c1baa5ddbab74de459a3efaac710be23.ppt
- Количество слайдов: 105
 Chemometrical Methods in Expert Systems for the Molecular Structure Elucidation. Mikhail Elyashberg Advanced Chemistry Development (ACD), Moscow-Toronto
Chemometrical Methods in Expert Systems for the Molecular Structure Elucidation. Mikhail Elyashberg Advanced Chemistry Development (ACD), Moscow-Toronto
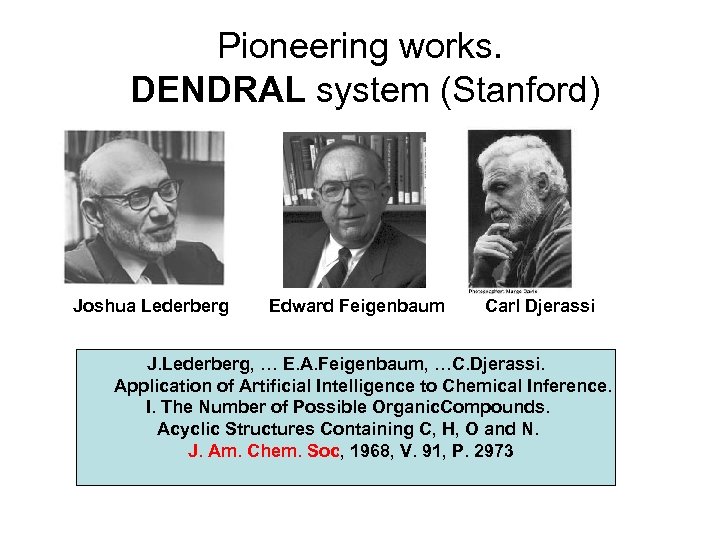 Pioneering works. DENDRAL system (Stanford) Joshua Lederberg Edward Feigenbaum Carl Djerassi J. Lederberg, … E. A. Feigenbaum, …C. Djerassi. Application of Artificial Intelligence to Chemical Inference. I. The Number of Possible Organic. Compounds. Acyclic Structures Containing C, H, O and N. J. Am. Chem. Soc, 1968, V. 91, P. 2973
Pioneering works. DENDRAL system (Stanford) Joshua Lederberg Edward Feigenbaum Carl Djerassi J. Lederberg, … E. A. Feigenbaum, …C. Djerassi. Application of Artificial Intelligence to Chemical Inference. I. The Number of Possible Organic. Compounds. Acyclic Structures Containing C, H, O and N. J. Am. Chem. Soc, 1968, V. 91, P. 2973
 Pioneering works. CASE system (Arizona) Morton Munk D. B. Nelson, M. E. Munk, K. B. Gasli, D. L. Horald. Alanylactinobicyclon. An Application of Computer Techniques to Structure Elucidation. J. Org. Chem. , 1969, V. 34, P. 3800
Pioneering works. CASE system (Arizona) Morton Munk D. B. Nelson, M. E. Munk, K. B. Gasli, D. L. Horald. Alanylactinobicyclon. An Application of Computer Techniques to Structure Elucidation. J. Org. Chem. , 1969, V. 34, P. 3800
 Pioneering works. CHEMICS system (Japan) Shin-Ichi Sasaki S. I. Sasaki, H. Abe, T. Ouki, M. Sakamoto and S. Ochiai. Automated Structure Elucidation of Several Kinds of Aliphatic and Alicyclic Compounds. Analytical Chemistry, 1968, V. 40, p. 2220
Pioneering works. CHEMICS system (Japan) Shin-Ichi Sasaki S. I. Sasaki, H. Abe, T. Ouki, M. Sakamoto and S. Ochiai. Automated Structure Elucidation of Several Kinds of Aliphatic and Alicyclic Compounds. Analytical Chemistry, 1968, V. 40, p. 2220
 Pioneering works. STREC system (Moscow) M. E. Elyashberg, L. A. Gribov Formal logical interpretation of IR spectra using characteristic frequencies. Zhurn. Appl. Spectrosc. (J. Appl. Spectrosc. ) 8, 1968, 998.
Pioneering works. STREC system (Moscow) M. E. Elyashberg, L. A. Gribov Formal logical interpretation of IR spectra using characteristic frequencies. Zhurn. Appl. Spectrosc. (J. Appl. Spectrosc. ) 8, 1968, 998.
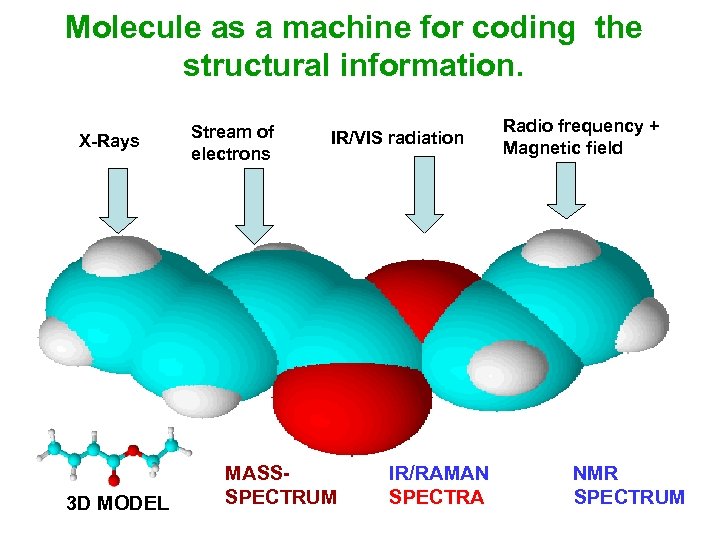 Molecule as a machine for coding the structural information. X-Rays 3 D MODEL Stream of electrons IR/VIS radiation MASSSPECTRUM Radio frequency + Magnetic field IR/RAMAN NMR SPECTRA SPECTRUM
Molecule as a machine for coding the structural information. X-Rays 3 D MODEL Stream of electrons IR/VIS radiation MASSSPECTRUM Radio frequency + Magnetic field IR/RAMAN NMR SPECTRA SPECTRUM
 Number of isomers of some natural products ~43 mln. are real
Number of isomers of some natural products ~43 mln. are real
 Properties of an isomer set • Isomer numbers for molecules of medium size are comparable with Avogadro’s number (1028). • Though the number of isomers is huge the isomers corresponding to the given molecular formula make up a countable and finite set.
Properties of an isomer set • Isomer numbers for molecules of medium size are comparable with Avogadro’s number (1028). • Though the number of isomers is huge the isomers corresponding to the given molecular formula make up a countable and finite set.
 General strategy of Computer-Aided Structure Elucidation (CASE) Elimination of “superfluous” isomers from the full set by imposing different structural constraints. Sources of structural constraints: Spectra, a priory information (sample origin, chemical rules, etc. )
General strategy of Computer-Aided Structure Elucidation (CASE) Elimination of “superfluous” isomers from the full set by imposing different structural constraints. Sources of structural constraints: Spectra, a priory information (sample origin, chemical rules, etc. )

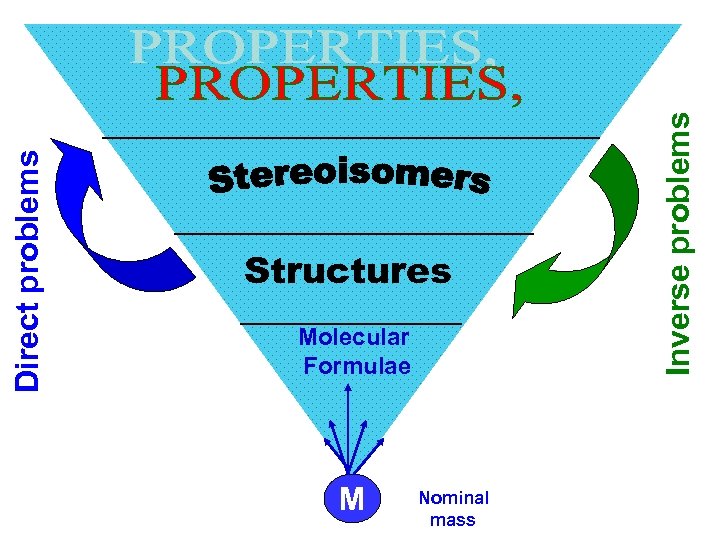 Molecular Formulae М Nominal mass Inverse problems Direct problems Structures
Molecular Formulae М Nominal mass Inverse problems Direct problems Structures
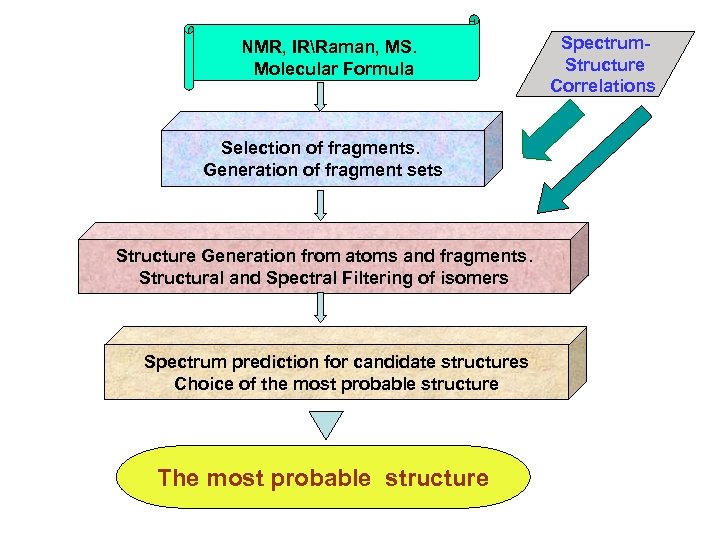 NMR, IRRaman, MS. Molecular Formula Selection of fragments. Generation of fragment sets Structure Generation from atoms and fragments. Structural and Spectral Filtering of isomers Spectrum prediction for candidate structures Choice of the most probable structure The most probable structure Spectrum. Structure Correlations
NMR, IRRaman, MS. Molecular Formula Selection of fragments. Generation of fragment sets Structure Generation from atoms and fragments. Structural and Spectral Filtering of isomers Spectrum prediction for candidate structures Choice of the most probable structure The most probable structure Spectrum. Structure Correlations
 Separate section Computer Techniques and Optimization
Separate section Computer Techniques and Optimization
 Prof. Jean-Thomas Clerc (1934 -1998) Chemometrics in Analytical Chemistry, CAC-1996 Tarragona, Spain Prominent scientist and vivid person.
Prof. Jean-Thomas Clerc (1934 -1998) Chemometrics in Analytical Chemistry, CAC-1996 Tarragona, Spain Prominent scientist and vivid person.
 L. A. Gribov, M. E. Elyashberg Computer-Assisted Identification of Organic Molecules by their Molecular Spectra. 1979. Monographic review.
L. A. Gribov, M. E. Elyashberg Computer-Assisted Identification of Organic Molecules by their Molecular Spectra. 1979. Monographic review.
 Achievements of the “Storm and Stress” period were generalized in monographs: • M. E. Elyashberg, L. A. Gribov, V. V. Serov. Molecular Spectral Analysis and Computer. Nauka, Moscow, 1980. • N. A. B. Gray. Computer-Assisted Structure Elucidation. Wiley, N. Y. 1986
Achievements of the “Storm and Stress” period were generalized in monographs: • M. E. Elyashberg, L. A. Gribov, V. V. Serov. Molecular Spectral Analysis and Computer. Nauka, Moscow, 1980. • N. A. B. Gray. Computer-Assisted Structure Elucidation. Wiley, N. Y. 1986
 Examples of structures identified with the aids of X-PERT program.
Examples of structures identified with the aids of X-PERT program.
