Chrom School Novosibirsk JLL.pptx
- Количество слайдов: 47

Chromatography School 2014 Novosibirsk Jean Luc Lognonné, Ph. D Proteomic Market Manager February 11, 2018

§ INTRODUCTION TO CHROMATOGRAPHY §Biomolecules Interaction Analysis §Regulatory & technical considerations for effective monitoring of host cell protein (HCP) impurities

What is Chromatography? Chromatography (from the Greek χρώμα: chroma = color and γραφειν: "grafein" = write) Collective term for a family of laboratory techniques for the separation of mixtures. An established technique established in 1906 by Mikhail Semenovich Tsvet (Russia). He used the technique to separate various plant pigments such as chlorophyll, xanthophylls, Carotenoids. Chlorophylla (Chromatography on thin layer)
What is Chromatography? § Chromatography is a Separative Technique that allows for the separation of molecules contained in a complex mixture § This is allowed by passing it through a medium in which components will move at different rate Separation through Medium Based on the Protein Properties: § Size, shape and conformation § Charge and isoelectric point § Hydrophobicity § Affinity to other molecules

Examples of Properties Exploitable on a Protein Molecule Positively-Charged basic residues Ion Exchange Hydrophobic « patch » Hydrophobic Interaction Macromolecular dimensions Gel Filtration, Size Exclusion Ligand binding pocket (active site) Affinity « tag » Affinity Negatively-Charged acidic residues Ion Exchange The charged groups, hydrophobic regions, size, solvation, tags affect the biophysical properties of the molecule and largely determine its purification behavior

§ § Liquid chromatography methods to isolate any type of protein Separating of biomolecules based on their physiochemical characteristics – polarity (hydrophobicity) HIC, RP – ionic characteristics (charge) IEX – size/mass (included/excluded from pores) GF, SEC – specificity (ligand binding, affinity) AC These properties allow molecules to become separated on the matrix/resin/support

Basic Chromatography Steps Pack & Equilibrate Column - mobile phase which will facilitate adsorption of the protein Apply Sample - mobile phase that will keep solubilize the protein & facilitate quantitative adsorption Wash off contaminants - mobile phase that will wash off unbound or weakly adsorbed protein Elute target protein - mobile phase that will increase dissociation constant (gradient or isocratic) Wash/Clean in Place - extreme p. H, ionic strength or organic solvents to remove strongly adsorbed components Regenerate column for reuse - convert adsorbent to reference state before a new separation Load Sample Elute Wash

ION EXCHANGE Buffer p. H/Protein p. I TITRATION CURVE + binds cation exchanger net protein COOH H 2 N +H 3 N 2 stability range COOH p. I H 2 N charge stability range denaturation - denaturation COO- binds anion exchanger 10 p. H

ION EXCHANGE Buffer p. H/Protein p. I binds cation exchanger + +H 3 N COOH net protein 2 10 p. H p. I charge - H 2 N COO- binds anion exchanger
What is the difference between FPLC and HPLC? § High Pressure Liquid Chromatography (HPLC) –Launched in the 60 s to separate and Qualify chemical molecules –Refered as « Analytical » § Fast Protein Liquid Chromatography (or Fast Performance Liquid Chromatography, as would say Pharmacia) –Launched in the 80 s to purify Recombinant Proteins –Referred to as « Preparative » and non-denaturating conditions –Highly efficient , easy to use and relatively inexpensive § The differences? –Solvent and column pressure –Type of Lab and applications: HPLC is for Chemist, FPLC is for Biochemist!

How to justify a FPLC system rather than Low Pressure? § It’s all about Resolution and Capacity –To increase the resolution of a separation (i. e. the « Quality » of the separation) using the same technique, one way is to decrease the size of the beads: More surface § More interaction spots § –Smaller beads = Higher Back-pressure –Typical techniques that have small beads are IEX, SEC and HIC

Solid Phase Particle Sizes Low Pressure : ~20 psi Low resolution Large beads : ~ 50 µm Medium Pressure : > 300 psi Medium resolution Medium beads : 10 - 15 µm High Pressure : > 1000 psi High resolution Small beads : 3 - 5 µm

