68cd94841a85bbc3b7af6fb5c05742ed.ppt
- Количество слайдов: 70
 Characterizing millisecond motions in proteins using CPMG-relaxation dispersion measurements Tony Mittermaier Mc. Gill Aug, 2007 CCPN
Characterizing millisecond motions in proteins using CPMG-relaxation dispersion measurements Tony Mittermaier Mc. Gill Aug, 2007 CCPN
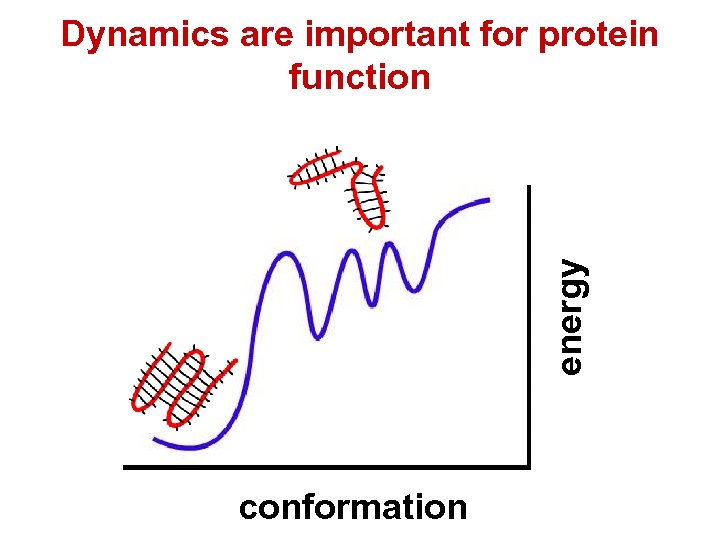 energy Dynamics are important for protein function conformation
energy Dynamics are important for protein function conformation
 Two-site conformational exchange • Weakly populated protein states are often not directly observable in NMR spectra.
Two-site conformational exchange • Weakly populated protein states are often not directly observable in NMR spectra.
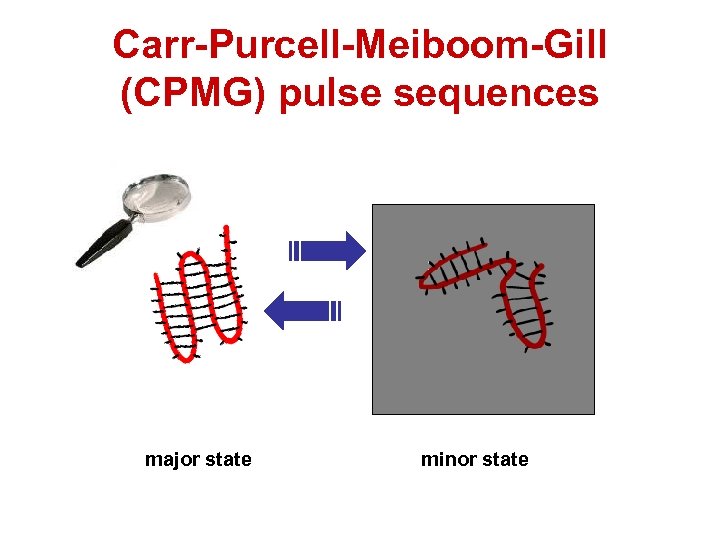 Carr-Purcell-Meiboom-Gill (CPMG) pulse sequences major state minor state
Carr-Purcell-Meiboom-Gill (CPMG) pulse sequences major state minor state
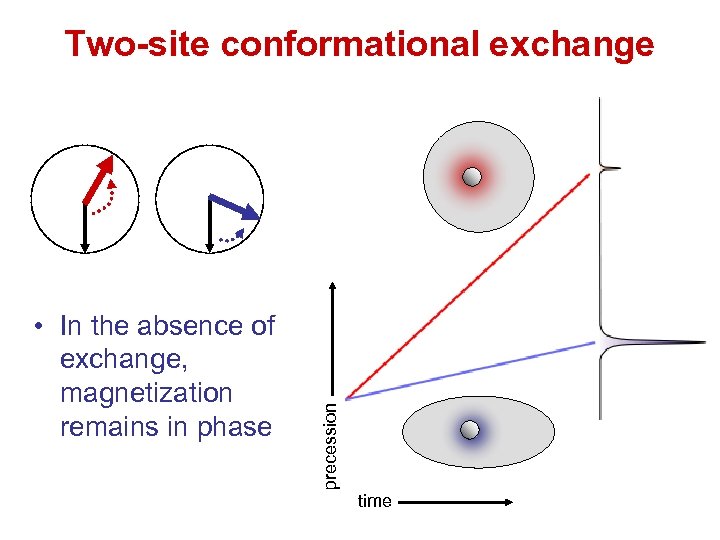 • In the absence of exchange, magnetization remains in phase precession Two-site conformational exchange time
• In the absence of exchange, magnetization remains in phase precession Two-site conformational exchange time
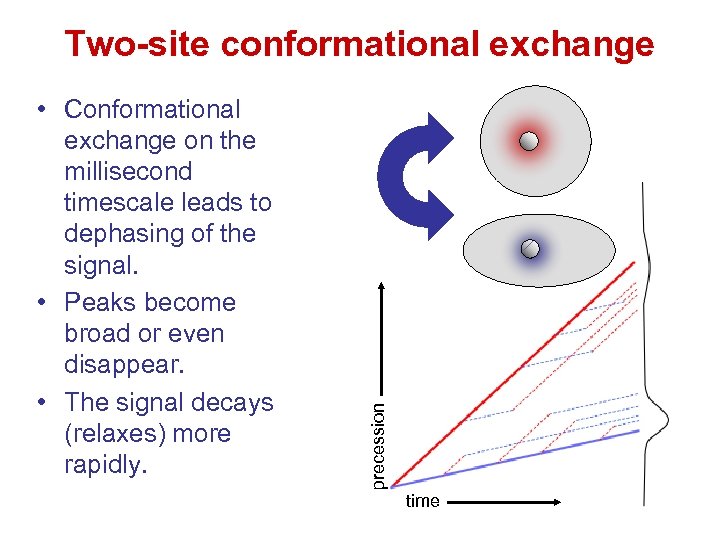 • Conformational exchange on the millisecond timescale leads to dephasing of the signal. • Peaks become broad or even disappear. • The signal decays (relaxes) more rapidly. precession Two-site conformational exchange time
• Conformational exchange on the millisecond timescale leads to dephasing of the signal. • Peaks become broad or even disappear. • The signal decays (relaxes) more rapidly. precession Two-site conformational exchange time