 Development of NMR techniques 1986 2006
Development of NMR techniques 1986 2006
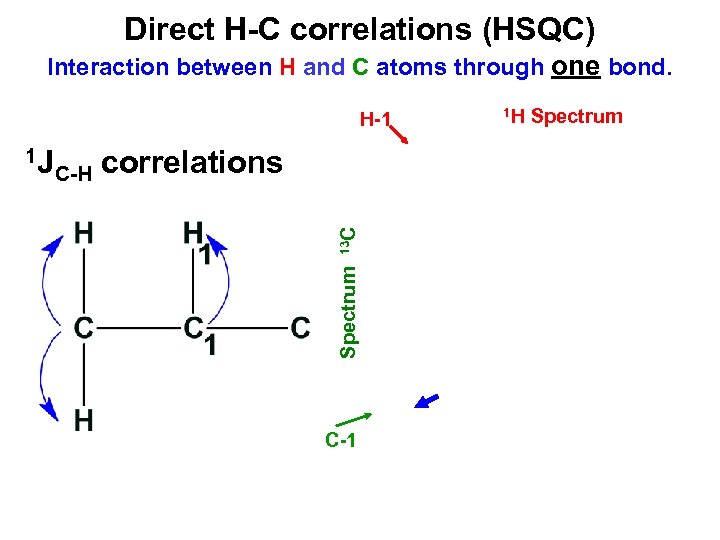 Direct H-C correlations (HSQC) Interaction between H and C atoms through one bond. H-1 C-H correlations Spectrum 13 C 1 J C-1 1 H Spectrum
Direct H-C correlations (HSQC) Interaction between H and C atoms through one bond. H-1 C-H correlations Spectrum 13 C 1 J C-1 1 H Spectrum
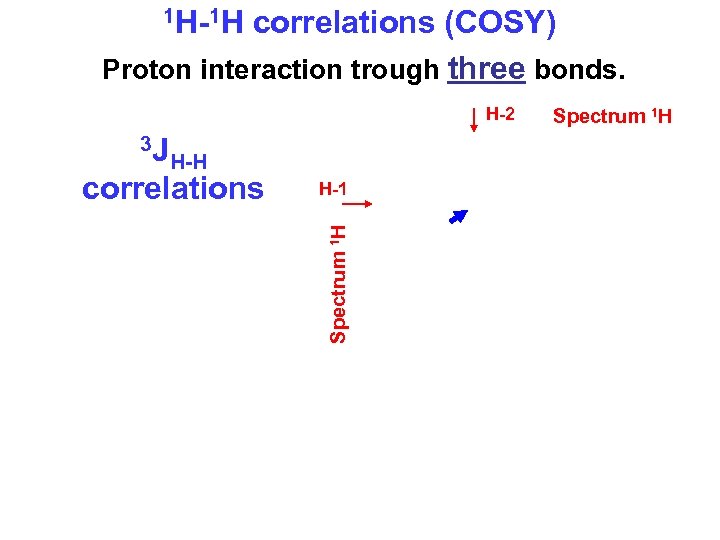 1 H-1 H correlations (COSY) Proton interaction trough three bonds. H-2 H-1 Spectrum 1 Н 3 JH-H correlations Spectrum 1 Н
1 H-1 H correlations (COSY) Proton interaction trough three bonds. H-2 H-1 Spectrum 1 Н 3 JH-H correlations Spectrum 1 Н
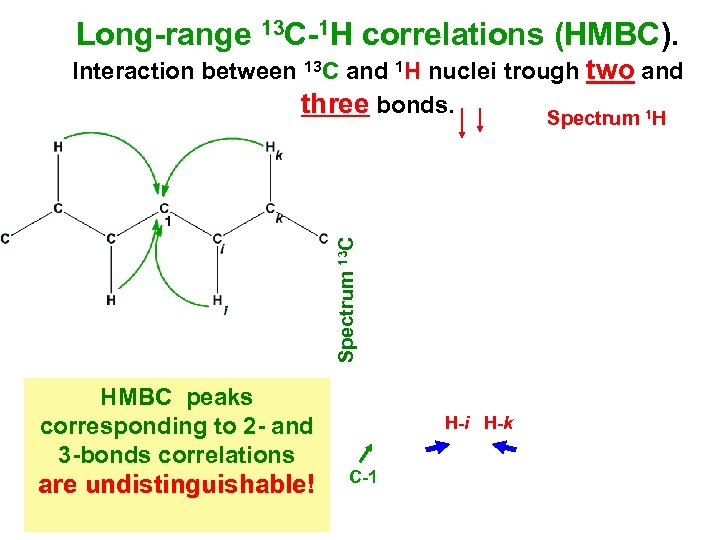 Long-range 13 C-1 H correlations (HMBC). Spectrum 13 С Interaction between 13 С and 1 Н nuclei trough two and three bonds. Spectrum 1 Н HMBC peaks corresponding to 2 - and 3 -bonds correlations are undistinguishable! H-i H-k С-1
Long-range 13 C-1 H correlations (HMBC). Spectrum 13 С Interaction between 13 С and 1 Н nuclei trough two and three bonds. Spectrum 1 Н HMBC peaks corresponding to 2 - and 3 -bonds correlations are undistinguishable! H-i H-k С-1
 Ratio Nobs Ntheor correlations for COSY and HMBC COSY HMBC
Ratio Nobs Ntheor correlations for COSY and HMBC COSY HMBC
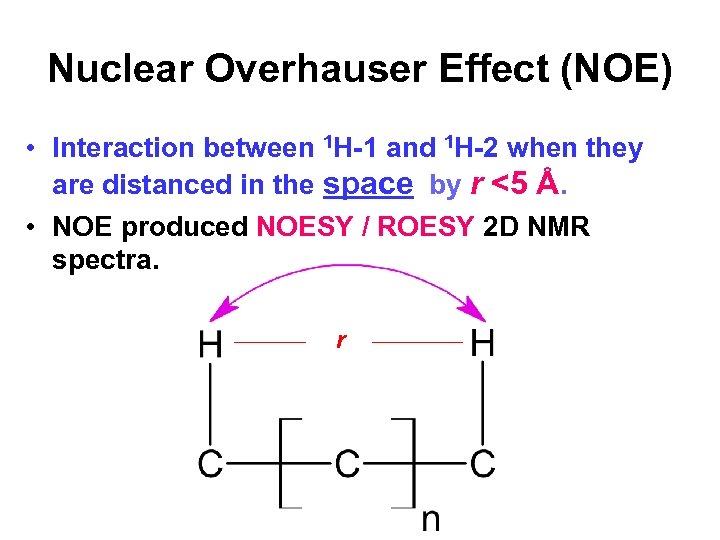 Nuclear Overhauser Effect (NOE) • Interaction between 1 H-1 and 1 H-2 when they are distanced in the space by r <5 Å. • NOE produced NOESY / ROESY 2 D NMR spectra. r
Nuclear Overhauser Effect (NOE) • Interaction between 1 H-1 and 1 H-2 when they are distanced in the space by r <5 Å. • NOE produced NOESY / ROESY 2 D NMR spectra. r
 Structural interpretation of 2 D NMR spectra. Main “axioms” COSY • If a peak (H-1, H-2) is observed in COSY, then a molecule contains the chemical bond (C-1) (C-2). HMBC • If a peak (H-1, C-2) is observed in HMBC, then atoms C-1 and C -2 are separated in the structure by ONE or TWO chemical bonds: (C-1) (C-2) or (C-1) (X) (C-2), X=C, O, N… NOESY • If a peak (H-1, H-2) is observed in NOESY (ROESY), then the distance between H-1 и H-2 in space is less than 5Å.
Structural interpretation of 2 D NMR spectra. Main “axioms” COSY • If a peak (H-1, H-2) is observed in COSY, then a molecule contains the chemical bond (C-1) (C-2). HMBC • If a peak (H-1, C-2) is observed in HMBC, then atoms C-1 and C -2 are separated in the structure by ONE or TWO chemical bonds: (C-1) (C-2) or (C-1) (X) (C-2), X=C, O, N… NOESY • If a peak (H-1, H-2) is observed in NOESY (ROESY), then the distance between H-1 и H-2 in space is less than 5Å.
 Interpretation of the Structure Elucidation problem in terms of an axiomatic theory. • Creation of the set of axioms and hypotheses necessary for solution of a given problem is equivalent to creation of some particular axiomatic theory. • To obtain a valid solution to the problem (i. e. manageable output file containing the correct structure) the set of axioms must be true, complete and consistent.
Interpretation of the Structure Elucidation problem in terms of an axiomatic theory. • Creation of the set of axioms and hypotheses necessary for solution of a given problem is equivalent to creation of some particular axiomatic theory. • To obtain a valid solution to the problem (i. e. manageable output file containing the correct structure) the set of axioms must be true, complete and consistent.
 Example of an expert system based on 1 D and 2 D NMR data. STRUCTURE ELUCIDATOR Advance Chemistry Development Ltd. , Moscow -Toronto K. A. Blinov, D. Carlson, M. E. Elyashberg et al. J. Magn. Reson. Chem. 2003, 41, 359 -372. M. E. Elyashberg, K. A. Blinov, S. G. Molodtsov et al. J. Chem. Inf. Model. 2004, 44, 771 -792
Example of an expert system based on 1 D and 2 D NMR data. STRUCTURE ELUCIDATOR Advance Chemistry Development Ltd. , Moscow -Toronto K. A. Blinov, D. Carlson, M. E. Elyashberg et al. J. Magn. Reson. Chem. 2003, 41, 359 -372. M. E. Elyashberg, K. A. Blinov, S. G. Molodtsov et al. J. Chem. Inf. Model. 2004, 44, 771 -792
 Knowledge of Structure Elucidator Factual knowledge: • Database of Structures (280, 000) and Fragments (1. 7 mln) with assigned NMR spectra (subspectra). Axiomatic Knowledge: • Correlation Tables for spectral structure filtering by NMR and IR spectra. • Atom Property Correlation Table (APCT). It is used for setting atom hybridization and possibility of neighboring with heteroatoms.
Knowledge of Structure Elucidator Factual knowledge: • Database of Structures (280, 000) and Fragments (1. 7 mln) with assigned NMR spectra (subspectra). Axiomatic Knowledge: • Correlation Tables for spectral structure filtering by NMR and IR spectra. • Atom Property Correlation Table (APCT). It is used for setting atom hybridization and possibility of neighboring with heteroatoms.
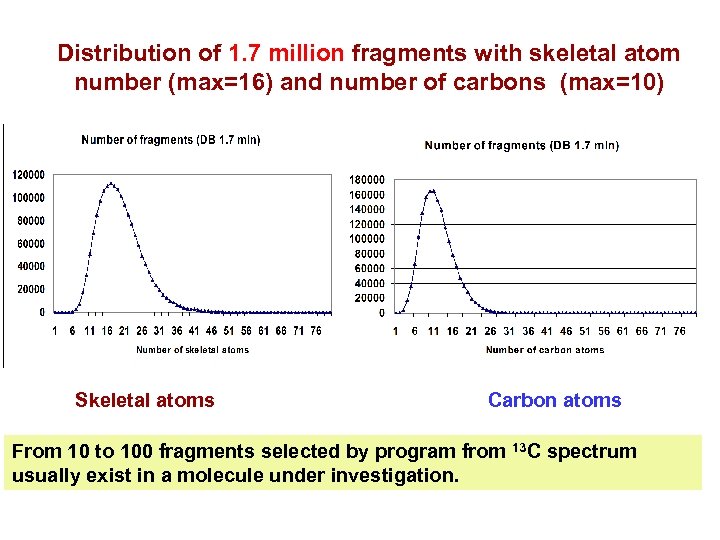 Distribution of 1. 7 million fragments with skeletal atom number (max=16) and number of carbons (max=10) Skeletal atoms Carbon atoms From 10 to 100 fragments selected by program from 13 C spectrum usually exist in a molecule under investigation.