The Chromatogram & Nomenclature Peak Elution volume First Peak, the void peak, Goes straight the column Without interacting with it. This is the unbound material. Amount of protein, as read by a UV monitor, is usually on the y-axis Vo Baseline Ve Volume Last Peak, this is the Total Volume of the column Vt Sometimes you have an axis on this side too, showing the gradient concentration Time or volume are always On the x-axis
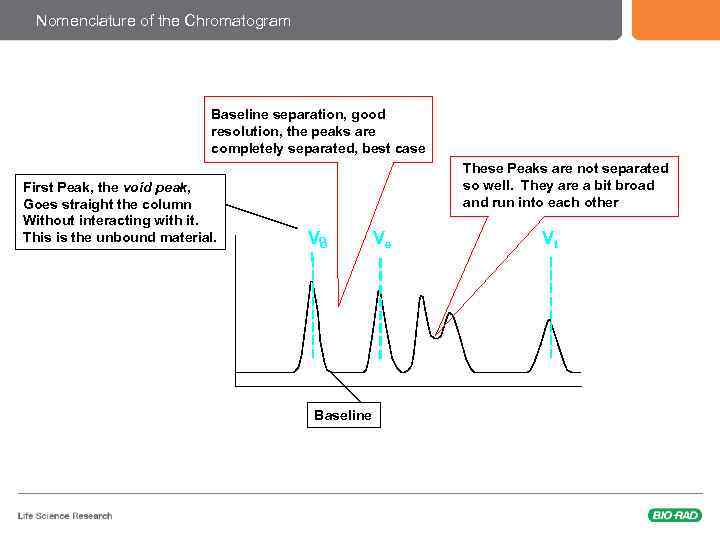
Nomenclature of the Chromatogram Baseline separation, good resolution, the peaks are completely separated, best case First Peak, the void peak, Goes straight the column Without interacting with it. This is the unbound material. These Peaks are not separated so well. They are a bit broad and run into each other Vo o Baseline Ve Vt
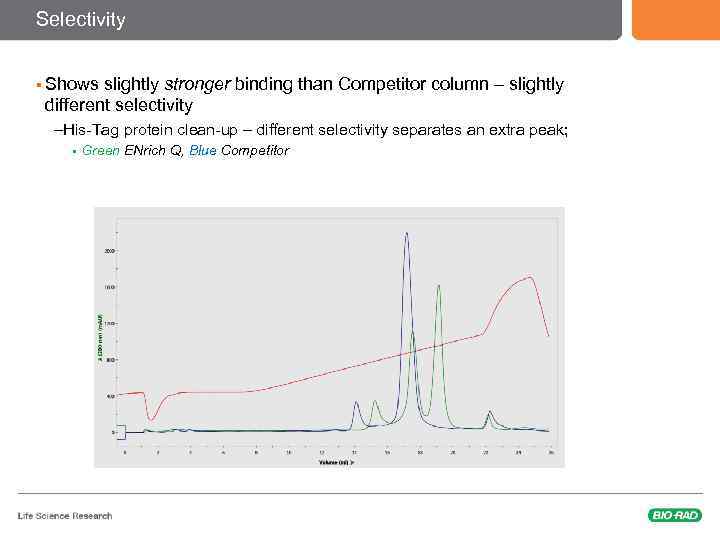
Selectivity § Shows slightly stronger binding than Competitor column – slightly different selectivity –His-Tag protein clean-up – different selectivity separates an extra peak; § Green ENrich Q, Blue Competitor

Liquid Chromatography: An Easy Principle Mobile Phase (Buffer) Sample Stationary Phase (Media)
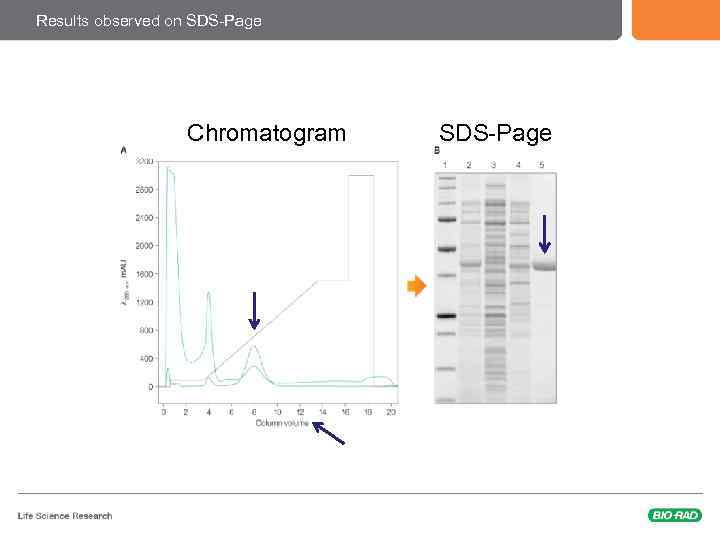
Results observed on SDS-Page Chromatogram SDS-Page
Biomolecules Interaction Analysis
Biomolecules Interaction Analysis: Brief Summary § Following an era dominated by Genomics and Gene regulation, a high interest is raising for Protein Regulation and Function § Protein Interactions are very difficult to predict – 20 different Aminoacids = Endless Configurations –Some known peptidic sequences § Multiple Methods have been developped to characterize Protein-Biomolecules Interactions