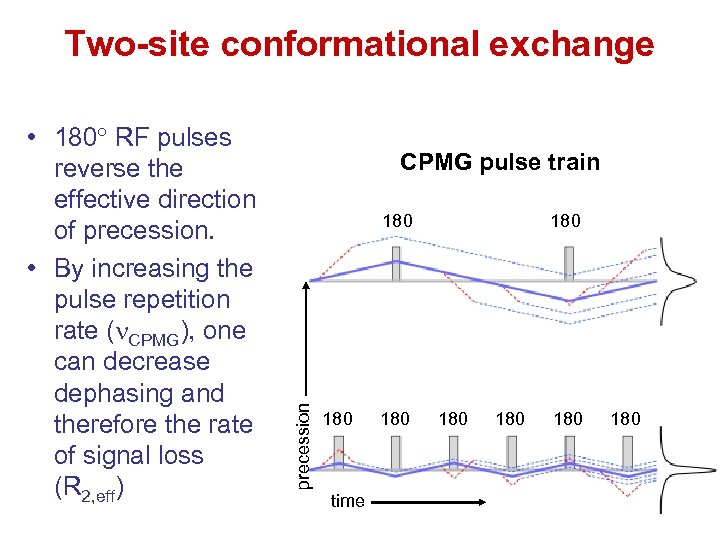 Two-site conformational exchange CPMG pulse train 180 precession • 180 RF pulses reverse the effective direction of precession. • By increasing the pulse repetition rate (n. CPMG), one can decrease dephasing and therefore the rate of signal loss (R 2, eff) 180 time 180 180 180
Two-site conformational exchange CPMG pulse train 180 precession • 180 RF pulses reverse the effective direction of precession. • By increasing the pulse repetition rate (n. CPMG), one can decrease dephasing and therefore the rate of signal loss (R 2, eff) 180 time 180 180 180
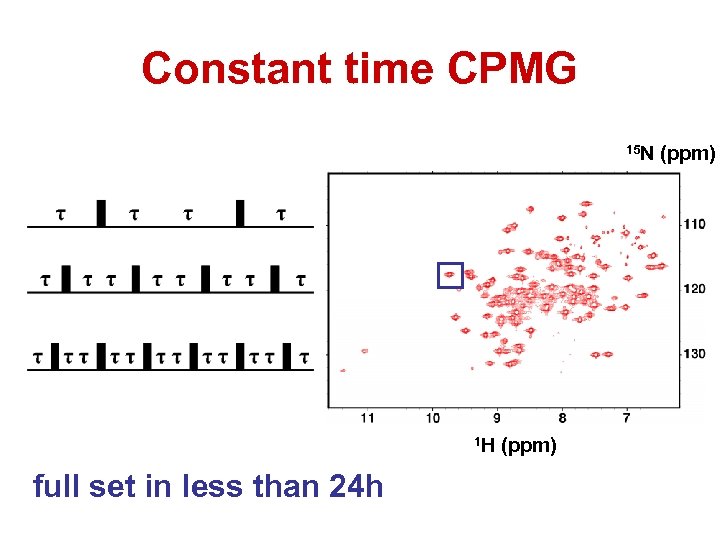 Constant time CPMG 15 N 1 H full set in less than 24 h (ppm)
Constant time CPMG 15 N 1 H full set in less than 24 h (ppm)
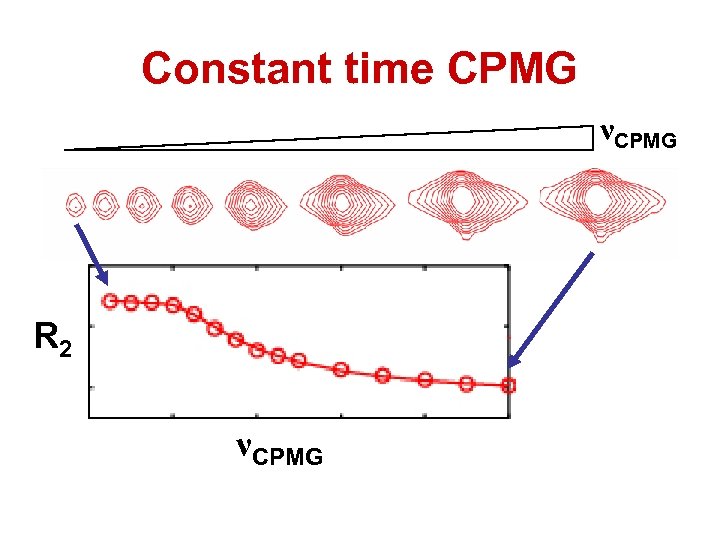 Constant time CPMG νCPMG R 2 νCPMG
Constant time CPMG νCPMG R 2 νCPMG
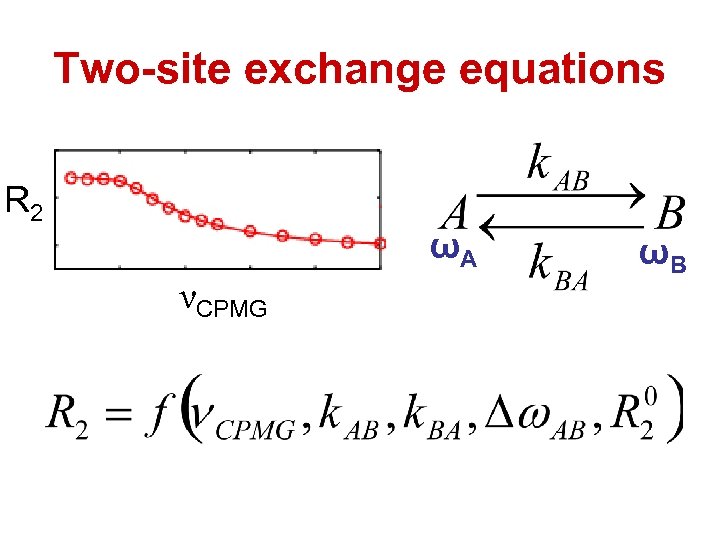 Two-site exchange equations R 2 ωA νCPMG ωB
Two-site exchange equations R 2 ωA νCPMG ωB
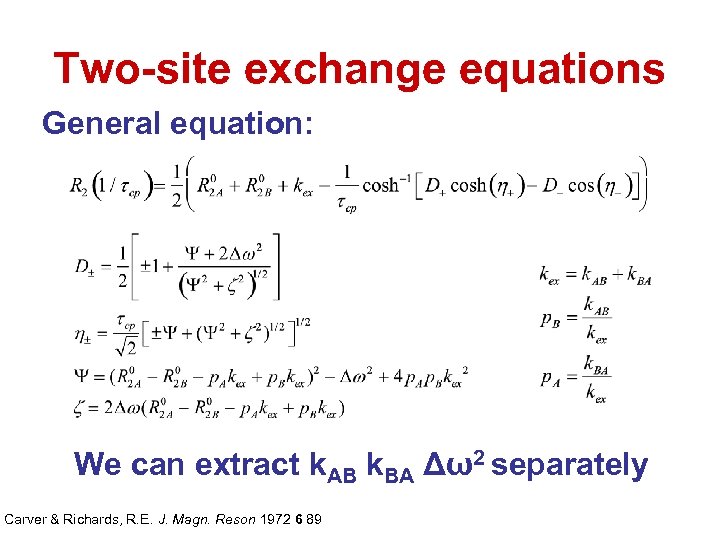 Two-site exchange equations General equation: We can extract k. AB k. BA Δω2 separately Carver & Richards, R. E. J. Magn. Reson 1972 6 89
Two-site exchange equations General equation: We can extract k. AB k. BA Δω2 separately Carver & Richards, R. E. J. Magn. Reson 1972 6 89
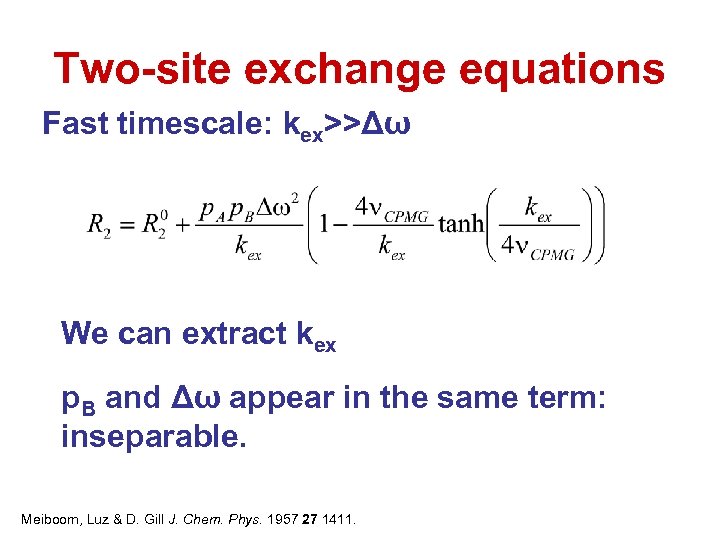 Two-site exchange equations Fast timescale: kex>>Δω We can extract kex p. B and Δω appear in the same term: inseparable. Meiboom, Luz & D. Gill J. Chem. Phys. 1957 27 1411.
Two-site exchange equations Fast timescale: kex>>Δω We can extract kex p. B and Δω appear in the same term: inseparable. Meiboom, Luz & D. Gill J. Chem. Phys. 1957 27 1411.
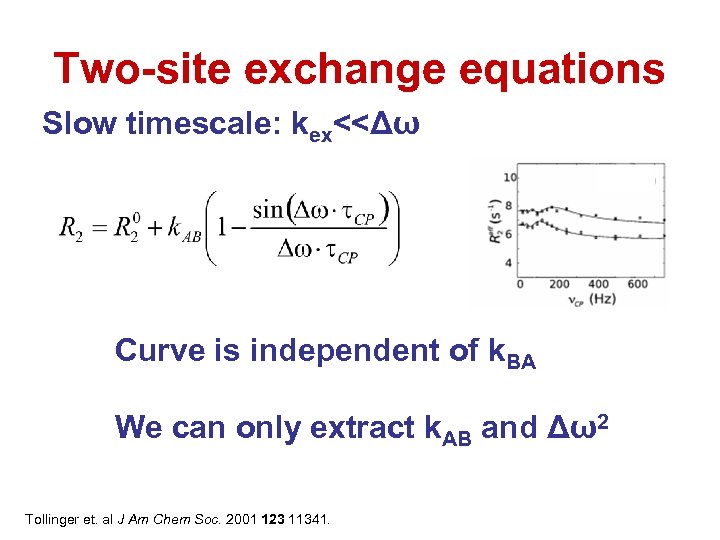 Two-site exchange equations Slow timescale: kex<<Δω Curve is independent of k. BA We can only extract k. AB and Δω2 Tollinger et. al J Am Chem Soc. 2001 123 11341.