Distribution of 1. 7 million fragments with skeletal atom number (max=16) and number of carbons (max=10) Skeletal atoms Carbon atoms From 10 to 100 fragments selected by program from 13 C spectrum usually exist in a molecule under investigation.
 Checking Knowledge reliability. • 98% of 280 000 structures passed checking by the Spectral Filter and Atom Property Correlation Tables. • 99. 8% of 17 000 natural product stood the same verification. Risk to lose the correct structure is minimal.
Checking Knowledge reliability. • 98% of 280 000 structures passed checking by the Spectral Filter and Atom Property Correlation Tables. • 99. 8% of 17 000 natural product stood the same verification. Risk to lose the correct structure is minimal.
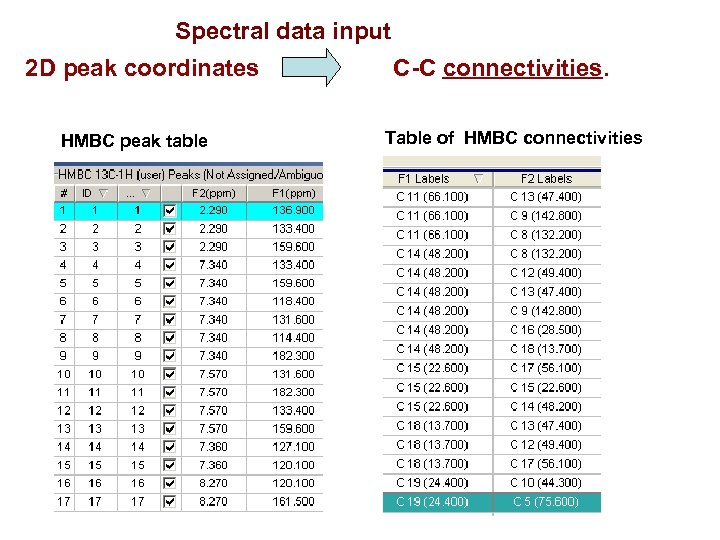 Spectral data input 2 D peak coordinates C-C connectivities. HMBC peak table Table of HMBC connectivities
Spectral data input 2 D peak coordinates C-C connectivities. HMBC peak table Table of HMBC connectivities
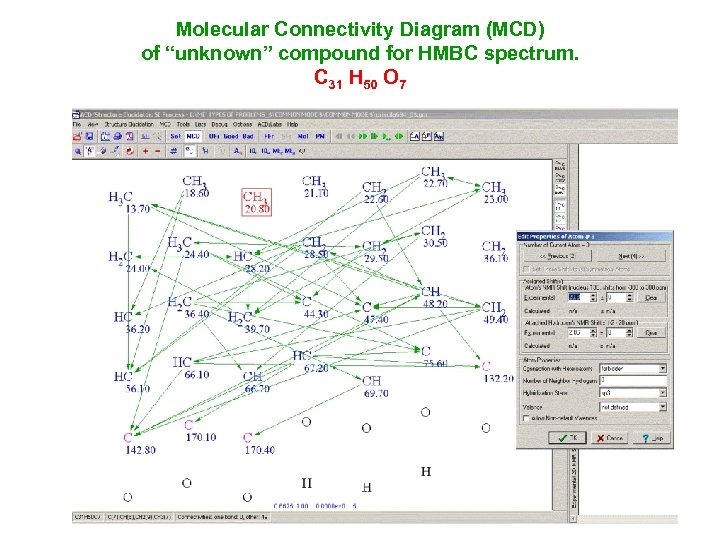 Molecular Connectivity Diagram (MCD) of “unknown” compound for НМВС spectrum. С 31 Н 50 О 7
Molecular Connectivity Diagram (MCD) of “unknown” compound for НМВС spectrum. С 31 Н 50 О 7
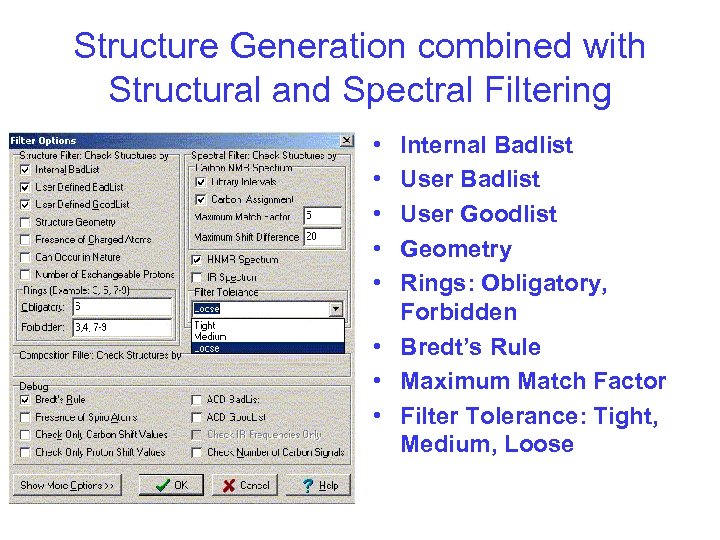 Structure Generation combined with Structural and Spectral Filtering • • • Internal Badlist User Goodlist Geometry Rings: Obligatory, Forbidden • Bredt’s Rule • Maximum Match Factor • Filter Tolerance: Tight, Medium, Loose
Structure Generation combined with Structural and Spectral Filtering • • • Internal Badlist User Goodlist Geometry Rings: Obligatory, Forbidden • Bredt’s Rule • Maximum Match Factor • Filter Tolerance: Tight, Medium, Loose
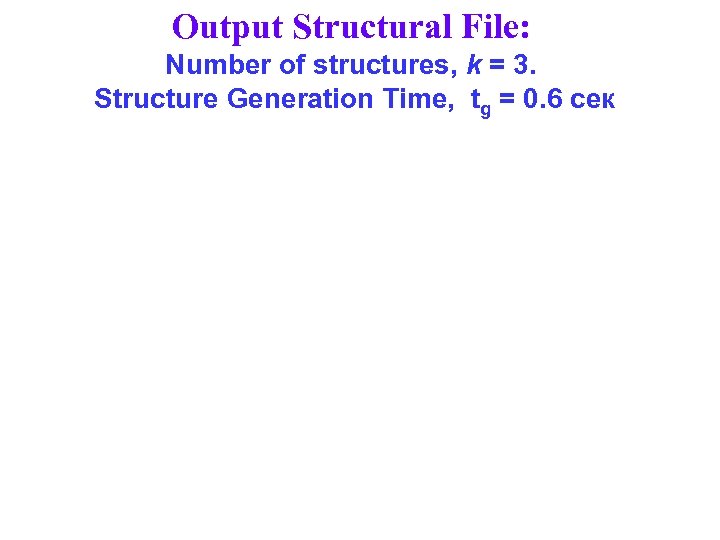 Output Structural File: Number of structures, k = 3. Structure Generation Time, tg = 0. 6 сек
Output Structural File: Number of structures, k = 3. Structure Generation Time, tg = 0. 6 сек
 Selection of the Preferable Structure 1. 13 C Chemical shift calculation for all structures of the output file. Removing duplicate structures. 2. Structure ranking in ascending order of average chemical shift deviation, d, found for calculated and experimental spectra. • A structure having minimum d value is declared as the most probable.
Selection of the Preferable Structure 1. 13 C Chemical shift calculation for all structures of the output file. Removing duplicate structures. 2. Structure ranking in ascending order of average chemical shift deviation, d, found for calculated and experimental spectra. • A structure having minimum d value is declared as the most probable.
 Methods of 13 C and 1 H spectrum prediction: 1. 2. 3. Fragment based approach Method of increments (PLS) Artificial neural nets. Recently speed and accuracy of Incremental Approach were significantly improved. Speed: 6000 -10000 13 C chemical shifts per second. For molecules С 20 -С 30: 200 -400 spectra per second. Accuracy: Average chemical shift deviation: 1. 8 ppm. Y. D. Smurnyy, K. A. Blinov, T. S. Churanova, M. E. Elyashberg, A. J. Williams. J. Chem. Inf. Model. 2008, 48, 128 -134
Methods of 13 C and 1 H spectrum prediction: 1. 2. 3. Fragment based approach Method of increments (PLS) Artificial neural nets. Recently speed and accuracy of Incremental Approach were significantly improved. Speed: 6000 -10000 13 C chemical shifts per second. For molecules С 20 -С 30: 200 -400 spectra per second. Accuracy: Average chemical shift deviation: 1. 8 ppm. Y. D. Smurnyy, K. A. Blinov, T. S. Churanova, M. E. Elyashberg, A. J. Williams. J. Chem. Inf. Model. 2008, 48, 128 -134
 The ranked output file r(all)=1
The ranked output file r(all)=1
 The higher speed and accuracy of chemical shift prediction influenced the system strategy. Then: Now: • Output file should be minimal. • For this goal, severe constraints (axioms) must be introduced. • Consequence: great risk to lose the correct structure. • Structural file is admitted to contain 105 and more structures (tcalc=5 -10 min). • Severe constraints may be removed. • Solutions became more reliable
The higher speed and accuracy of chemical shift prediction influenced the system strategy. Then: Now: • Output file should be minimal. • For this goal, severe constraints (axioms) must be introduced. • Consequence: great risk to lose the correct structure. • Structural file is admitted to contain 105 and more structures (tcalc=5 -10 min). • Severe constraints may be removed. • Solutions became more reliable
 Acceleration of Structure Generation • The Structure Generation algorithm first produces substructures which are then complemented by new bonds until full structures are generated. • We suggested that fast 13 C chemical shift prediction for incomplete structures would prevent generation of such structural branches that contradict experimental 13 C NMR spectrum. • The expected result : significant acceleration of the Structure Generation.
Acceleration of Structure Generation • The Structure Generation algorithm first produces substructures which are then complemented by new bonds until full structures are generated. • We suggested that fast 13 C chemical shift prediction for incomplete structures would prevent generation of such structural branches that contradict experimental 13 C NMR spectrum. • The expected result : significant acceleration of the Structure Generation.