Protein Interaction Analysis: Protein-Antibody Interaction §Protein-Antibody interaction characterization is critical for: –Biotech developing Western Blot antibodies or ELISA –Pharma developing Therapeutics antibodies –Labs developing their own antibodies §Current methods of screening include: –ELISA –Western Blots / Dot Blots… • Time Consuming • Only end-point results = Interaction at equilibrium

Protein Interaction Analysis: Protein-Protein or Protein-DNA Interaction § Common Standard Techniques for Protein-Protein Interactions Analysis: –FRET (Fluorescence Resonance Energy Transfer) § Powerful because can be done in vivo § No standardize instrument, difficult referencing § Necessitate Labelling with a fluorophore –Co-IP/Pulldown § Necessitate good antibodies: For Immunoprecipitation and for Western Blot § Could add a Tag to Pulldown the Bait § Alternative: Coupling to Mass Spectrometry to « fish » potential partners • Need to add a Tag or have Good Antibodies • Only end-point results = interaction at equilibrium • Long Time to Result
Protein Interaction Analysis: Protein-Small Molecule § Essential for Drug Discovery, in Pharma, CROs and Research Institutes § Can be done By Fluorescence: –Requires a Fluorophore on the small Molecule (direct or indirect), and a microarrayer –Modifying the Small Molecule may change the function –Modifying the Small Molecule might not be possible l • Requires a Tag or Fluorescent Properties • No/not accurate ka and kd data
What Solution could be proposed? § Many drawbacks in Biomolecule interaction analysis Techniques: –Long and Tedious –Only Equilibrium –Need to Tag the Protein (Fluorophore or for Pulldown) –Low Throughput… § Needed a Technique that could improve the means of Understanding Biomolecules Interactions Biomolecule Interaction Analysis: The Surface Plasmon Resonance Solution

How Can this Principles be Used for Biomolecule interactions? The Molecule attached to the surface is called the « Ligand » The Molecule that flows in the Microfluidics is called the « Analyte » § Step 1: Immobilisation of the Ligand Baseline § Step 2: Injection of the Analyte – Using Microfluidics Association § Step 3: Wash the Analyte away Dissociation Response 2 1 3 Time
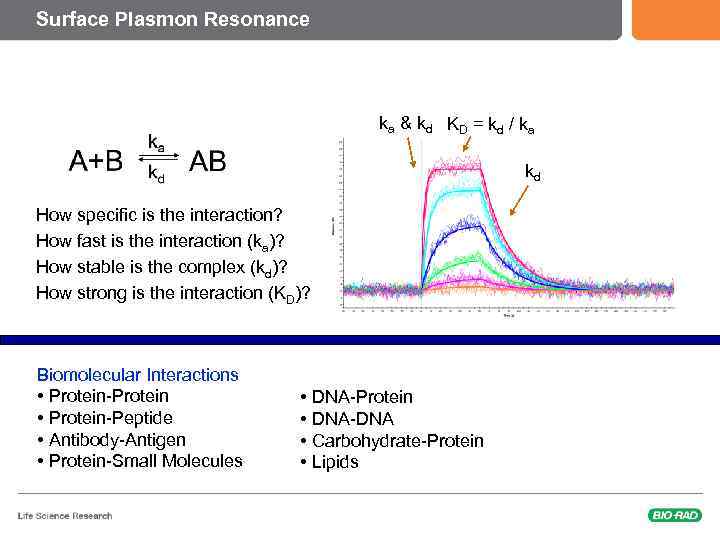
Surface Plasmon Resonance ka & kd KD = kd / ka kd How specific is the interaction? How fast is the interaction (ka)? How stable is the complex (kd)? How strong is the interaction (KD)? Biomolecular Interactions • Protein-Protein • Protein-Peptide • Antibody-Antigen • Protein-Small Molecules • DNA-Protein • DNA-DNA • Carbohydrate-Protein • Lipids
![Why Kinetics? Association Dissociation KD = 1. 0 n. M [A] = 10 n. Why Kinetics? Association Dissociation KD = 1. 0 n. M [A] = 10 n.](https://present5.com/presentation/120440918_331068995/image-26.jpg)
Why Kinetics? Association Dissociation KD = 1. 0 n. M [A] = 10 n. M 5 Interactions Identical KD Different ka and kd ka (M-1 s-1) kd (s-1) 1. 00 E+03 1. 00 E-06 1. 00 E+04 1. 00 E-05 1. 00 E+05 1. 00 E-04 1. 00 E+06 1. 00 E-03 1. 00 E+07 1. 00 E-02