Two-site exchange equations Slow timescale: kex<<Δω Curve is independent of k. BA We can only extract k. AB and Δω2 Tollinger et. al J Am Chem Soc. 2001 123 11341.
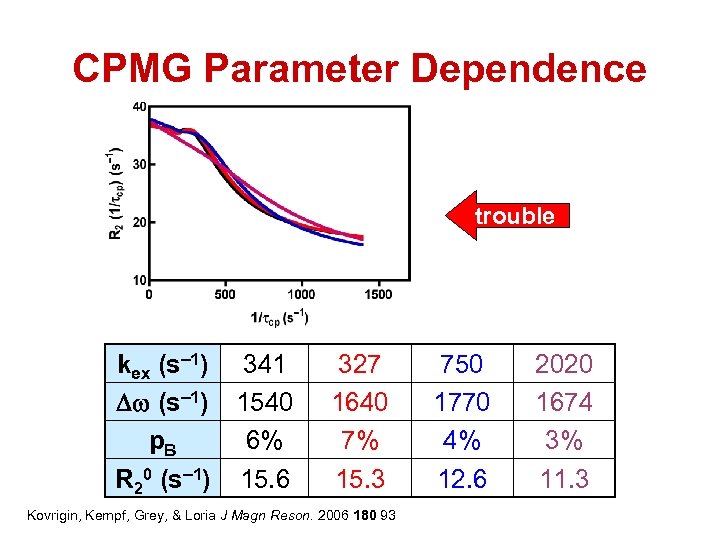 CPMG Parameter Dependence trouble kex (s– 1) 341 Dw (s– 1) 1540 p. B 6% R 20 (s– 1) 15. 6 327 1640 7% 15. 3 Kovrigin, Kempf, Grey, & Loria J Magn Reson. 2006 180 93 750 1770 4% 12. 6 2020 1674 3% 11. 3
CPMG Parameter Dependence trouble kex (s– 1) 341 Dw (s– 1) 1540 p. B 6% R 20 (s– 1) 15. 6 327 1640 7% 15. 3 Kovrigin, Kempf, Grey, & Loria J Magn Reson. 2006 180 93 750 1770 4% 12. 6 2020 1674 3% 11. 3
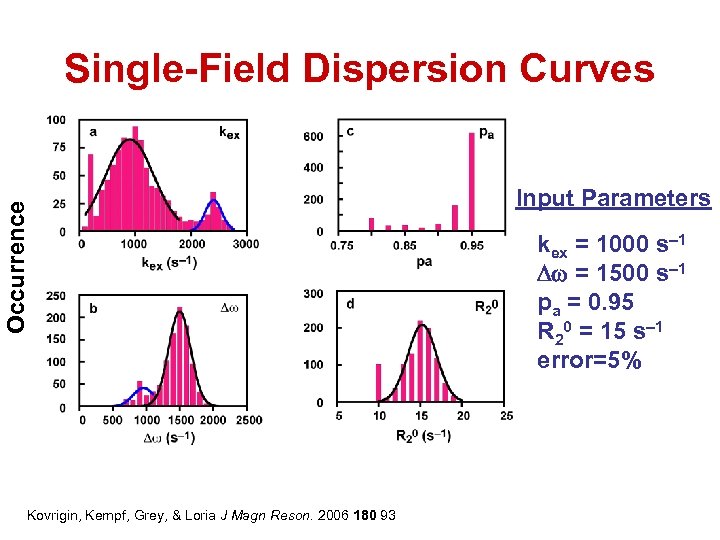 Occurrence Single-Field Dispersion Curves Kovrigin, Kempf, Grey, & Loria J Magn Reson. 2006 180 93 Input Parameters kex = 1000 s– 1 Dw = 1500 s– 1 pa = 0. 95 R 20 = 15 s– 1 error=5%
Occurrence Single-Field Dispersion Curves Kovrigin, Kempf, Grey, & Loria J Magn Reson. 2006 180 93 Input Parameters kex = 1000 s– 1 Dw = 1500 s– 1 pa = 0. 95 R 20 = 15 s– 1 error=5%
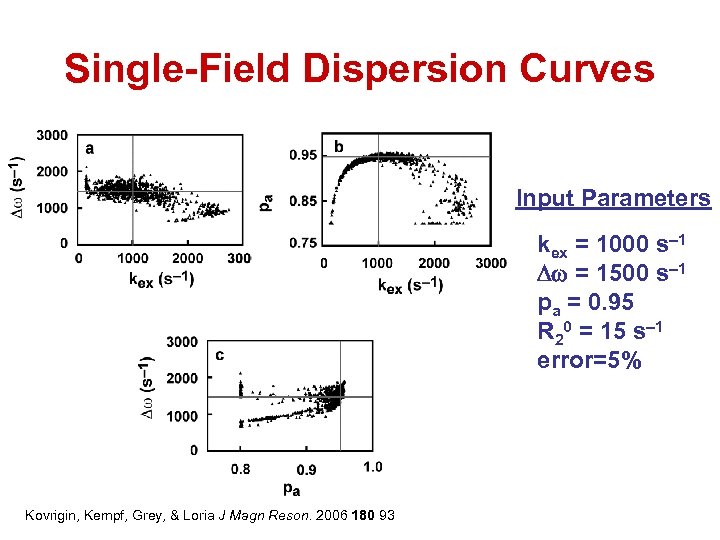 Single-Field Dispersion Curves Input Parameters kex = 1000 s– 1 Dw = 1500 s– 1 pa = 0. 95 R 20 = 15 s– 1 error=5% Kovrigin, Kempf, Grey, & Loria J Magn Reson. 2006 180 93
Single-Field Dispersion Curves Input Parameters kex = 1000 s– 1 Dw = 1500 s– 1 pa = 0. 95 R 20 = 15 s– 1 error=5% Kovrigin, Kempf, Grey, & Loria J Magn Reson. 2006 180 93
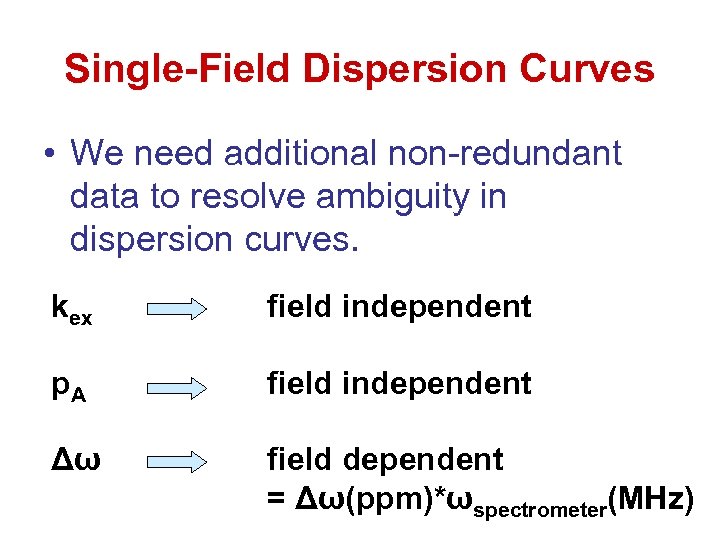 Single-Field Dispersion Curves • We need additional non-redundant data to resolve ambiguity in dispersion curves. kex field independent p. A field independent Δω field dependent = Δω(ppm)*ωspectrometer(MHz)
Single-Field Dispersion Curves • We need additional non-redundant data to resolve ambiguity in dispersion curves. kex field independent p. A field independent Δω field dependent = Δω(ppm)*ωspectrometer(MHz)
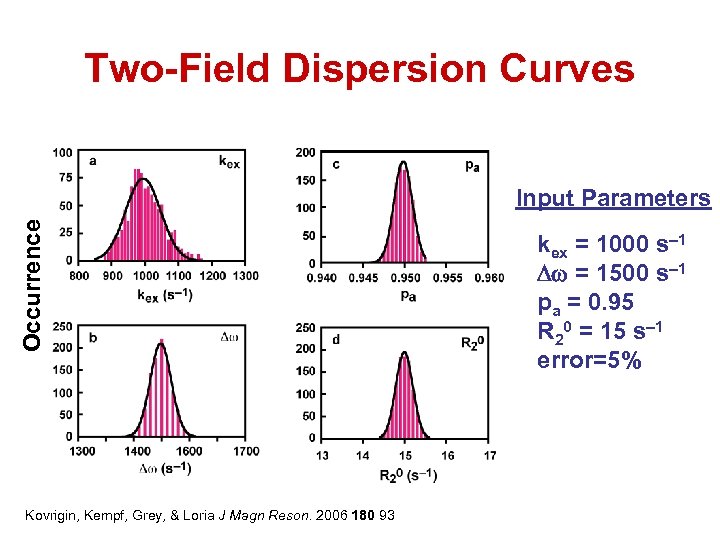 Two-Field Dispersion Curves Occurrence Input Parameters Kovrigin, Kempf, Grey, & Loria J Magn Reson. 2006 180 93 kex = 1000 s– 1 Dw = 1500 s– 1 pa = 0. 95 R 20 = 15 s– 1 error=5%
Two-Field Dispersion Curves Occurrence Input Parameters Kovrigin, Kempf, Grey, & Loria J Magn Reson. 2006 180 93 kex = 1000 s– 1 Dw = 1500 s– 1 pa = 0. 95 R 20 = 15 s– 1 error=5%