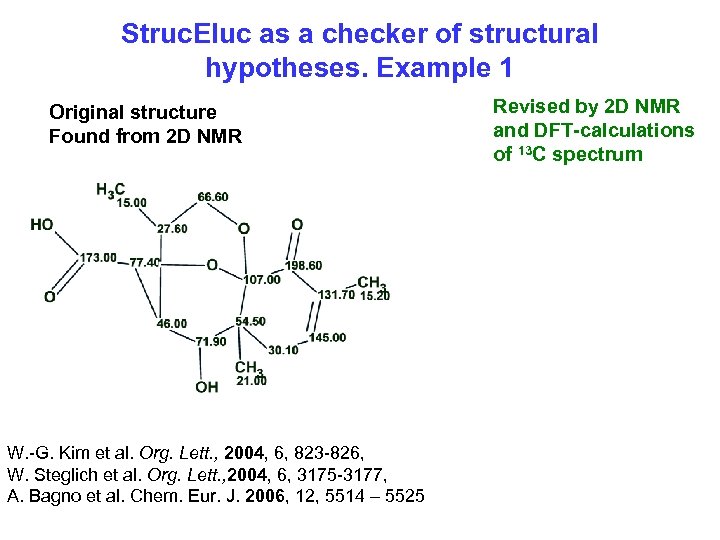 Struc. Eluc as a checker of structural hypotheses. Example 1 Original structure Found from 2 D NMR W. -G. Kim et al. Org. Lett. , 2004, 6, 823 -826, W. Steglich et al. Org. Lett. , 2004, 6, 3175 -3177, A. Bagno et al. Chem. Eur. J. 2006, 12, 5514 – 5525 Revised by 2 D NMR and DFT-calculations of 13 C spectrum
Struc. Eluc as a checker of structural hypotheses. Example 1 Original structure Found from 2 D NMR W. -G. Kim et al. Org. Lett. , 2004, 6, 823 -826, W. Steglich et al. Org. Lett. , 2004, 6, 3175 -3177, A. Bagno et al. Chem. Eur. J. 2006, 12, 5514 – 5525 Revised by 2 D NMR and DFT-calculations of 13 C spectrum
 The top of ranked output file found by Struc. Eluc: k=37176 Filter 149 Remove Dupl. 135, tg=1 m 40 s d. NN=2. 17 d. NN=3. 08
The top of ranked output file found by Struc. Eluc: k=37176 Filter 149 Remove Dupl. 135, tg=1 m 40 s d. NN=2. 17 d. NN=3. 08
 Struc. Eluc as a checker of structural hypotheses. Example 2 M=262, C 16 H 10 N 2 O 2 A. Balandina et al, J. Mol. Struct. 791, 2006, 77 -81
Struc. Eluc as a checker of structural hypotheses. Example 2 M=262, C 16 H 10 N 2 O 2 A. Balandina et al, J. Mol. Struct. 791, 2006, 77 -81
 Structural Hypotheses to be checked by DFT 13 C prediction C
Structural Hypotheses to be checked by DFT 13 C prediction C
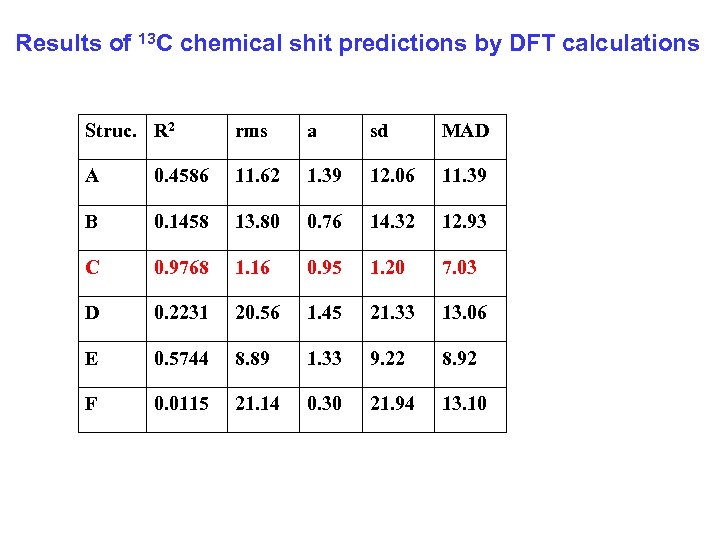 Results of 13 C chemical shit predictions by DFT calculations Struc. R 2 rms a sd MAD A 0. 4586 11. 62 1. 39 12. 06 11. 39 B 0. 1458 13. 80 0. 76 14. 32 12. 93 C 0. 9768 1. 16 0. 95 1. 20 7. 03 D 0. 2231 20. 56 1. 45 21. 33 13. 06 E 0. 5744 8. 89 1. 33 9. 22 8. 92 F 0. 0115 21. 14 0. 30 21. 94 13. 10
Results of 13 C chemical shit predictions by DFT calculations Struc. R 2 rms a sd MAD A 0. 4586 11. 62 1. 39 12. 06 11. 39 B 0. 1458 13. 80 0. 76 14. 32 12. 93 C 0. 9768 1. 16 0. 95 1. 20 7. 03 D 0. 2231 20. 56 1. 45 21. 33 13. 06 E 0. 5744 8. 89 1. 33 9. 22 8. 92 F 0. 0115 21. 14 0. 30 21. 94 13. 10
 Molecular Connectivity Diagram M=262, C 16 H 10 N 2 O 2 A. Balandina et al, J. Mol. Struct. 791, 2006, 77 -81
Molecular Connectivity Diagram M=262, C 16 H 10 N 2 O 2 A. Balandina et al, J. Mol. Struct. 791, 2006, 77 -81
 Solution to the problem by Structure Elucidator Structure Generation and Filtering: k=247 Filter 16 Duplicates 4 tg= 1 s 434 ms Expected by authors
Solution to the problem by Structure Elucidator Structure Generation and Filtering: k=247 Filter 16 Duplicates 4 tg= 1 s 434 ms Expected by authors
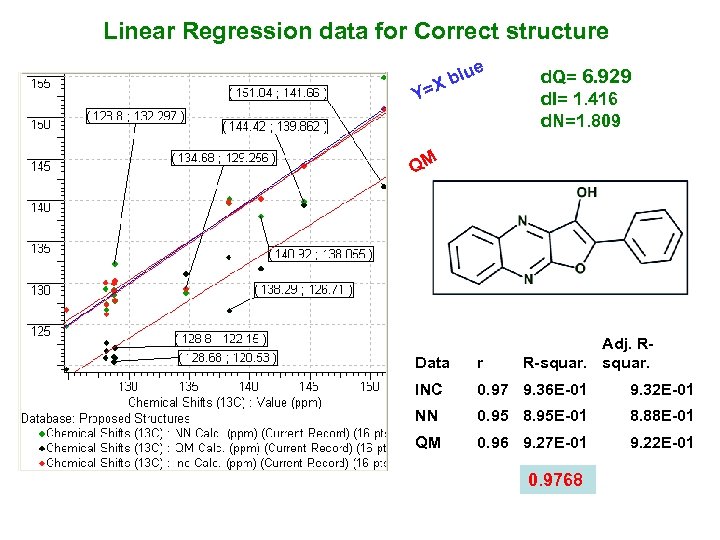 Linear Regression data for Correct structure blu X Y= e d. Q= 6. 929 d. I= 1. 416 d. N=1. 809 QM Adj. RR-squar. Data r INC 0. 97 9. 36 E-01 9. 32 E-01 NN 0. 95 8. 95 E-01 8. 88 E-01 QM 0. 96 9. 27 E-01 9. 22 E-01 0. 9768
Linear Regression data for Correct structure blu X Y= e d. Q= 6. 929 d. I= 1. 416 d. N=1. 809 QM Adj. RR-squar. Data r INC 0. 97 9. 36 E-01 9. 32 E-01 NN 0. 95 8. 95 E-01 8. 88 E-01 QM 0. 96 9. 27 E-01 9. 22 E-01 0. 9768
 Example 3. Inconsistent structural hypotheses were checked by DFT calculations. Measured accurate mass produced MF = C 27 H 22 N 4 O 3 A. Balandina et al. Rus. Chem. Bul. , Int. Ed. , 2006, 55, 2256 -2264
Example 3. Inconsistent structural hypotheses were checked by DFT calculations. Measured accurate mass produced MF = C 27 H 22 N 4 O 3 A. Balandina et al. Rus. Chem. Bul. , Int. Ed. , 2006, 55, 2256 -2264
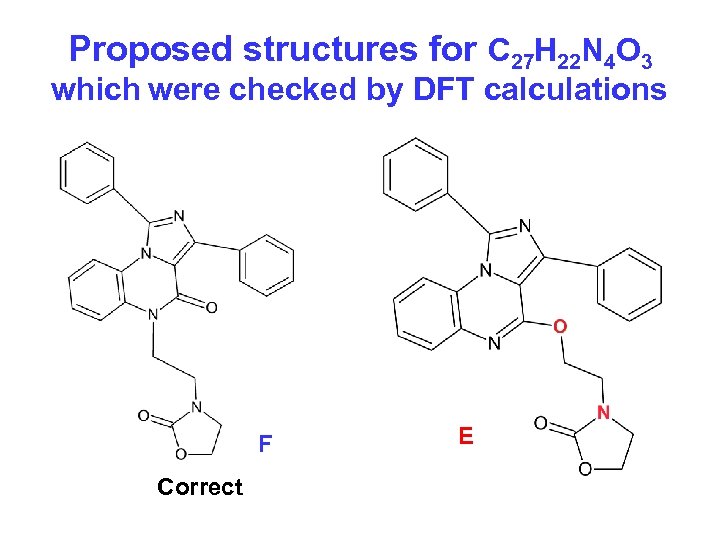 Proposed structures for C 27 H 22 N 4 O 3 which were checked by DFT calculations F Correct E
Proposed structures for C 27 H 22 N 4 O 3 which were checked by DFT calculations F Correct E
 Proposed structures with different MFs which were checked by DFT calculations. Experimental MF=C 27 H 22 N 4 O 3 C 27 H 23 N 4 O 2 Doublet! 154 ~sp 3! C 27 H 22 N 4 O 2
Proposed structures with different MFs which were checked by DFT calculations. Experimental MF=C 27 H 22 N 4 O 3 C 27 H 23 N 4 O 2 Doublet! 154 ~sp 3! C 27 H 22 N 4 O 2
 Structure Generation was run from MCD
Structure Generation was run from MCD
 Result: k=44 25, tg=0 s 891 ms
Result: k=44 25, tg=0 s 891 ms
 Nonstandard correlations (NSCs) a=2 • If the axioms upon correlation length are violated, the data become contradictory. a=1 COSY a=1 a=2 HMBC
Nonstandard correlations (NSCs) a=2 • If the axioms upon correlation length are violated, the data become contradictory. a=1 COSY a=1 a=2 HMBC
 Automatic removing contradictions from 2 D NMR data. Case when a=1. 1. Logical analysis of integrated 2 D NMR data is performed. Such atoms are detected at which nonstandard connectivities can present. 2. All connectivities at suspicious atoms are lengthened by one bond (a=1). • Structure Generation is performed from the modified connectivity set. S. G. Molodtsov, M. E. Elyashberg, K. A. Blinov et al. J. Chem. Inf. Model. 2004, 44, 1737 -1751.
Automatic removing contradictions from 2 D NMR data. Case when a=1. 1. Logical analysis of integrated 2 D NMR data is performed. Such atoms are detected at which nonstandard connectivities can present. 2. All connectivities at suspicious atoms are lengthened by one bond (a=1). • Structure Generation is performed from the modified connectivity set. S. G. Molodtsov, M. E. Elyashberg, K. A. Blinov et al. J. Chem. Inf. Model. 2004, 44, 1737 -1751.
