5 8. 65 53. 9 43. 0 42. 5 40. 7 42. 0 40. 0 44. 9 38. 4 35. 0 36. 5 38. 0 31. 0 33. 0 29. 0 27. 0 28. 0 25. 0 23. 5 23. 0 22. 5 22. 3 22 20 17 18 4. 0 6. 0 13. 0 2. 0 1. 2 2. 0 0 1 Mbase
Sequence coverage of chromosome 7 Centromere Short arm Euchromatin Pericentromeric heterochromatin Long arm Euchromatin Mbases Sequenced 1. 33 Mbases Estimation 6. 2 To be sequenced 4. 87 4. 68 18. 9 30% of the sequences belong to heterochromatin 10. 25 EU-SOL 2008 November 13 -16, Toulouse, FRANCE PAG January 11 th, 2009 95. 0 44. 3 44. 6 46 51 56 63. 0 68. 0 72. 5 8. 65 53. 9 43. 0 42. 5 40. 7 42. 0 40. 0 44. 9 38. 4 35. 0 36. 5 38. 0 31. 0 33. 0 29. 0 27. 0 28. 0 25. 0 23. 5 23. 0 22. 5 22. 3 22 20 17 18 4. 0 6. 0 13. 0 2. 0 1. 2 2. 0 0 1 Mbase
 Generation of 3 D-DNA pools and Macroarray filters from BAC and Fosmids libraries These resources were generated in collaboration with the French Plant Genomic Resource Centre (http: //cnrgv. toulouse. inra. fr/) 3 D DNA pools - Half of the Hind. III BAC library : 7. 8 x - The entire Mbo. I BAC library : 7. 5 x Available for the community and already distributed to Spain, Italy, UK, USA, Israel, India, Germany Macroarray filters - The entire Eco. RI BAC library : 8 x - Half of the Fosmids library (150000 including 50000 end sequenced) : 6. 1 x Available for the community : the filters can be either sent for hybridization or the CNRGV can proceed to the hybridizations EU-SOL 2008 November 13 -16, Toulouse, FRANCE
Generation of 3 D-DNA pools and Macroarray filters from BAC and Fosmids libraries These resources were generated in collaboration with the French Plant Genomic Resource Centre (http: //cnrgv. toulouse. inra. fr/) 3 D DNA pools - Half of the Hind. III BAC library : 7. 8 x - The entire Mbo. I BAC library : 7. 5 x Available for the community and already distributed to Spain, Italy, UK, USA, Israel, India, Germany Macroarray filters - The entire Eco. RI BAC library : 8 x - Half of the Fosmids library (150000 including 50000 end sequenced) : 6. 1 x Available for the community : the filters can be either sent for hybridization or the CNRGV can proceed to the hybridizations EU-SOL 2008 November 13 -16, Toulouse, FRANCE
 Details on the Macroarray filters (high density) • 108 plates 384 spotted on 1 membrane (22 x 22 cm) with duplicated spots on a 6 x 6 pattern. • Eco. RI BAC library has been spotted on 2 filters • Fosmids library has been spotted on 4 filters • Image analysis software needed : HDFR 3 by INCOGEN EU-SOL 2008 November 13 -16, Toulouse, FRANCE
Details on the Macroarray filters (high density) • 108 plates 384 spotted on 1 membrane (22 x 22 cm) with duplicated spots on a 6 x 6 pattern. • Eco. RI BAC library has been spotted on 2 filters • Fosmids library has been spotted on 4 filters • Image analysis software needed : HDFR 3 by INCOGEN EU-SOL 2008 November 13 -16, Toulouse, FRANCE