 From CPMG data to protein motions R 2, eff νCPMG p. B kex
From CPMG data to protein motions R 2, eff νCPMG p. B kex
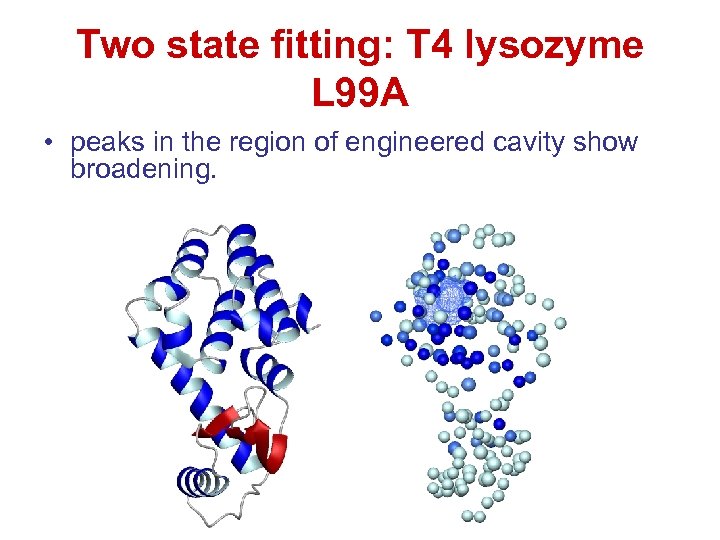 Two state fitting: T 4 lysozyme L 99 A • peaks in the region of engineered cavity show broadening.
Two state fitting: T 4 lysozyme L 99 A • peaks in the region of engineered cavity show broadening.
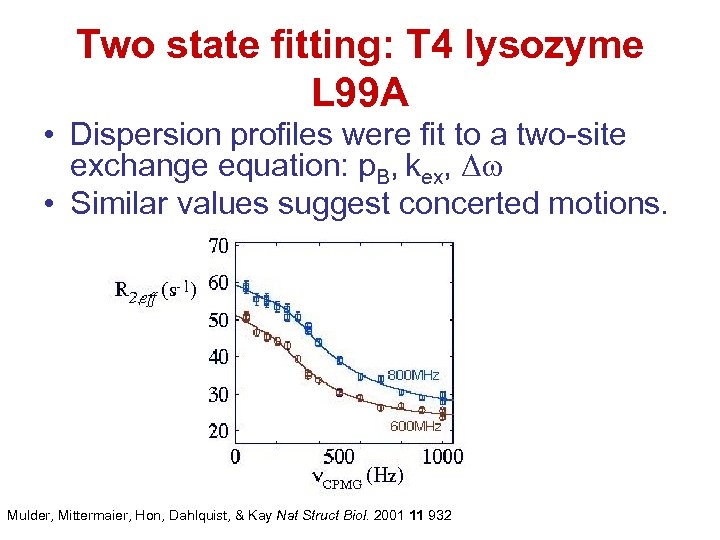 Two state fitting: T 4 lysozyme L 99 A • Dispersion profiles were fit to a two-site exchange equation: p. B, kex, Δω • Similar values suggest concerted motions. Mulder, Mittermaier, Hon, Dahlquist, & Kay Nat Struct Biol. 2001 11 932
Two state fitting: T 4 lysozyme L 99 A • Dispersion profiles were fit to a two-site exchange equation: p. B, kex, Δω • Similar values suggest concerted motions. Mulder, Mittermaier, Hon, Dahlquist, & Kay Nat Struct Biol. 2001 11 932
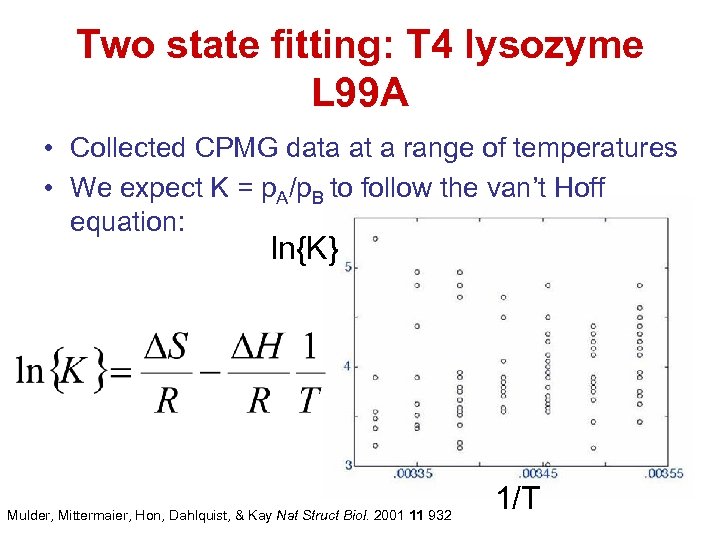 Two state fitting: T 4 lysozyme L 99 A • Collected CPMG data at a range of temperatures • We expect K = p. A/p. B to follow the van’t Hoff equation: ln{K} Mulder, Mittermaier, Hon, Dahlquist, & Kay Nat Struct Biol. 2001 11 932 1/T
Two state fitting: T 4 lysozyme L 99 A • Collected CPMG data at a range of temperatures • We expect K = p. A/p. B to follow the van’t Hoff equation: ln{K} Mulder, Mittermaier, Hon, Dahlquist, & Kay Nat Struct Biol. 2001 11 932 1/T
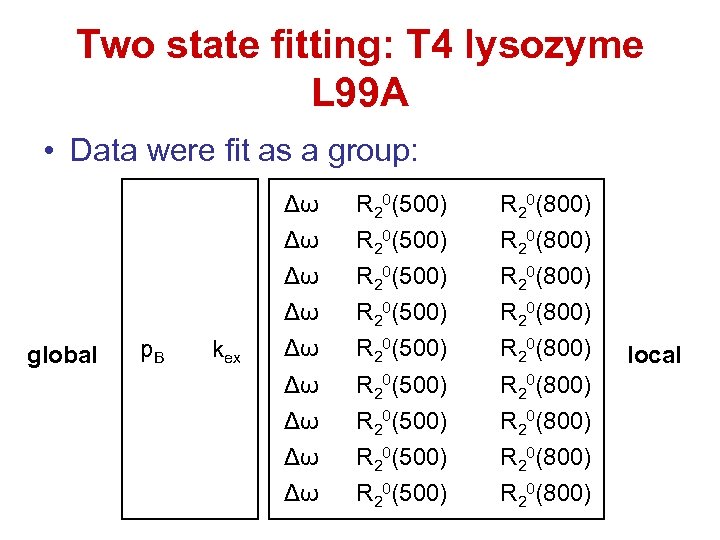 Two state fitting: T 4 lysozyme L 99 A • Data were fit as a group: p. B Δω R 20(500) R 20(800) p. B kex Δω R 20(500) R 20(800) p. B global kex Δω R 20(500) R 20(800) p. B kex Δω R 20(500) R 20(800) local
Two state fitting: T 4 lysozyme L 99 A • Data were fit as a group: p. B Δω R 20(500) R 20(800) p. B kex Δω R 20(500) R 20(800) p. B global kex Δω R 20(500) R 20(800) p. B kex Δω R 20(500) R 20(800) local
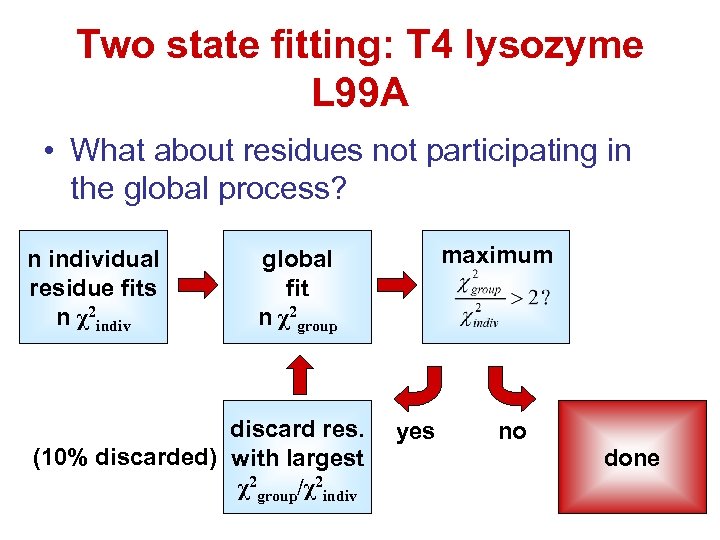 Two state fitting: T 4 lysozyme L 99 A • What about residues not participating in the global process? n individual residue fits n χ2 indiv maximum global fit n χ2 group discard res. (10% discarded) with largest χ2 group/χ2 indiv yes no done