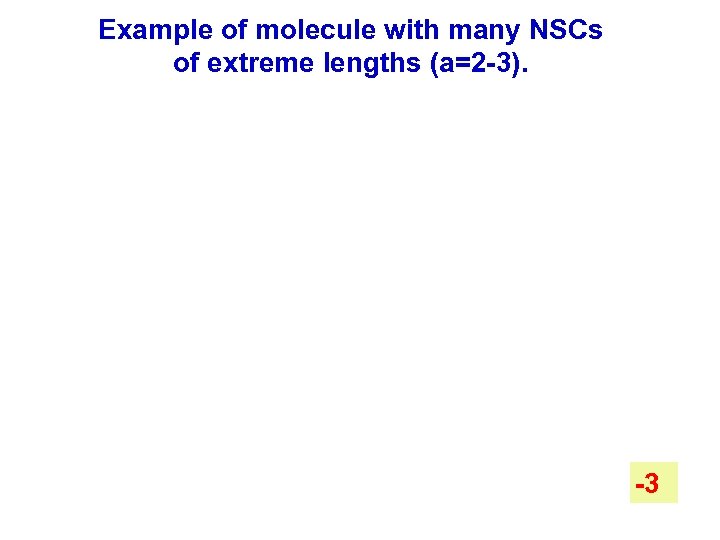 Example of molecule with many NSCs of extreme lengths (a=2 -3). m=15, a=1 -3
Example of molecule with many NSCs of extreme lengths (a=2 -3). m=15, a=1 -3
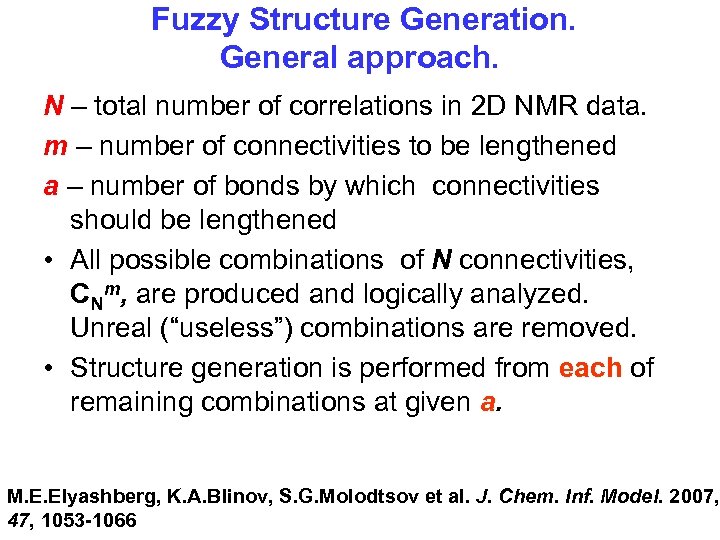 Fuzzy Structure Generation. General approach. N – total number of correlations in 2 D NMR data. m – number of connectivities to be lengthened а – number of bonds by which connectivities should be lengthened • All possible combinations of N connectivities, CNm, are produced and logically analyzed. Unreal (“useless”) combinations are removed. • Structure generation is performed from each of remaining combinations at given a. M. E. Elyashberg, K. A. Blinov, S. G. Molodtsov et al. J. Chem. Inf. Model. 2007, 47, 1053 -1066
Fuzzy Structure Generation. General approach. N – total number of correlations in 2 D NMR data. m – number of connectivities to be lengthened а – number of bonds by which connectivities should be lengthened • All possible combinations of N connectivities, CNm, are produced and logically analyzed. Unreal (“useless”) combinations are removed. • Structure generation is performed from each of remaining combinations at given a. M. E. Elyashberg, K. A. Blinov, S. G. Molodtsov et al. J. Chem. Inf. Model. 2007, 47, 1053 -1066
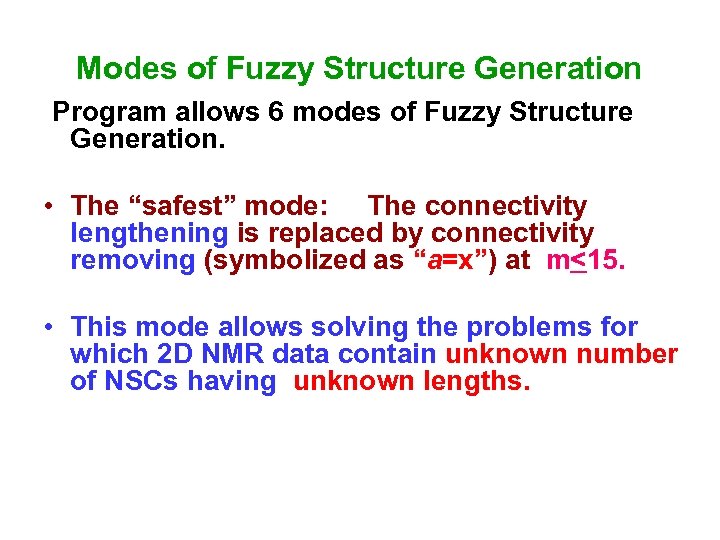 Modes of Fuzzy Structure Generation Program allows 6 modes of Fuzzy Structure Generation. • The “safest” mode: The connectivity lengthening is replaced by connectivity removing (symbolized as “а=x”) at m<15. • This mode allows solving the problems for which 2 D NMR data contain unknown number of NSCs having unknown lengths.
Modes of Fuzzy Structure Generation Program allows 6 modes of Fuzzy Structure Generation. • The “safest” mode: The connectivity lengthening is replaced by connectivity removing (symbolized as “а=x”) at m<15. • This mode allows solving the problems for which 2 D NMR data contain unknown number of NSCs having unknown lengths.
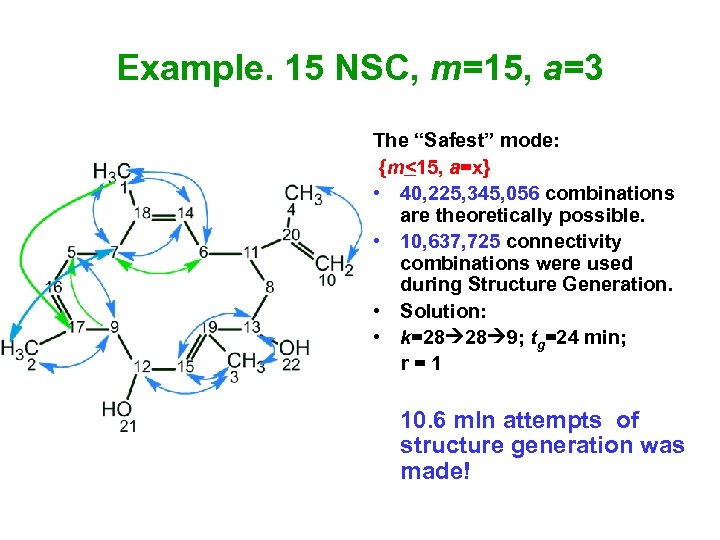 Example. 15 NSC, m=15, а=3 The “Safest” mode: {m<15, a=x} • 40, 225, 345, 056 combinations are theoretically possible. • 10, 637, 725 connectivity combinations were used during Structure Generation. • Solution: • k=28 28 9; tg=24 min; r = 1 10. 6 mln attempts of structure generation was made!
Example. 15 NSC, m=15, а=3 The “Safest” mode: {m<15, a=x} • 40, 225, 345, 056 combinations are theoretically possible. • 10, 637, 725 connectivity combinations were used during Structure Generation. • Solution: • k=28 28 9; tg=24 min; r = 1 10. 6 mln attempts of structure generation was made!
 About 15 000 of ~ 200 000 natural products posses symmetry. Peculiarities of structure generation of symmetric molecules from 2 D NMR data were not investigated. Structure generation was stopped after 44(!) h of program running. New algorithm of structure generation reveals symmetry in NMR data. Algorithm is capable of automatic adjusting to generation of symmetric molecules.
About 15 000 of ~ 200 000 natural products posses symmetry. Peculiarities of structure generation of symmetric molecules from 2 D NMR data were not investigated. Structure generation was stopped after 44(!) h of program running. New algorithm of structure generation reveals symmetry in NMR data. Algorithm is capable of automatic adjusting to generation of symmetric molecules.
 Example: C 44 H 72 O 16, n=60 There are 2 NSCs in HMBC. FUZZY STRUCTURE GENERATION: m=0 15, a=x. RESULT: k=5304 174 139; tg=4 m 30 s; r=1
Example: C 44 H 72 O 16, n=60 There are 2 NSCs in HMBC. FUZZY STRUCTURE GENERATION: m=0 15, a=x. RESULT: k=5304 174 139; tg=4 m 30 s; r=1
 Ionic structures
Ionic structures
 Properties of information obtained from 2 D NMR data • Information is fuzzy by the nature (2 or 3 bonds between H and C in НМВС). • Not all possible correlations are observed in spectra, i. e. information is incomplete. • Presence of nonstandard correlations frequently makes information contradictory. • Number of NSCs and they lengths are unknown. Signal overlapping leads to appearance of ambiguous correlations. Information is else indefinite.
Properties of information obtained from 2 D NMR data • Information is fuzzy by the nature (2 or 3 bonds between H and C in НМВС). • Not all possible correlations are observed in spectra, i. e. information is incomplete. • Presence of nonstandard correlations frequently makes information contradictory. • Number of NSCs and they lengths are unknown. Signal overlapping leads to appearance of ambiguous correlations. Information is else indefinite.
 Когда б вы знали, из какого сора Растут стихи. . . O, if you knew from which rubbish Poetry grows… Anna Akhmatova
Когда б вы знали, из какого сора Растут стихи. . . O, if you knew from which rubbish Poetry grows… Anna Akhmatova
 To overcome the lack of information, Database Fragments (1. 7 mln) or/and User’s Fragments are used. Introduction of fragments is necessary IF: 1. Number of observed 2 D NMR correlations is markedly smaller than theoretically expected one. 2. Deficit of hydrogen atoms has place. As a result even theoretically expected number of correlations is too small. • Taking this into account an algorithm of fragment “implantation” into MCD was developed.
To overcome the lack of information, Database Fragments (1. 7 mln) or/and User’s Fragments are used. Introduction of fragments is necessary IF: 1. Number of observed 2 D NMR correlations is markedly smaller than theoretically expected one. 2. Deficit of hydrogen atoms has place. As a result even theoretically expected number of correlations is too small. • Taking this into account an algorithm of fragment “implantation” into MCD was developed.
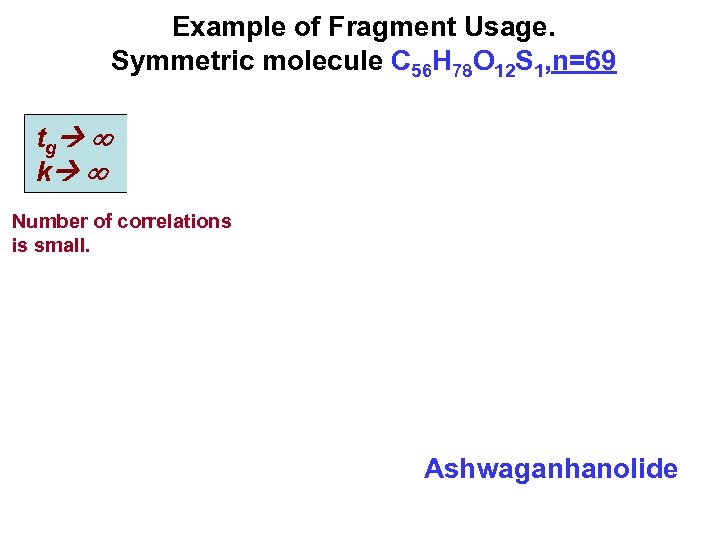 Example of Fragment Usage. Symmetric molecule C 56 H 78 O 12 S 1, n=69 tg k Number of correlations is small. Ashwaganhanolide
Example of Fragment Usage. Symmetric molecule C 56 H 78 O 12 S 1, n=69 tg k Number of correlations is small. Ashwaganhanolide
 Fragments were found in DB from 13 C NMR search. Number of Found Fragments L=5524. Fragment # 1 С 17 Н 22 О 2 Mol. Frag.
Fragments were found in DB from 13 C NMR search. Number of Found Fragments L=5524. Fragment # 1 С 17 Н 22 О 2 Mol. Frag.
 Solution • • 960 MCDs were created from the fragment #1 Structure Generation from 960 MCDs: k=960 24 6 tg= 29 m 30 s
Solution • • 960 MCDs were created from the fragment #1 Structure Generation from 960 MCDs: k=960 24 6 tg= 29 m 30 s
 Ashwaganhanolide. Output file.
Ashwaganhanolide. Output file.