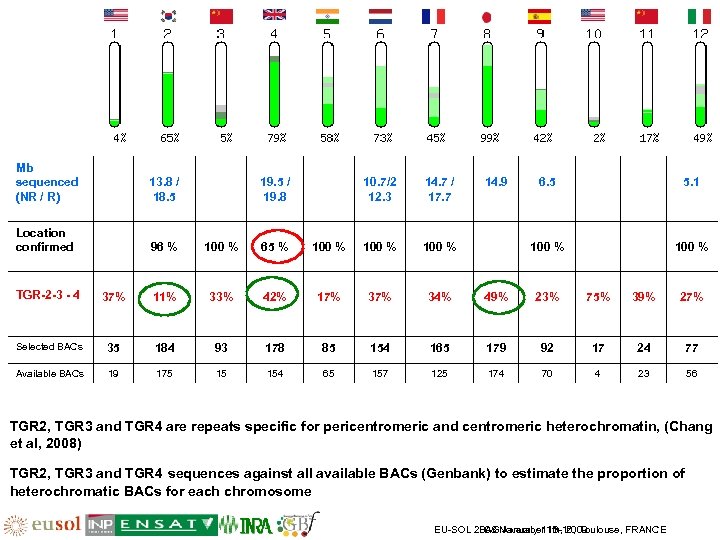 Mb sequenced (NR / R) 13. 8 / 18. 5 Location confirmed 96 % 100 % 65 % 19. 5 / 19. 8 10. 7/2 12. 3 14. 7 / 17. 7 100 % 14. 9 100 % 6. 5 5. 1 100 % TGR-2 -3 - 4 37% 11% 33% 42% 17% 34% 49% 23% 75% 39% 27% Selected BACs 35 184 93 178 85 154 165 179 92 17 24 77 Available BACs 19 175 15 154 65 157 125 174 70 4 23 56 TGR 2, TGR 3 and TGR 4 are repeats specific for pericentromeric and centromeric heterochromatin, (Chang et al, 2008) TGR 2, TGR 3 and TGR 4 sequences against all available BACs (Genbank) to estimate the proportion of heterochromatic BACs for each chromosome EU-SOL 2008 November 13 -16, Toulouse, FRANCE PAG January 11 th, 2009
Mb sequenced (NR / R) 13. 8 / 18. 5 Location confirmed 96 % 100 % 65 % 19. 5 / 19. 8 10. 7/2 12. 3 14. 7 / 17. 7 100 % 14. 9 100 % 6. 5 5. 1 100 % TGR-2 -3 - 4 37% 11% 33% 42% 17% 34% 49% 23% 75% 39% 27% Selected BACs 35 184 93 178 85 154 165 179 92 17 24 77 Available BACs 19 175 15 154 65 157 125 174 70 4 23 56 TGR 2, TGR 3 and TGR 4 are repeats specific for pericentromeric and centromeric heterochromatin, (Chang et al, 2008) TGR 2, TGR 3 and TGR 4 sequences against all available BACs (Genbank) to estimate the proportion of heterochromatic BACs for each chromosome EU-SOL 2008 November 13 -16, Toulouse, FRANCE PAG January 11 th, 2009
 Next. Gen Sequencing strategy • France plan to achieve a 10 x coverage by 454 Titanum with PET (starting in February) Long- • During this we will continue sequencing of batches of BACs and Fosmids by 454 • After achieving the WGS we will sequence more BACs and Fos to fill the gaps on chromosome 7 gene-rich regions • Need for tight coordination between different inititives (NL, Italy, France, USA, EU 6 SOL) • Use of the same DNA sample to be sequenced by all partners ? EU-SOL 2008 November 13 -16, Toulouse, FRANCE PAG January 11 th, 2009
Next. Gen Sequencing strategy • France plan to achieve a 10 x coverage by 454 Titanum with PET (starting in February) Long- • During this we will continue sequencing of batches of BACs and Fosmids by 454 • After achieving the WGS we will sequence more BACs and Fos to fill the gaps on chromosome 7 gene-rich regions • Need for tight coordination between different inititives (NL, Italy, France, USA, EU 6 SOL) • Use of the same DNA sample to be sequenced by all partners ? EU-SOL 2008 November 13 -16, Toulouse, FRANCE PAG January 11 th, 2009
 Contributors GBF (INRA/INP-ENSAT) Cogenics CNRGV Murielle Philippot Pierre Frasse Mohamed Zouine Farid Regad Mondher Bouzayen Stéphanie Penaud Hervé Duborjal Marcel de. Leeuw Diliana Dimova Réjane Beugnot François Pons Hélène Bergès Sonia Vautrin Elisa Prat Wageningen (NL) Colorado (USA) Hans De Jong Dorà Szinay Steve Stack EU-SOL 2008 November 13 -16, Toulouse, FRANCE
Contributors GBF (INRA/INP-ENSAT) Cogenics CNRGV Murielle Philippot Pierre Frasse Mohamed Zouine Farid Regad Mondher Bouzayen Stéphanie Penaud Hervé Duborjal Marcel de. Leeuw Diliana Dimova Réjane Beugnot François Pons Hélène Bergès Sonia Vautrin Elisa Prat Wageningen (NL) Colorado (USA) Hans De Jong Dorà Szinay Steve Stack EU-SOL 2008 November 13 -16, Toulouse, FRANCE