Two state fitting: T 4 lysozyme L 99 A • What about residues not participating in the global process? n individual residue fits n χ2 indiv maximum global fit n χ2 group discard res. (10% discarded) with largest χ2 group/χ2 indiv yes no done
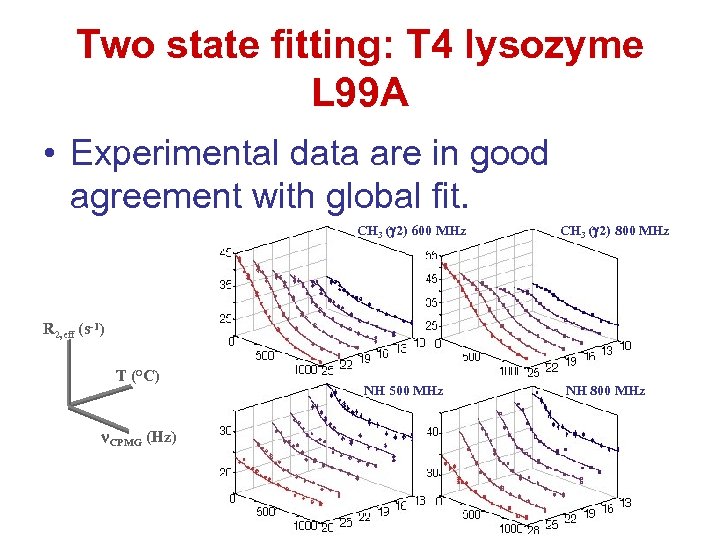 Two state fitting: T 4 lysozyme L 99 A • Experimental data are in good agreement with global fit. CH 3 ( 2) 600 MHz CH 3 ( 2) 800 MHz R 2, eff (s-1) T (°C) CPMG (Hz) NH 500 MHz NH 800 MHz
Two state fitting: T 4 lysozyme L 99 A • Experimental data are in good agreement with global fit. CH 3 ( 2) 600 MHz CH 3 ( 2) 800 MHz R 2, eff (s-1) T (°C) CPMG (Hz) NH 500 MHz NH 800 MHz
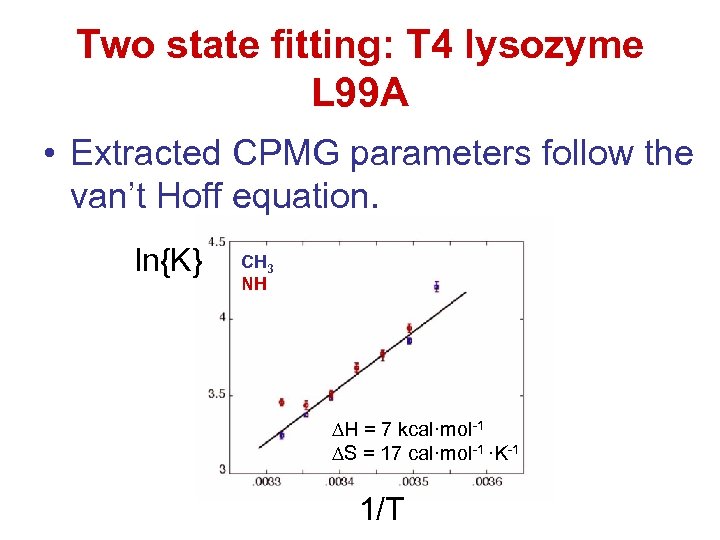 Two state fitting: T 4 lysozyme L 99 A • Extracted CPMG parameters follow the van’t Hoff equation. ln{K} CH 3 NH H = 7 kcal·mol-1 S = 17 cal·mol-1 ·K-1 1/T
Two state fitting: T 4 lysozyme L 99 A • Extracted CPMG parameters follow the van’t Hoff equation. ln{K} CH 3 NH H = 7 kcal·mol-1 S = 17 cal·mol-1 ·K-1 1/T
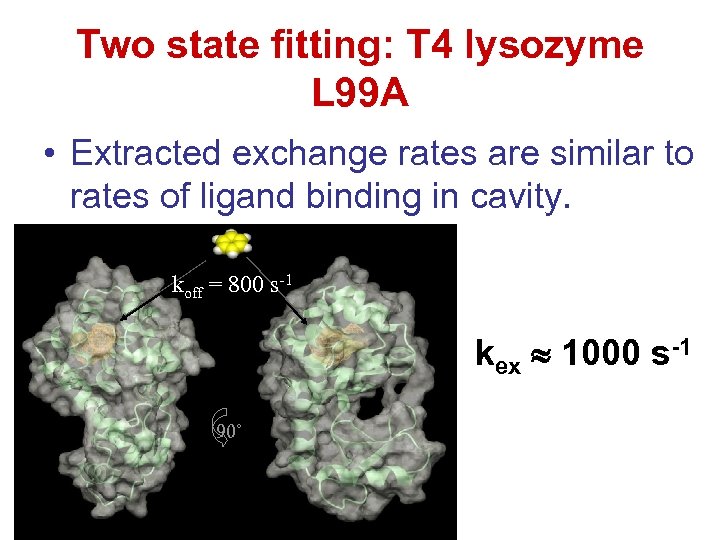 Two state fitting: T 4 lysozyme L 99 A • Extracted exchange rates are similar to rates of ligand binding in cavity. koff = 800 s-1 kex 1000 s-1 90˚
Two state fitting: T 4 lysozyme L 99 A • Extracted exchange rates are similar to rates of ligand binding in cavity. koff = 800 s-1 kex 1000 s-1 90˚
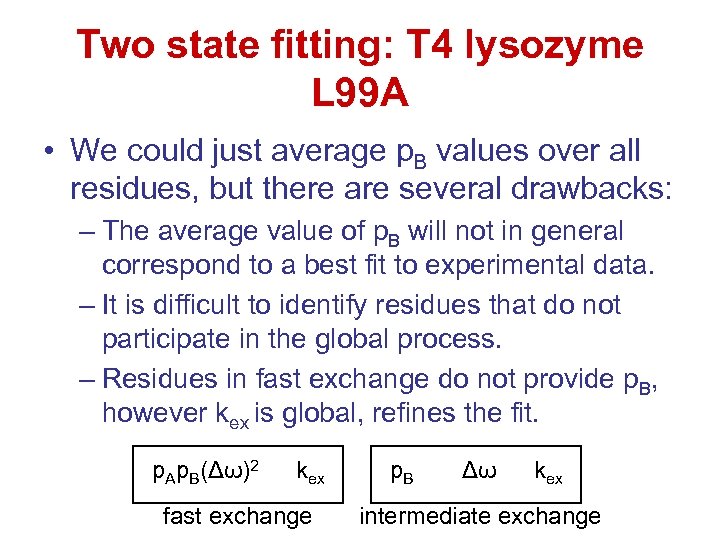 Two state fitting: T 4 lysozyme L 99 A • We could just average p. B values over all residues, but there are several drawbacks: – The average value of p. B will not in general correspond to a best fit to experimental data. – It is difficult to identify residues that do not participate in the global process. – Residues in fast exchange do not provide p. B, however kex is global, refines the fit. p. Ap. B(Δω)2 kex fast exchange p. B Δω kex intermediate exchange
Two state fitting: T 4 lysozyme L 99 A • We could just average p. B values over all residues, but there are several drawbacks: – The average value of p. B will not in general correspond to a best fit to experimental data. – It is difficult to identify residues that do not participate in the global process. – Residues in fast exchange do not provide p. B, however kex is global, refines the fit. p. Ap. B(Δω)2 kex fast exchange p. B Δω kex intermediate exchange