 C 42 H 28 О 10, n=52 Common Mode, k= 8 1, t= 8 сек
C 42 H 28 О 10, n=52 Common Mode, k= 8 1, t= 8 сек
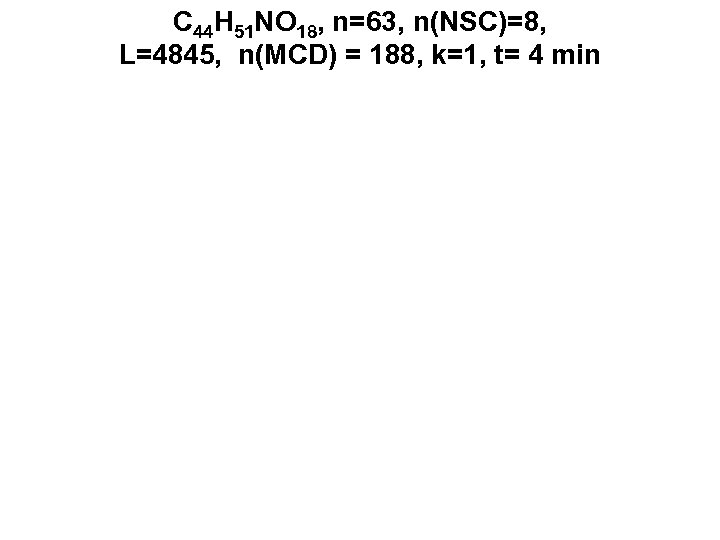 C 44 H 51 NO 18, n=63, n(NSC)=8, L=4845, n(MCD) = 188, k=1, t= 4 min
C 44 H 51 NO 18, n=63, n(NSC)=8, L=4845, n(MCD) = 188, k=1, t= 4 min
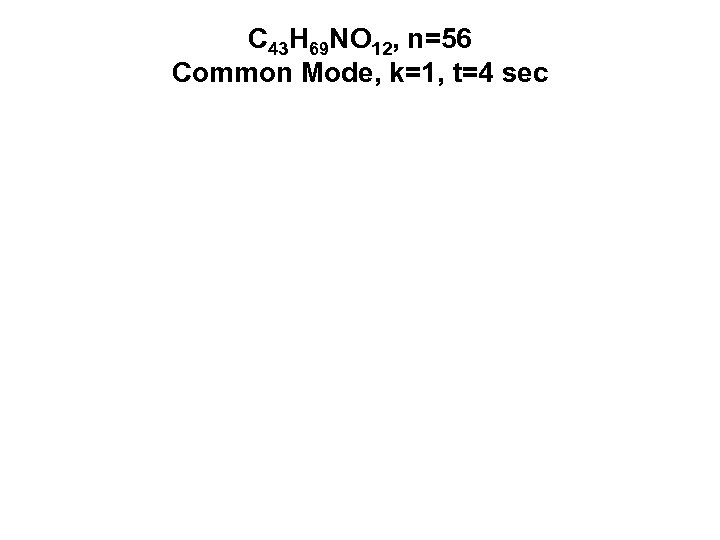 C 43 H 69 NO 12, n=56 Common Mode, k=1, t=4 sec
C 43 H 69 NO 12, n=56 Common Mode, k=1, t=4 sec
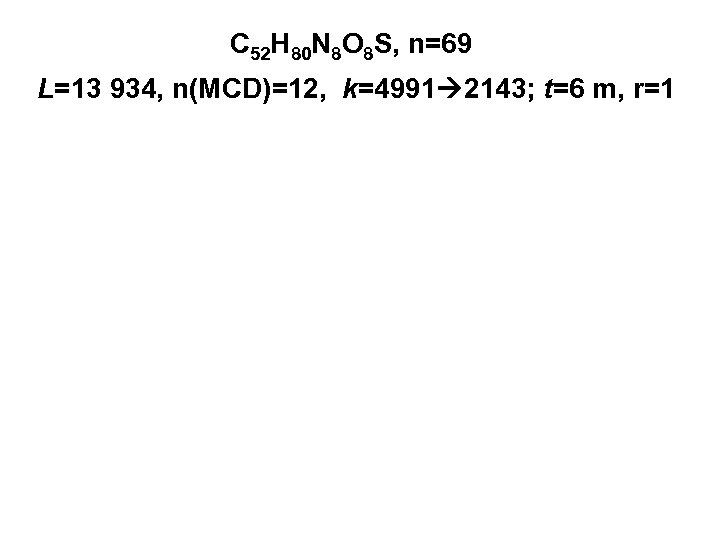 C 52 H 80 N 8 O 8 S, n=69 L=13 934, n(MCD)=12, k=4991 2143; t=6 m, r=1
C 52 H 80 N 8 O 8 S, n=69 L=13 934, n(MCD)=12, k=4991 2143; t=6 m, r=1
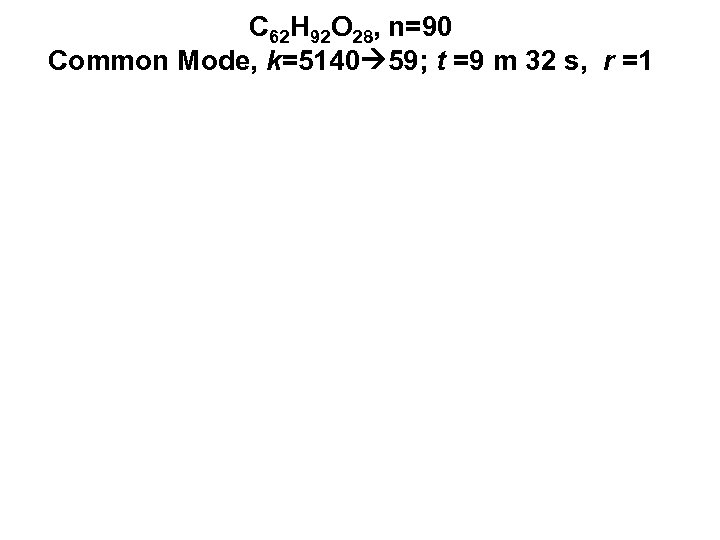 C 62 H 92 O 28, n=90 Common Mode, k=5140 59; t =9 m 32 s, r =1
C 62 H 92 O 28, n=90 Common Mode, k=5140 59; t =9 m 32 s, r =1
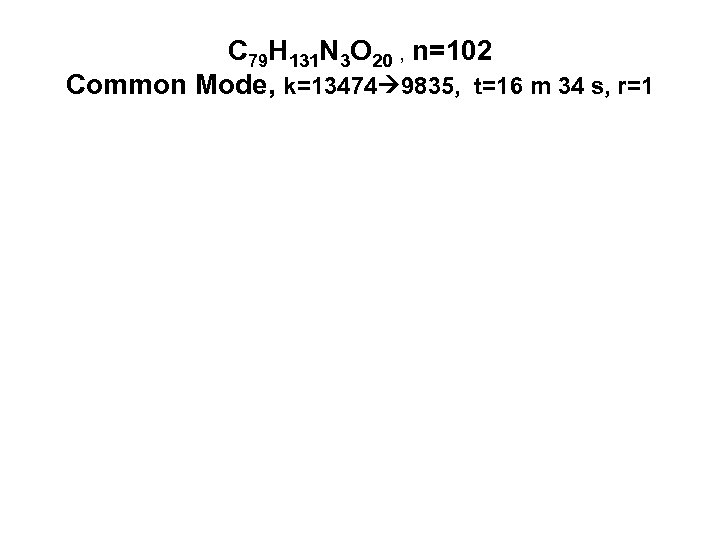 С 79 Н 131 N 3 O 20 , n=102 Common Mode, k=13474 9835, t=16 m 34 s, r=1
С 79 Н 131 N 3 O 20 , n=102 Common Mode, k=13474 9835, t=16 m 34 s, r=1
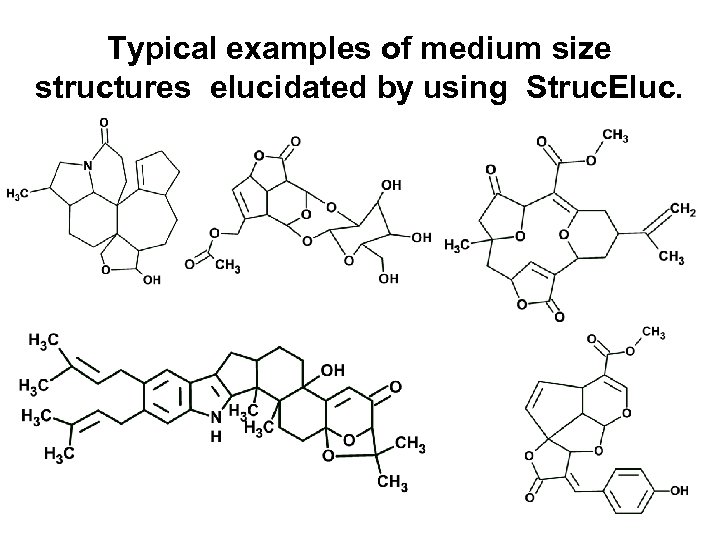 Typical examples of medium size structures elucidated by using Struc. Eluc.
Typical examples of medium size structures elucidated by using Struc. Eluc.
 Usage of fragments is not panacea for all cases. Possible causes of failures: • Large fragments capable of helping to solve a problem are absent from DB of the system. • Appropriate fragments are found or introduced by chemist, but the number of possible shift assignments is so huge (more than 100 million), that CPU resources fail (combinatorial explosion). • Number of MCDs created by program is huge. Structure generation CPU time becomes not acceptable.
Usage of fragments is not panacea for all cases. Possible causes of failures: • Large fragments capable of helping to solve a problem are absent from DB of the system. • Appropriate fragments are found or introduced by chemist, but the number of possible shift assignments is so huge (more than 100 million), that CPU resources fail (combinatorial explosion). • Number of MCDs created by program is huge. Structure generation CPU time becomes not acceptable.
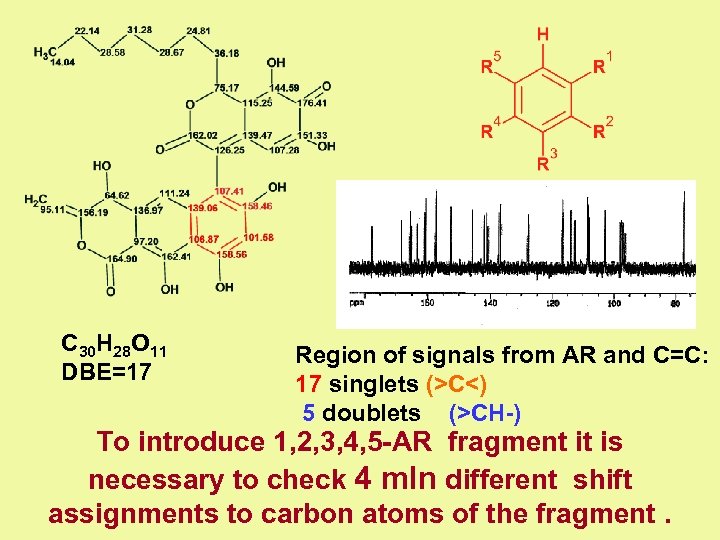 C 30 H 28 O 11 DBE=17 Region of signals from AR and С=С: 17 singlets (>C<) 5 doublets (>CH-) To introduce 1, 2, 3, 4, 5 -AR fragment it is necessary to check 4 mln different shift assignments to carbon atoms of the fragment.
C 30 H 28 O 11 DBE=17 Region of signals from AR and С=С: 17 singlets (>C<) 5 doublets (>CH-) To introduce 1, 2, 3, 4, 5 -AR fragment it is necessary to check 4 mln different shift assignments to carbon atoms of the fragment.
 “Between two combinatorial explosions…” • Attempt of structure generation from free atoms (Common Mode) leads to combinatorial explosion (too many structures). • Introducing large fragments to overcome the explosion leads to another combinatorial explosion (too many assignments) In this situation User Database can help.
“Between two combinatorial explosions…” • Attempt of structure generation from free atoms (Common Mode) leads to combinatorial explosion (too many structures). • Introducing large fragments to overcome the explosion leads to another combinatorial explosion (too many assignments) In this situation User Database can help.