Application Exemple: Antibody Screening with Capturing • Goal of the Experiment: • • To Screen and Determine the Full Kinetics of Many Antibodies binding a unique Antigen Material used: • • • 250 different Hybridoma Supernatants (each expressing a different monoclonal Antibody raised against the Antigen) Enough Recombinant Antigen One single Prote. On Amine Coupling Chip with low binding Capacity

Application Exemple: Antibody Screening with Capturing - Workflow Anti Ig. G Immobilized 1 Antibody from Supernatants Captured 1 2 3 4 5 6 4 2 Injection of 5 Concentrations of Antigen Buffer Interspot reference Regeneration 3

Application Exemple: Antibody Screening with Capturing - Ranking Iso-Affinity lines High Affinity Binders 0. 01 n. M 0. 1 n. M 100 n. M Low Affinity Binders 10 n. M

Pharmaceutical Applications • Antibody Kinetic Screening üEfficient experiment optimization üAccurate kinetic analysis üCompatible with crude samples üHigh screening throughput • Epitope Mapping / Epitope Binning üFlexible experiment configuration üAvailable for “Sandwich” assays • Drug Compound Screening üHigh sensitivity (>95 Dalton) üHigh screening throughput üAvailable for fragment screening

Ab Kinetic Screening – Bayer, USA Femtomolar Fab binding affinities to a protein target by alternative CDR residue co-optimization strategies without phage or cell surface display. Votsmeier C, et al. , m. Abs 4, 341 -348, 2012. Expression of soluble Fab library variants with CDR diversification Screening of soluble Fab library variants Recombination of gathered mutants New round of expression and screening Affinity maturation of adalimumab using: (1) Quantitative screening of soluble Fab fragments with diversification to complementarity-determining region (CDR); (2) Alternative recombination to co-optimize large sets of the found improving mutations.