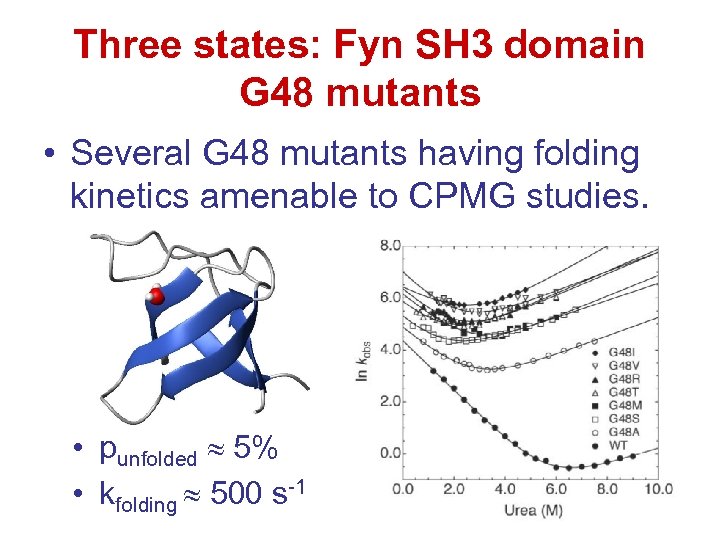 Three states: Fyn SH 3 domain G 48 mutants • Several G 48 mutants having folding kinetics amenable to CPMG studies. • punfolded 5% • kfolding 500 s-1
Three states: Fyn SH 3 domain G 48 mutants • Several G 48 mutants having folding kinetics amenable to CPMG studies. • punfolded 5% • kfolding 500 s-1
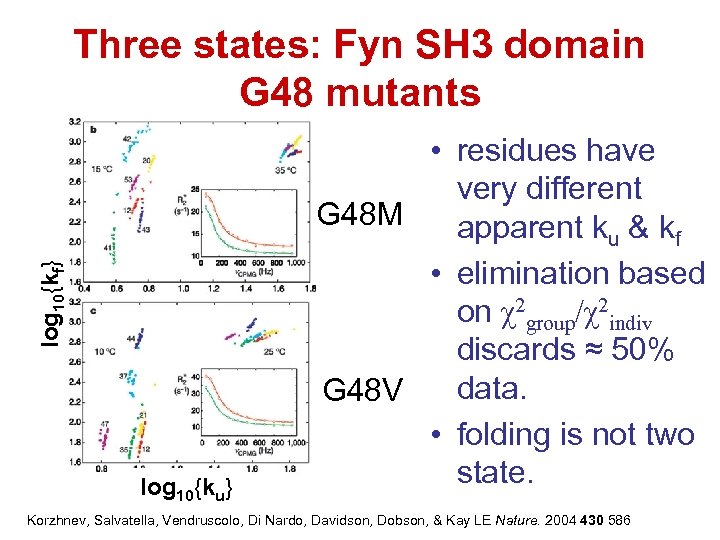 log 10{kf} Three states: Fyn SH 3 domain G 48 mutants log 10{ku} • residues have very different G 48 M apparent ku & kf • elimination based on χ2 group/χ2 indiv discards ≈ 50% data. G 48 V • folding is not two state. Korzhnev, Salvatella, Vendruscolo, Di Nardo, Davidson, Dobson, & Kay LE Nature. 2004 430 586
log 10{kf} Three states: Fyn SH 3 domain G 48 mutants log 10{ku} • residues have very different G 48 M apparent ku & kf • elimination based on χ2 group/χ2 indiv discards ≈ 50% data. G 48 V • folding is not two state. Korzhnev, Salvatella, Vendruscolo, Di Nardo, Davidson, Dobson, & Kay LE Nature. 2004 430 586
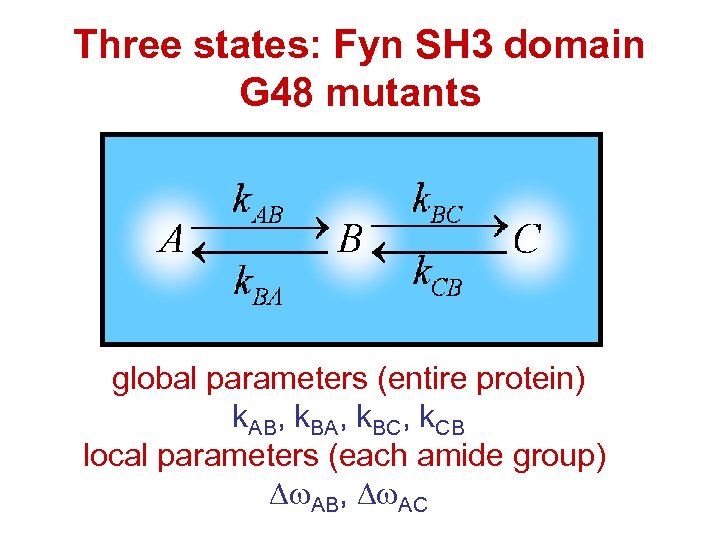 Three states: Fyn SH 3 domain G 48 mutants global parameters (entire protein) k. AB, k. BA, k. BC, k. CB local parameters (each amide group) AB, AC
Three states: Fyn SH 3 domain G 48 mutants global parameters (entire protein) k. AB, k. BA, k. BC, k. CB local parameters (each amide group) AB, AC
 Three-state dispersion profiles • Two-state exchange described by analytical expressions. • Three-state exchange profiles can be calculated numerically using modified Bloch-Mc. Connell equations.
Three-state dispersion profiles • Two-state exchange described by analytical expressions. • Three-state exchange profiles can be calculated numerically using modified Bloch-Mc. Connell equations.
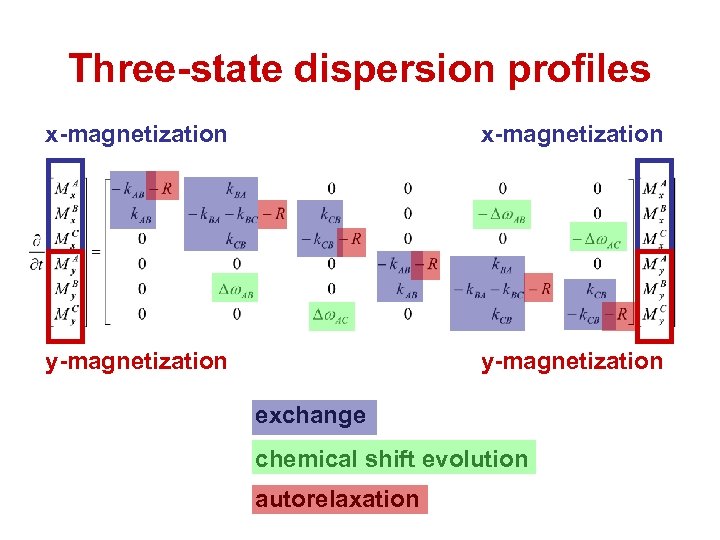 Three-state dispersion profiles x-magnetization y-magnetization exchange chemical shift evolution autorelaxation
Three-state dispersion profiles x-magnetization y-magnetization exchange chemical shift evolution autorelaxation
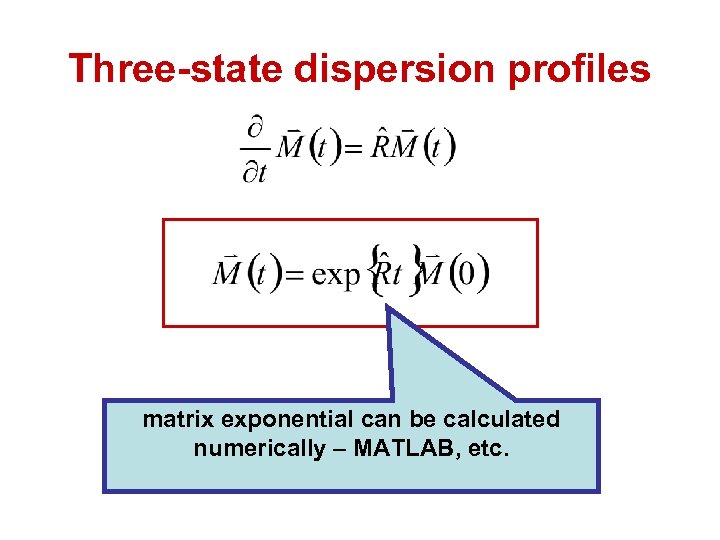 Three-state dispersion profiles matrix exponential can be calculated numerically – MATLAB, etc.