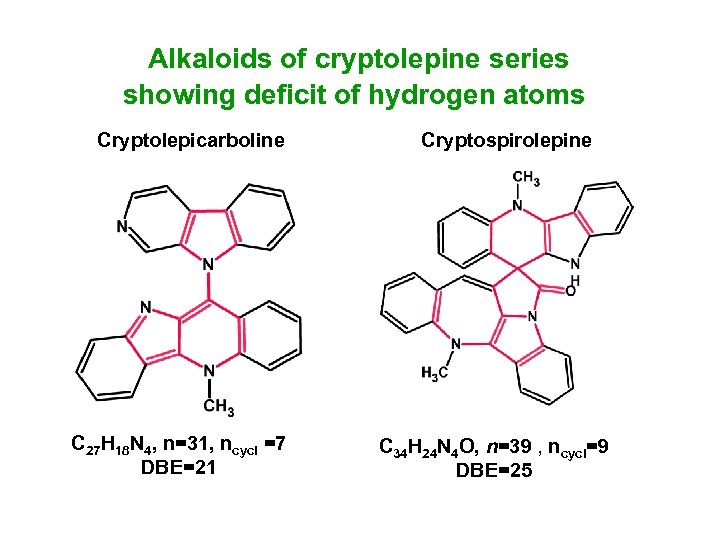 Alkaloids of cryptolepine series showing deficit of hydrogen atoms Cryptolepicarboline C 27 H 18 N 4, n=31, ncycl =7 DBE=21 Cryptospirolepine C 34 H 24 N 4 O, n=39 , ncycl=9 DBE=25
Alkaloids of cryptolepine series showing deficit of hydrogen atoms Cryptolepicarboline C 27 H 18 N 4, n=31, ncycl =7 DBE=21 Cryptospirolepine C 34 H 24 N 4 O, n=39 , ncycl=9 DBE=25
 Alkaloids of cryptolepine series for which signals in 13 C и 1 H NMR are assigned. User Fragment Data Base (UDB) was created. UDB contains 342 fragments.
Alkaloids of cryptolepine series for which signals in 13 C и 1 H NMR are assigned. User Fragment Data Base (UDB) was created. UDB contains 342 fragments.
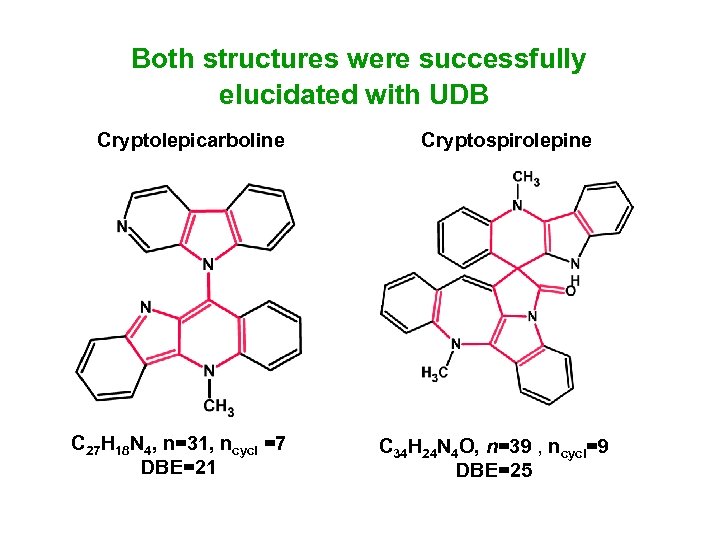 Both structures were successfully elucidated with UDB Cryptolepicarboline C 27 H 18 N 4, n=31, ncycl =7 DBE=21 Cryptospirolepine C 34 H 24 N 4 O, n=39 , ncycl=9 DBE=25
Both structures were successfully elucidated with UDB Cryptolepicarboline C 27 H 18 N 4, n=31, ncycl =7 DBE=21 Cryptospirolepine C 34 H 24 N 4 O, n=39 , ncycl=9 DBE=25
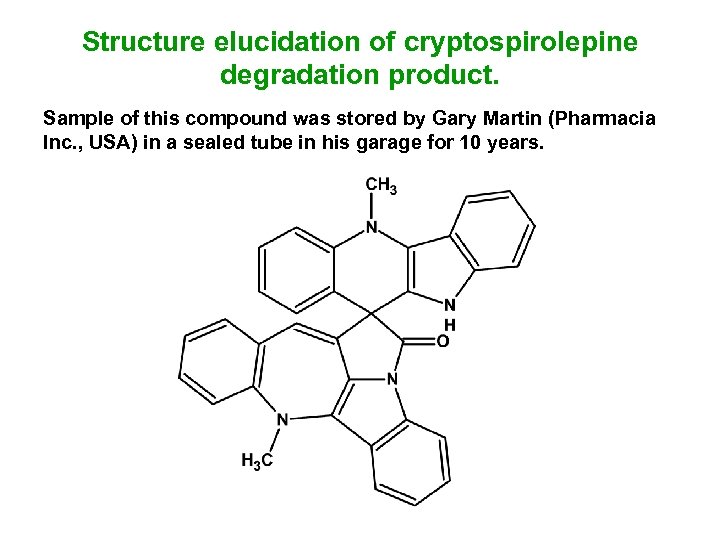 Structure elucidation of cryptospirolepine degradation product. Sample of this compound was stored by Gary Martin (Pharmacia Inc. , USA) in a sealed tube in his garage for 10 years.
Structure elucidation of cryptospirolepine degradation product. Sample of this compound was stored by Gary Martin (Pharmacia Inc. , USA) in a sealed tube in his garage for 10 years.
 LC chromatogram of degradation products (26 peaks). 35 % 16 %
LC chromatogram of degradation products (26 peaks). 35 % 16 %
 DP-2 separation and spectra registration were performed by several groups in USA. • DP-1 (35%, 1. 1 mg), • DP-2 (16%, 200 g). • ЯМР DP-2: solution of 100 g in 150 l of D-DMSO; ampoule 3 mm, Т=25 К, • HSQC (17 h), HMBC (17 h), • 1 H-15 N HMBC (72 h), sensitivity to 15 N is 50 times lower than to 13 C • Н-Н ROESY • It was found from MS: • MSMS : MH+=479, C 32 H 22 N 4 O
DP-2 separation and spectra registration were performed by several groups in USA. • DP-1 (35%, 1. 1 mg), • DP-2 (16%, 200 g). • ЯМР DP-2: solution of 100 g in 150 l of D-DMSO; ampoule 3 mm, Т=25 К, • HSQC (17 h), HMBC (17 h), • 1 H-15 N HMBC (72 h), sensitivity to 15 N is 50 times lower than to 13 C • Н-Н ROESY • It was found from MS: • MSMS : MH+=479, C 32 H 22 N 4 O
 DP-2. Solution to the problem. From MS/MS: C 32 H 22 N 4 O 101 fragments were selected from UDB by NMR 13 C. 1376 MCDs were created from the fragments Structure generation from 1376 MCDs. Results: k=785 75, tgen = 6 min.
DP-2. Solution to the problem. From MS/MS: C 32 H 22 N 4 O 101 fragments were selected from UDB by NMR 13 C. 1376 MCDs were created from the fragments Structure generation from 1376 MCDs. Results: k=785 75, tgen = 6 min.
 First 8 structures of ranked output file.
First 8 structures of ranked output file.
 COST OF THE VICTORY Martin, G. E. ; Hadden, B. D. ; Russell, C. E. ; Kaluzny, D. J. ; Guido, J. E. ; Duholke, W. K; Stiemsma, B. A. ; Thamann, T. J. ; Crouch, R. C. ; Blinov, K. A. ; Elyashberg, M. E. ; Martirosian, E. R. ; Molodtsov, S. G. ; Williams, A. J. ; Schiff, P. L. Jr. Identification of Degradants of a Complex Alkaloid Using NMR Cryoprobe Technology and ACD/Structure Elucidator. J. Het. Chem. 2002, 39, 1241 -1250. Iliya Repin. Barge haulers on Volga. 1872
COST OF THE VICTORY Martin, G. E. ; Hadden, B. D. ; Russell, C. E. ; Kaluzny, D. J. ; Guido, J. E. ; Duholke, W. K; Stiemsma, B. A. ; Thamann, T. J. ; Crouch, R. C. ; Blinov, K. A. ; Elyashberg, M. E. ; Martirosian, E. R. ; Molodtsov, S. G. ; Williams, A. J. ; Schiff, P. L. Jr. Identification of Degradants of a Complex Alkaloid Using NMR Cryoprobe Technology and ACD/Structure Elucidator. J. Het. Chem. 2002, 39, 1241 -1250. Iliya Repin. Barge haulers on Volga. 1872
 ТС-6. The greatest challenge for CASE systems Gary Martin’s group has separated unknown alkaloid ТС-6 of cryptolepine series. Martin, a prominent expert in NMR and the structure elucidation, failed to determine structure of this compound during 10 years (since 90 th). Solution was found using Struc. Eluc in interactive mode. Initial MCD was transformed into the final one by spectroscopist during 12 hours of program operating.
ТС-6. The greatest challenge for CASE systems Gary Martin’s group has separated unknown alkaloid ТС-6 of cryptolepine series. Martin, a prominent expert in NMR and the structure elucidation, failed to determine structure of this compound during 10 years (since 90 th). Solution was found using Struc. Eluc in interactive mode. Initial MCD was transformed into the final one by spectroscopist during 12 hours of program operating.
 SOLUTION: k=353 266, tgen=2 s The first 8 structures of the output file.
SOLUTION: k=353 266, tgen=2 s The first 8 structures of the output file.
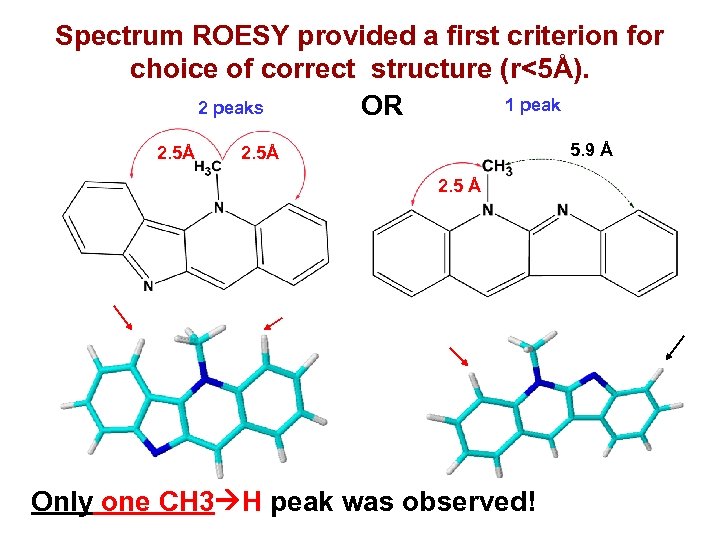 Spectrum ROESY provided a first criterion for choice of correct structure (r<5Å). 1 peak 2 peaks OR 2. 5Å 5. 9 Å 2. 5 Å Only one CH 3 H peak was observed!
Spectrum ROESY provided a first criterion for choice of correct structure (r<5Å). 1 peak 2 peaks OR 2. 5Å 5. 9 Å 2. 5 Å Only one CH 3 H peak was observed!
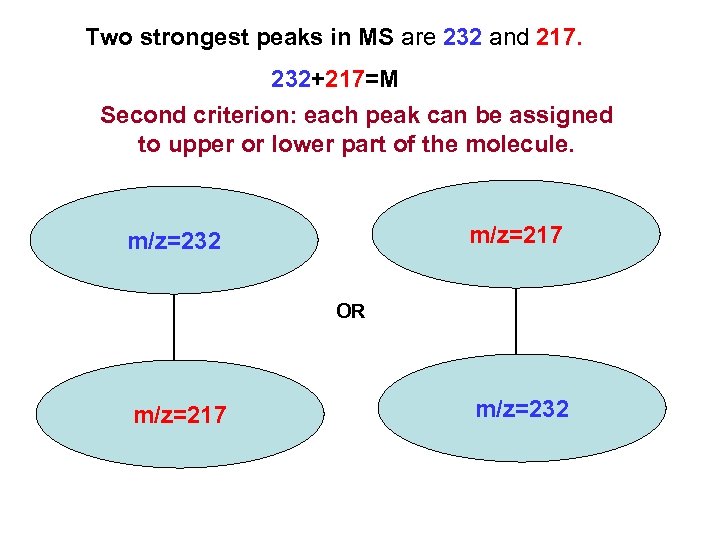 Two strongest peaks in MS are 232 and 217. 232+217=M Second criterion: each peak can be assigned to upper or lower part of the molecule. m/z=217 m/z=232 OR m/z=217 m/z=232
Two strongest peaks in MS are 232 and 217. 232+217=M Second criterion: each peak can be assigned to upper or lower part of the molecule. m/z=217 m/z=232 OR m/z=217 m/z=232
 Top of the output file Only structure #2 meets MS and ROESY constraints.