Academic Applications • Structural Biology üHighly efficient mutagenesis workflow üAvailable for thermodynamics üCompatible with crude samples • Thermodynamics üHighly efficient thermodynamics workflow üStable performance on reproducibility • Histidine-Tagged Protein Analysis üHis-tagged protein capturing stability for accurate analysis üSelectivity for simple online purification process üRegeneration capability for low cost • Lipid Membrane / Membrane Protein Analysis üHydrophilic surface chemistry allowing high performance for capturing lipid assemblies üReal-time comparison of different conditions
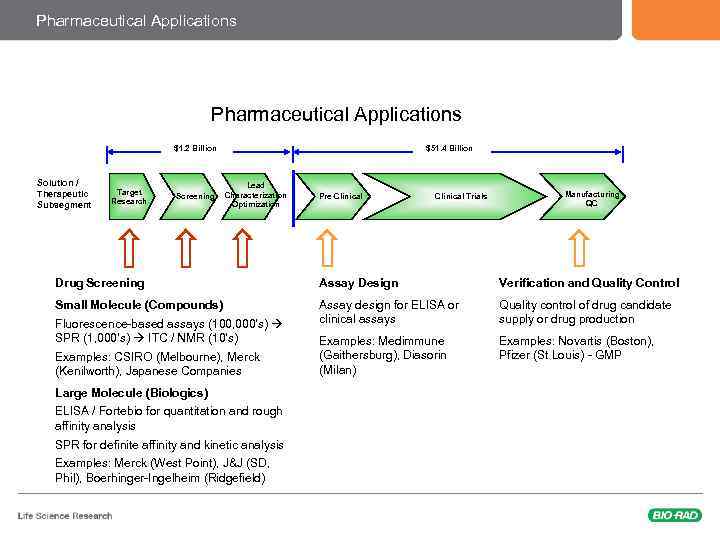
Pharmaceutical Applications $1. 2 Billion Solution / Therapeutic Subsegment Target Research Screening $51. 4 Billion Lead Characterization Optimization Pre-Clinical Trials Manufacturing QC Drug Screening Assay Design Verification and Quality Control Small Molecule (Compounds) Assay design for ELISA or clinical assays Quality control of drug candidate supply or drug production Examples: Medimmune (Gaithersburg), Diasorin (Milan) Examples: Novartis (Boston), Pfizer (St Louis) - GMP Fluorescence-based assays (100, 000’s) SPR (1, 000’s) ITC / NMR (10’s) Examples: CSIRO (Melbourne), Merck (Kenilworth), Japanese Companies Large Molecule (Biologics) ELISA / Fortebio for quantitation and rough affinity analysis SPR for definite affinity and kinetic analysis Examples: Merck (West Point), J&J (SD, Phil), Boerhinger-Ingelheim (Ridgefield)
Regulatory & technical considerations for effective monitoring of host cell protein (HCP) impurities
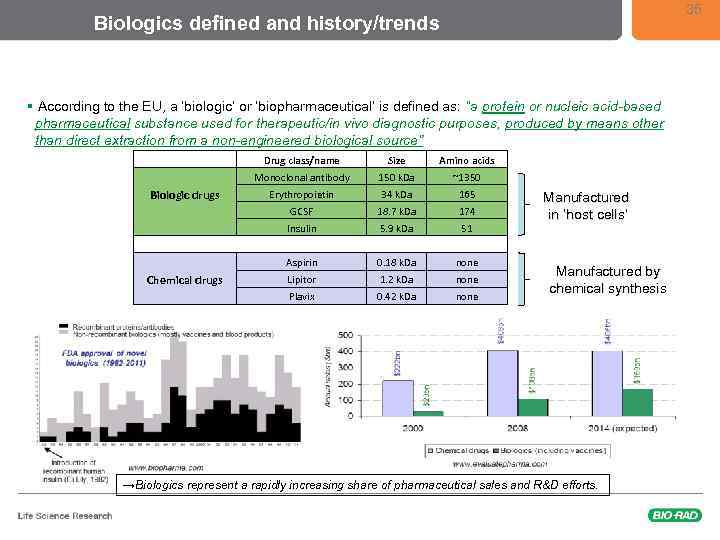
35 Biologics defined and history/trends § According to the EU, a ‘biologic’ or ‘biopharmaceutical’ is defined as: “a protein or nucleic acid-based pharmaceutical substance used for therapeutic/in vivo diagnostic purposes, produced by means other than direct extraction from a non-engineered biological source” Drug class/name Size Amino acids Monoclonal antibody 150 k. Da ~1350 Erythropoietin 34 k. Da 165 GCSF 18. 7 k. Da 174 Insulin 5. 9 k. Da 51 Biologic drugs Aspirin Chemical drugs Manufactured in ‘host cells’ 0. 18 k. Da none Lipitor 1. 2 k. Da none Plavix 0. 42 k. Da none Manufactured by chemical synthesis www. biopharma. com →Biologics represent a rapidly increasing share of pharmaceutical sales and R&D efforts.
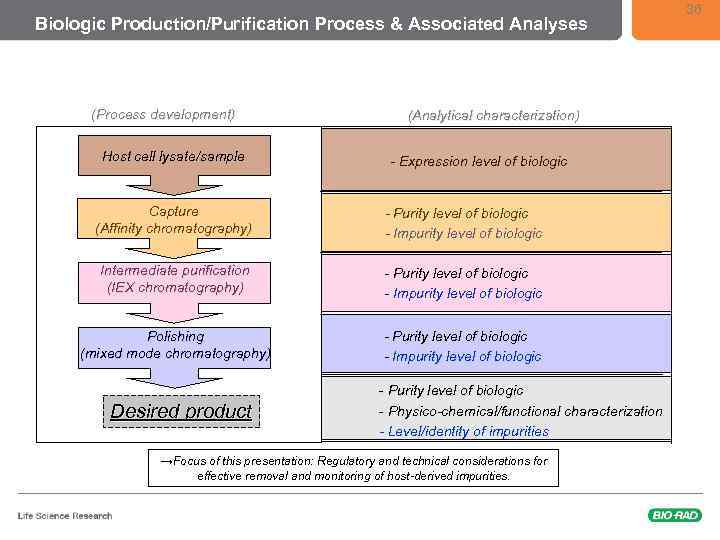
Biologic Production/Purification Process & Associated Analyses (Process development) Host cell lysate/sample (Analytical characterization) - Expression level of biologic Capture (Affinity chromatography) - Purity level of biologic - Impurity level of biologic Intermediate purification (IEX chromatography) - Purity level of biologic - Impurity level of biologic Polishing (mixed mode chromatography) - Purity level of biologic - Impurity level of biologic Desired product - Purity level of biologic - Physico-chemical/functional characterization - Level/identity of impurities →Focus of this presentation: Regulatory and technical considerations for effective removal and monitoring of host-derived impurities. 36