Three-state dispersion profiles matrix exponential can be calculated numerically – MATLAB, etc.
 Three-state dispersion profiles τ 180 τ n
Three-state dispersion profiles τ 180 τ n
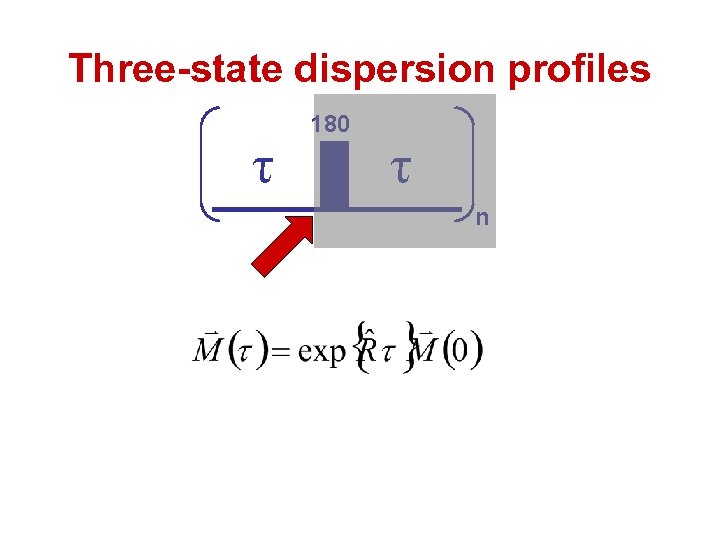 Three-state dispersion profiles τ 180 τ n
Three-state dispersion profiles τ 180 τ n
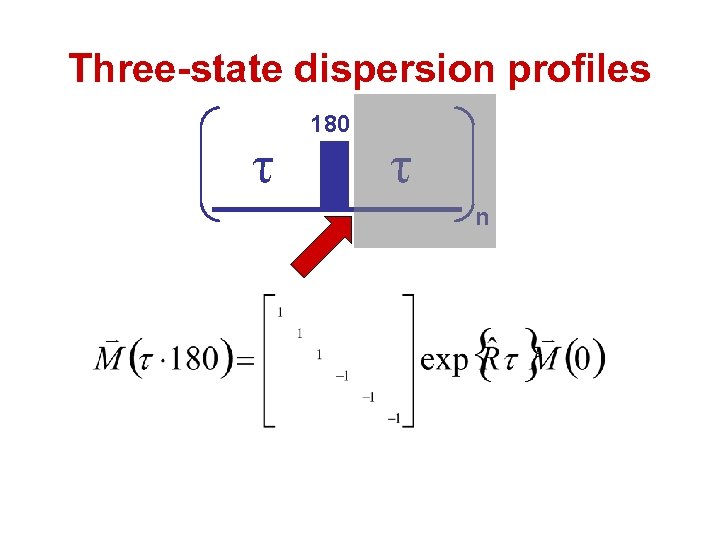 Three-state dispersion profiles τ 180 τ n
Three-state dispersion profiles τ 180 τ n
 Three-state dispersion profiles τ 180 τ n
Three-state dispersion profiles τ 180 τ n
 Three-state dispersion profiles τ 180 τ n
Three-state dispersion profiles τ 180 τ n
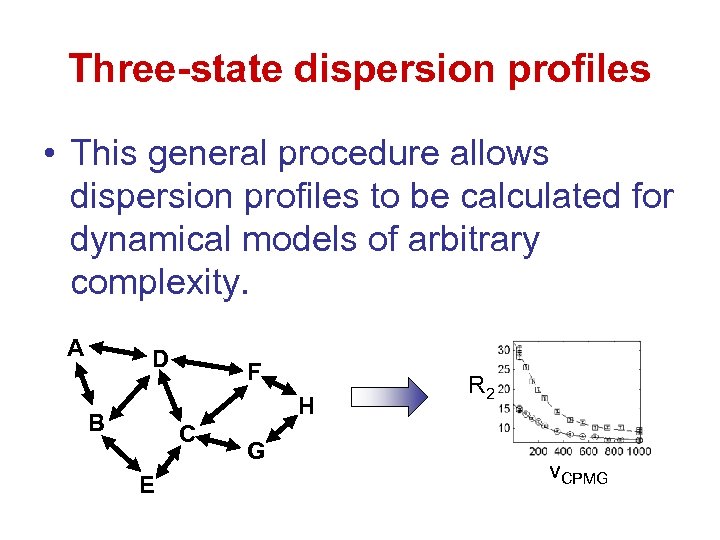 Three-state dispersion profiles • This general procedure allows dispersion profiles to be calculated for dynamical models of arbitrary complexity. A D F H B C E G R 2 v. CPMG
Three-state dispersion profiles • This general procedure allows dispersion profiles to be calculated for dynamical models of arbitrary complexity. A D F H B C E G R 2 v. CPMG
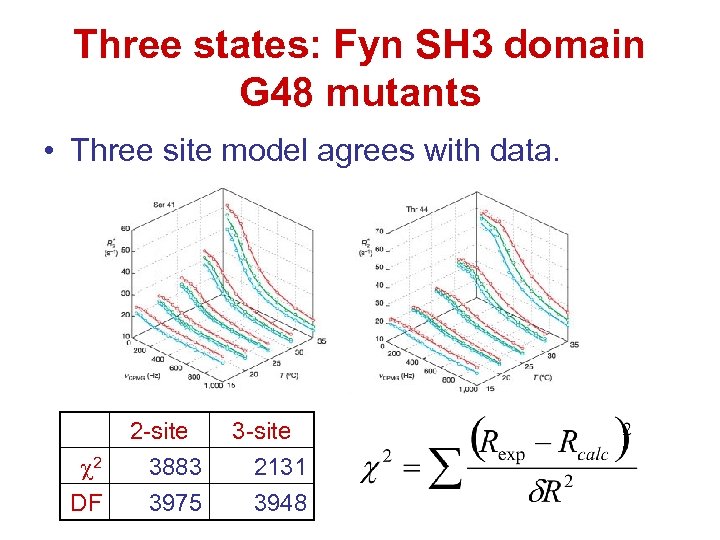 Three states: Fyn SH 3 domain G 48 mutants • Three site model agrees with data. 2 DF 2 -site 3883 3975 3 -site 2131 3948
Three states: Fyn SH 3 domain G 48 mutants • Three site model agrees with data. 2 DF 2 -site 3883 3975 3 -site 2131 3948
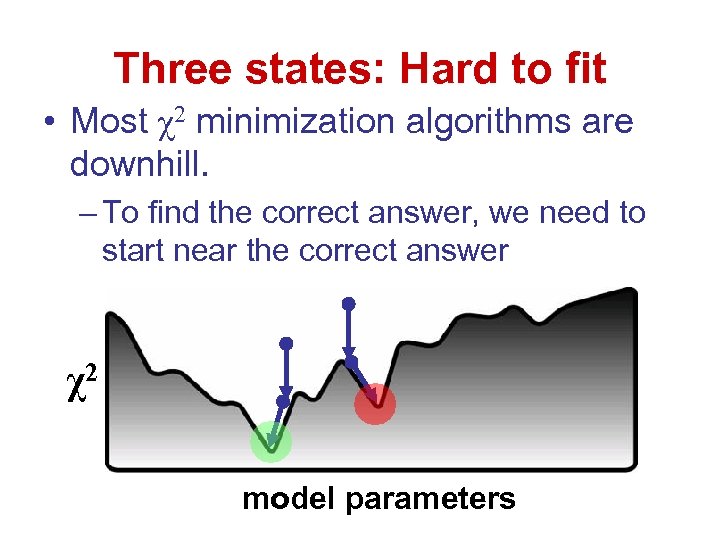 Three states: Hard to fit • Most χ2 minimization algorithms are downhill. – To find the correct answer, we need to start near the correct answer χ2 model parameters
Three states: Hard to fit • Most χ2 minimization algorithms are downhill. – To find the correct answer, we need to start near the correct answer χ2 model parameters
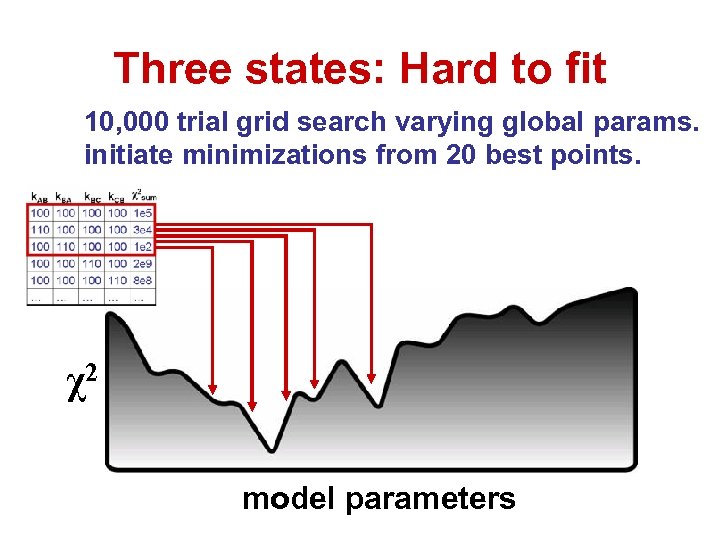 Three states: Hard to fit 10, 000 trial grid search varying global params. initiate minimizations from 20 best points. χ2 model parameters