Top of the output file Only structure #2 meets MS and ROESY constraints.
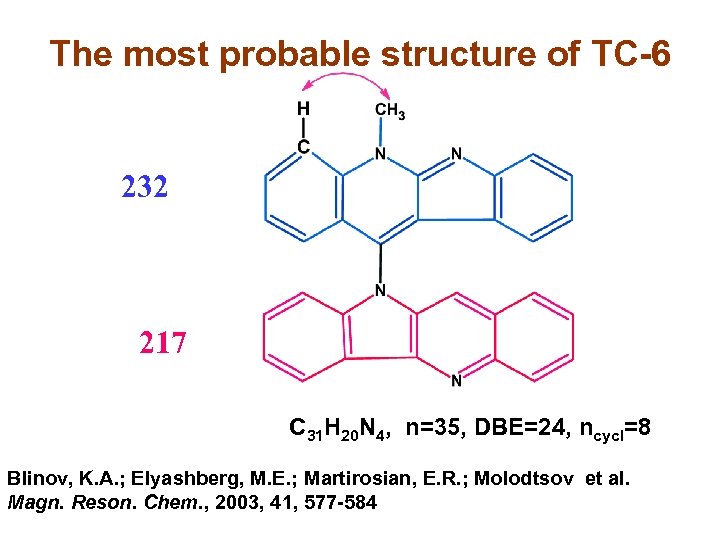 The most probable structure of ТС-6 232 217 C 31 H 20 N 4, n=35, DBE=24, ncycl=8 Blinov, K. A. ; Elyashberg, M. E. ; Martirosian, E. R. ; Molodtsov et al. Magn. Reson. Chem. , 2003, 41, 577 -584
The most probable structure of ТС-6 232 217 C 31 H 20 N 4, n=35, DBE=24, ncycl=8 Blinov, K. A. ; Elyashberg, M. E. ; Martirosian, E. R. ; Molodtsov et al. Magn. Reson. Chem. , 2003, 41, 577 -584
 For the first time, application of ES allowed solving a structural problem, which a prominent expert in NMR spectroscopy and structure elucidation failed to solve.
For the first time, application of ES allowed solving a structural problem, which a prominent expert in NMR spectroscopy and structure elucidation failed to solve.
 One more challenge. . . • MW = 1515. 38 Da for (M+H)+ • Raw spectra: 1 М: 13 C NMR , 13 C NMR DEPT , 1 H NMR, 2 М: 1 H/13 C HSQC, 1 H/13 C HMBC, 1 H/1 H COSY, 1 H/1 H TOCSY. • From 13 C NMR: C 69 • From 1 H NMR and 1 H/13 C HSQC: H 66
One more challenge. . . • MW = 1515. 38 Da for (M+H)+ • Raw spectra: 1 М: 13 C NMR , 13 C NMR DEPT , 1 H NMR, 2 М: 1 H/13 C HSQC, 1 H/13 C HMBC, 1 H/1 H COSY, 1 H/1 H TOCSY. • From 13 C NMR: C 69 • From 1 H NMR and 1 H/13 C HSQC: H 66
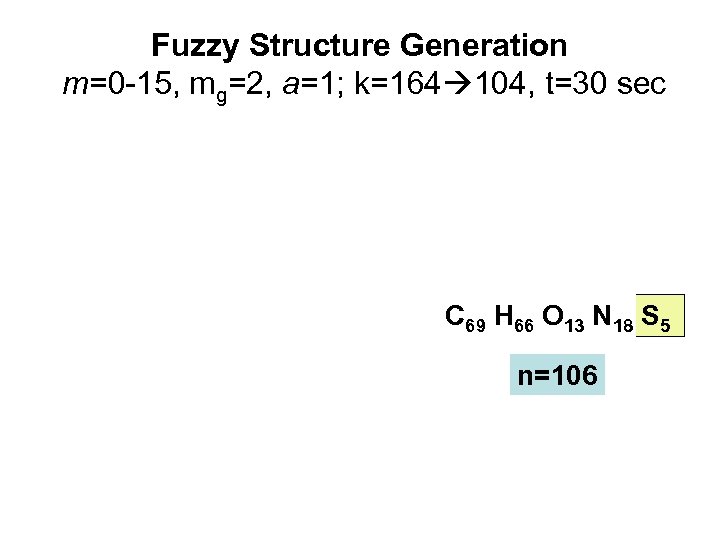 Fuzzy Structure Generation m=0 -15, mg=2, a=1; k=164 104, t=30 sec C 69 H 66 O 13 N 18 S 5 n=106
Fuzzy Structure Generation m=0 -15, mg=2, a=1; k=164 104, t=30 sec C 69 H 66 O 13 N 18 S 5 n=106
 Determination of relative stereochemistry of identified structures. • Biological activity of substances depends on their stereochemistry. • Struc. Eluc was enhanced by algorithm of determining the most probable relative stereochemistry of rigid structures. . Stereochemistry is determined using NOESY ROESY data. For structures having more than 7 stereocenters, optimization of geometry is performed by means of Genetic Algorithm (GA).
Determination of relative stereochemistry of identified structures. • Biological activity of substances depends on their stereochemistry. • Struc. Eluc was enhanced by algorithm of determining the most probable relative stereochemistry of rigid structures. . Stereochemistry is determined using NOESY ROESY data. For structures having more than 7 stereocenters, optimization of geometry is performed by means of Genetic Algorithm (GA).
 Brevetoxin B Number of stereocenters: N=23 Number of stereoisomers ~ 8, 400 000 CPU time necessary for optimizing geometry of all 8. 4 mln stereoisomers ~ 1 month Configuration of all 23 stereocenters was correctly determined by GA in 2 h 50 m.
Brevetoxin B Number of stereocenters: N=23 Number of stereoisomers ~ 8, 400 000 CPU time necessary for optimizing geometry of all 8. 4 mln stereoisomers ~ 1 month Configuration of all 23 stereocenters was correctly determined by GA in 2 h 50 m.
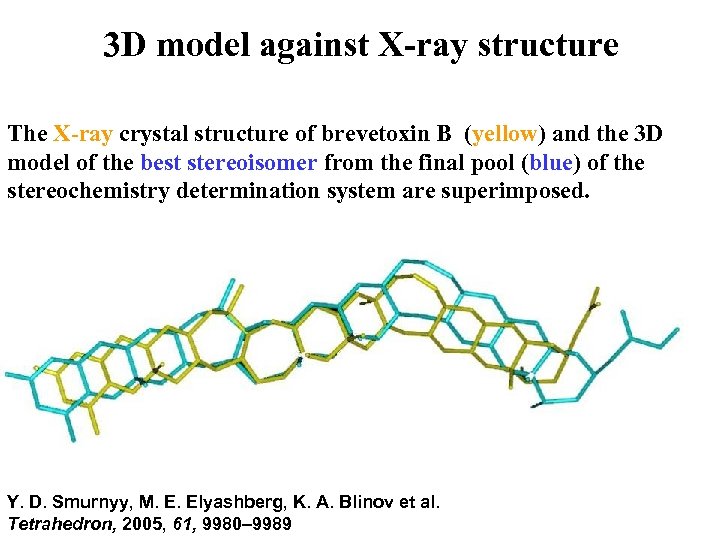 3 D model against X-ray structure The X-ray crystal structure of brevetoxin B (yellow) and the 3 D model of the best stereoisomer from the final pool (blue) of the stereochemistry determination system are superimposed. Y. D. Smurnyy, M. E. Elyashberg, K. A. Blinov et al. Tetrahedron, 2005, 61, 9980– 9989
3 D model against X-ray structure The X-ray crystal structure of brevetoxin B (yellow) and the 3 D model of the best stereoisomer from the final pool (blue) of the stereochemistry determination system are superimposed. Y. D. Smurnyy, M. E. Elyashberg, K. A. Blinov et al. Tetrahedron, 2005, 61, 9980– 9989
 Efficiency of Structure Elucidator • System efficiency was proved by structure elucidation of ~300 natural products. • Permanent solving new complicated problems is a basis for creation and further development of the Structure Elucidator.
Efficiency of Structure Elucidator • System efficiency was proved by structure elucidation of ~300 natural products. • Permanent solving new complicated problems is a basis for creation and further development of the Structure Elucidator.
 Other CASE systems • • • SESAMI (USA) CISOC-SES (USA) LSD (France) COCON (Germany) SENECA (Germany) All system have no Database containing Structures and Fragments with assigned NMR spectra. • All systems cannot do with nonstandard correlations. • Only “ideal” 2 D NMR data can be processed. • Some of these systems are used by authors. M. E. Elyashberg, A. J. Williams and G. E. Martin. Computer-Assisted Structure Verification and Elucidation Tools in NMR-Based Structure Elucidation. Progress in NMR Spectroscopy, 2008, No 2. Monographic review.
Other CASE systems • • • SESAMI (USA) CISOC-SES (USA) LSD (France) COCON (Germany) SENECA (Germany) All system have no Database containing Structures and Fragments with assigned NMR spectra. • All systems cannot do with nonstandard correlations. • Only “ideal” 2 D NMR data can be processed. • Some of these systems are used by authors. M. E. Elyashberg, A. J. Williams and G. E. Martin. Computer-Assisted Structure Verification and Elucidation Tools in NMR-Based Structure Elucidation. Progress in NMR Spectroscopy, 2008, No 2. Monographic review.
 Struc. Eluc is used in ca. 100 organizations in many countries. • • • Pfizer Roche Eli Lilly Novartis Astra. Zeneca Merck Bayer Mitsubishi Chemical Shell Chimie Samsung Electronics • Schering-Plough • Microbial Screening Technologies • Crompton Corporation • MNL Pharma • Fujisawa Pharm. Co • Amgen Inc • Sankyo Co. Ltd • Astellas Pharma Inc • Biovitrum AB • NCI-FRED CANCER • INOVACIA SWEDEN • Janssen Pharm.
Struc. Eluc is used in ca. 100 organizations in many countries. • • • Pfizer Roche Eli Lilly Novartis Astra. Zeneca Merck Bayer Mitsubishi Chemical Shell Chimie Samsung Electronics • Schering-Plough • Microbial Screening Technologies • Crompton Corporation • MNL Pharma • Fujisawa Pharm. Co • Amgen Inc • Sankyo Co. Ltd • Astellas Pharma Inc • Biovitrum AB • NCI-FRED CANCER • INOVACIA SWEDEN • Janssen Pharm.
 Expert system as a kernel of research center • It should be expected that an expert system similar to Structure Elucidator can serve as a kernel of a research center intended for molecular structure elucidation and investigation.
Expert system as a kernel of research center • It should be expected that an expert system similar to Structure Elucidator can serve as a kernel of a research center intended for molecular structure elucidation and investigation.
 • Expert systems like the Struc. Eluc will be used widespread in the nearest 5 -10 years. • They will become a routine tool in laboratories engaged in spectroscopy, organic chemistry, chemistry of natural products and analytical chemistry.
• Expert systems like the Struc. Eluc will be used widespread in the nearest 5 -10 years. • They will become a routine tool in laboratories engaged in spectroscopy, organic chemistry, chemistry of natural products and analytical chemistry.
 Structure Elucidator Team Sergey Molodtsov, Mikhail Elyashberg, Tatiyana Churanova, Kirill Blinov
Structure Elucidator Team Sergey Molodtsov, Mikhail Elyashberg, Tatiyana Churanova, Kirill Blinov