Regulations & Guidelines “… HCPs from the cell substrate co-purified with the active substance could induce immune response against themselves…” or function as “adjuvants for the protein of interest. ” “Whenever possible, contaminants introduced by the recovery and purification process should be below detectable levels using a highly sensitive analytical method. ” Points to Consider in the Manufacture & Testing of Monoclonal Products for Human Use, US Food and Drug Administration, 1997 EMEA guidelines CHMP/BMWP/14327/2006 “…. . Whatever the product…residual HCP have to be tested for on a routine basis… …. results from batch to batch should be consistent and meet specification limits” EMEA guidelines CPMP/BWP/382/97 “…Differences in purity/impurity profiles should be evaluated to assess potential impact on safety and efficacy… The methods should be validated in accordance with ICH guidelines… ” Regulatory Requirements for Marketing Authorization in India, 2012 “For HCPs, a sensitive assay, e. g. , immunoassay, capable of detecting a wide range of protein impurities is generally utilized. . a polyclonal antibody used …is generated by immunization with a preparation of a production cell minus the product-coding gene, fusion partners or other appropriate cell lines ICH Q 6 B; Test Procedures & Acceptance Criteria For Biotechnological/Biological Products 37

38 Effect of residual HCPs on approvals of biologics/biosimilars Omnitrope (r. HGH biosimilar); 2006 Inspite of efficacy, application initially rejected by European regulators (EMEA) due to excess host cell protein associated immunogenicity; subsequently resolved with “a modified downstream manufacturing process with significantly enhanced host cell protein (HCP) clearance. ” http: //www. ema. europa. eu/docs/en_GB/document_library/EPAR_-_Scientific_Discussion/human/000607/WC 500043692. pdf Alpheon (interferon alfa-2 a biosimilar); 2008 Rejected by EMEA owing to concerns regarding comparability to Roferon-A (reference product), because of differences “such as impurities”…. inadequate process validation. . and insufficient validation of immunogenicity testing. http: //www. ema. europa. eu/docs/en_GB/document_library/Summary_of_opinion_-_Initial_authorisation/human/000585/WC 500017451. pdf IB 1001/IXinity (recombinant factor IX); 2012, 2013 2012 -US regulators (FDA) placed on hold clinical trials after participants developed antibodies to proteins from CHO (host) cells at a higher level than was expected from earlier studies. 2013 -Drug application withdrawn from EMEA http: //www. inspirationbio. com/files/994/76221. pdf; http: //www. ema. europa. eu/ema/index. jsp? curl=pages/medicines/ human/medicines/002349/wapp/Initial_authorisation/human_wapp_000167. jsp&mid=WC 0 b 01 ac 058001 d 128)

HCP monitoring questions §Product: –Nature/identity and level of HCPs in DS/DP. –Batch to batch monitoring for total/specific HCPs. §Chromatography Protocol and Process: –Influence of process change(s) on residual HCPs. –Match purification protocol / process to physico-chemical properties of total/specific HCPs. 39

40 Approaches for HCP Analyses Approach Sensitivity SDS-PAGE/HPLC/CE 2 DE Followed by sensitive protein stain Medium sensitivity approaches (~mg level impurities /mg biologic) Mass Spectrometry (MS) Salient features Used as a complement to ELISA/immunoassays (not as a stand-alone method) Valuable for identifying effects of process changes on product quality and/or impurity profiles Industry standard Immunoassay/ELISA (using polyclonal antibodies to HCPs) High sensitivity approach (~ng level impurities /mg biologic) Anti-HCP antibodies must be ‘qualified’ for ability to detect wide range/all of HCPs →Due to high sensitivity and broad reactivity ELISA is the mainstay; other approaches mainly used in a complementary manner

Reactivity of HCP ELISA to potential impurities must be qualified § Critical HCP ELISA reagent →anti-HCP antibodies. - ELISA is ‘blind’ to HCPs not recognized by antibodies. - Different methods exist to determine breadth of reactivity (qualification) of HCP ELISA Qualification method & depiction Krishgen Biosystems Salient features Reactivity potentially determined against protein complexes & not individual proteins Limited resolution Gold standard method Q: What regulatory guidance exists on most suitable approach for ELISA qualification? 41
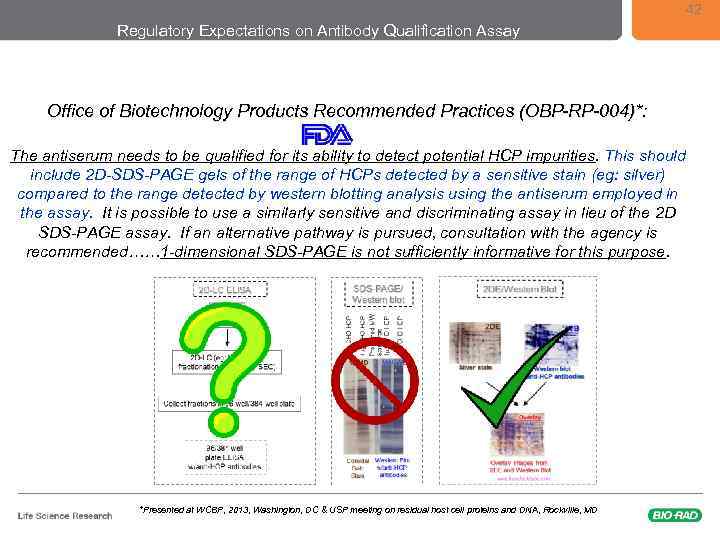
42 Regulatory Expectations on Antibody Qualification Assay Office of Biotechnology Products Recommended Practices (OBP-RP-004)*: The antiserum needs to be qualified for its ability to detect potential HCP impurities. This should include 2 D-SDS-PAGE gels of the range of HCPs detected by a sensitive stain (eg: silver) compared to the range detected by western blotting analysis using the antiserum employed in the assay. It is possible to use a similarly sensitive and discriminating assay in lieu of the 2 D SDS-PAGE assay. If an alternative pathway is pursued, consultation with the agency is recommended…… 1 -dimensional SDS-PAGE is not sufficiently informative for this purpose. *Presented at WCBP, 2013, Washington, DC & USP meeting on residual host cell proteins and DNA, Rockville, MD
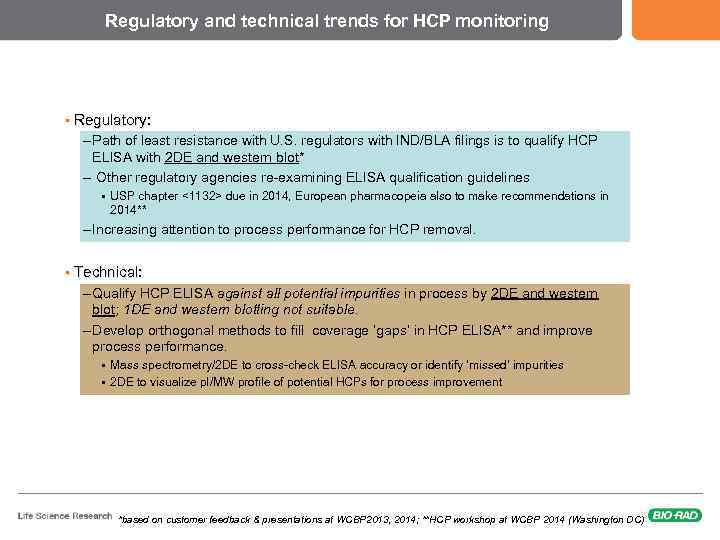
Regulatory and technical trends for HCP monitoring § Regulatory: – Path of least resistance with U. S. regulators with IND/BLA filings is to qualify HCP ELISA with 2 DE and western blot* – Other regulatory agencies re-examining ELISA qualification guidelines § USP chapter <1132> due in 2014, European pharmacopeia also to make recommendations in 2014** – Increasing attention to process performance for HCP removal. § Technical: – Qualify HCP ELISA against all potential impurities in process by 2 DE and western blot; 1 DE and western blotting not suitable. – Develop orthogonal methods to fill coverage ‘gaps’ in HCP ELISA** and improve process performance. § § Mass spectrometry/2 DE to cross-check ELISA accuracy or identify ‘missed’ impurities 2 DE to visualize p. I/MW profile of potential HCPs for process improvement *based on customer feedback & presentations at WCBP 2013, 2014; **HCP workshop at WCBP 2014 (Washington DC)

44 Considerations for confidence in 2 DE/western qualification of HCP ELISA Parameter Optimization p. H gradient Sample preparation/components Electrophoretic conditions Workflow 2 DE (IEF & MW Separation) Result Effective, reproducible sample separation SDS-PAGE Transfer method PAGE chemistry Protein size/charge Protein Blotting High-efficiency protein transfer across p. I/MW range ‘Blotting’ membranes Stain sensitivity & dynamic range Gel/Blot stain Blocking conditions Antibody concentration, detection format (direct/sandwich, chemiluminescent/fluorescent) Image capture options Overlay analysis software Protein Staining & Western Analysis Image Overlay Reliability & high sensitivity comparison feasible Confidence in Results

45 Bio-Rad’s 2 DE and Western Blotting Workflow Solution for your HCP ELISA qualification Parameter(s) p. H gradient Sample preparation/components Electrophoretic conditions Workflow Solution 2 DE (IEF & MW Separation) Protean i 12 IEF system Result Effective, reproducible sample separation SDS-PAGE Criterion ® TGX™ SDS-PAGE Gels Transfer method PAGE chemistry Protein size/charge Protein Blotting Trans-blot® Turbo™ RTA Midi LF PVDF Trans-blot® Turbo™ ‘Blotting’ membranes Stain sensitivity & dynamic range Gel/Blot stain Blocking conditions Antibody concentration, detection format (direct/sandwich, chemiluminescent, fluorescent) High-efficiency protein transfer across p. I/MW range Protein Staining & Western Analysis Your anti-HCP antibodies Same membrane protein staining and western SYPRO Ruby analysis Protein Blot Stain PDQuest™ 2 -D Analysis Software Image capture options Overlay analysis software Image Overlay Chemi. Doc™ MP System Reliability & high sensitivity comparison feasible Confidence in Results

References of interest § 2 DE/western blotting workflow considerations: - Berkelman T et al. , 2013 Bioprocess International http: //www. bioprocessintl. com/multimedia/archive/00229/BPI_A_131108 AR 05_O_229801 a. pdf - Berkelman T et al. , 2013 Technote 6393 Rev A http: //www. bio-rad. com/webroot/web/pdf/lsr/literature/Bulletin_6393. pdf § 2 DE electrophoresis: - Workflow details: http: //www. bio-rad. com/en-us/product/2 -d-electrophoresis-workflow - 2 D Tips and Techniques Webinar: www. bio-rad. com/info/view 2 Dwebinar - Inter-lab reproducibility of 2 DE using Protean i 12 http: //www. ncbi. nlm. nih. gov/pubmed/23679042; Posch A et al. , 2013 Arch Physiol Biochem § Effectiveness of 2 DE/western blotting over 1 DE/western blotting for anti-HCP qualification – Rusbuldt J et al. , 2013 Technote 6405 Rev A http: //www. bio-rad. com/webroot/web/pdf/lsr/literature/Bulletin_6405. pdf 46

THANKS
Chrom School Novosibirsk JLL.pptx