Three states: Hard to fit 10, 000 trial grid search varying global params. initiate minimizations from 20 best points. χ2 model parameters
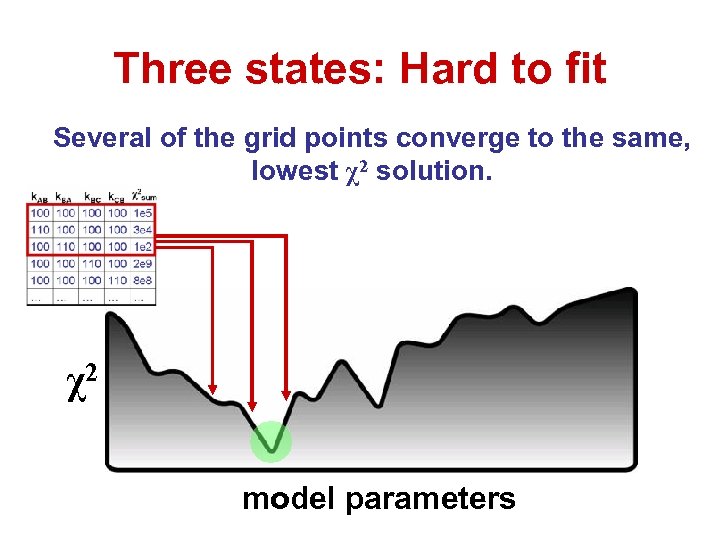 Three states: Hard to fit Several of the grid points converge to the same, lowest χ2 solution. χ2 model parameters
Three states: Hard to fit Several of the grid points converge to the same, lowest χ2 solution. χ2 model parameters
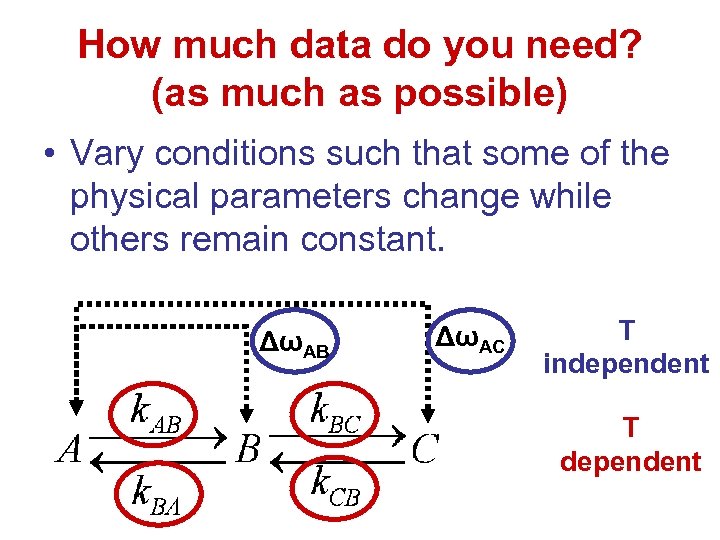 How much data do you need? (as much as possible) • Vary conditions such that some of the physical parameters change while others remain constant. ΔωAB ΔωAC T independent T dependent
How much data do you need? (as much as possible) • Vary conditions such that some of the physical parameters change while others remain constant. ΔωAB ΔωAC T independent T dependent
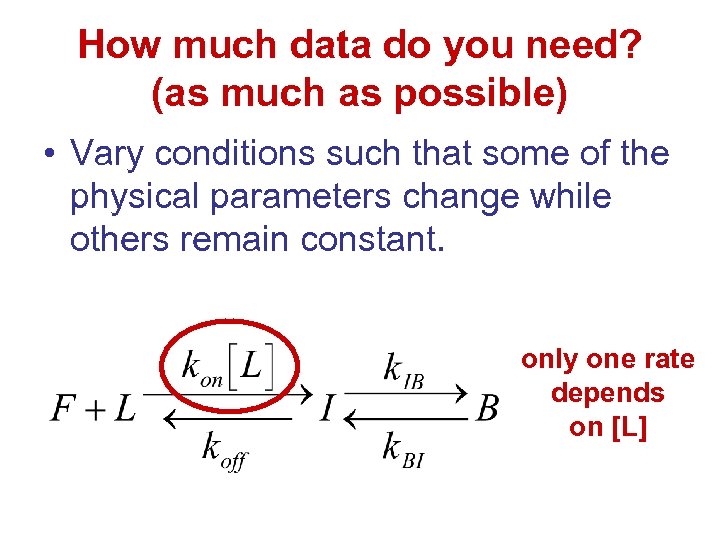 How much data do you need? (as much as possible) • Vary conditions such that some of the physical parameters change while others remain constant. only one rate depends on [L]
How much data do you need? (as much as possible) • Vary conditions such that some of the physical parameters change while others remain constant. only one rate depends on [L]
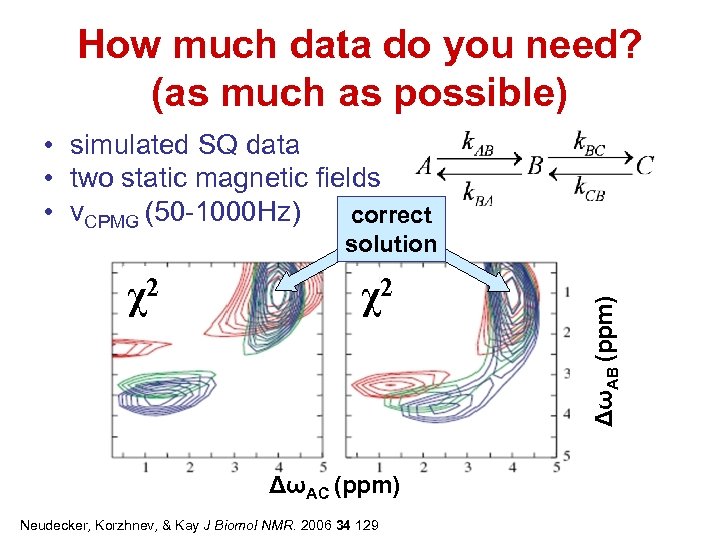 How much data do you need? (as much as possible) • simulated SQ data • two static magnetic fields • νCPMG (50 -1000 Hz) correct χ2 χ2 ΔωAC (ppm) Neudecker, Korzhnev, & Kay J Biomol NMR. 2006 34 129 ΔωAB (ppm) solution
How much data do you need? (as much as possible) • simulated SQ data • two static magnetic fields • νCPMG (50 -1000 Hz) correct χ2 χ2 ΔωAC (ppm) Neudecker, Korzhnev, & Kay J Biomol NMR. 2006 34 129 ΔωAB (ppm) solution
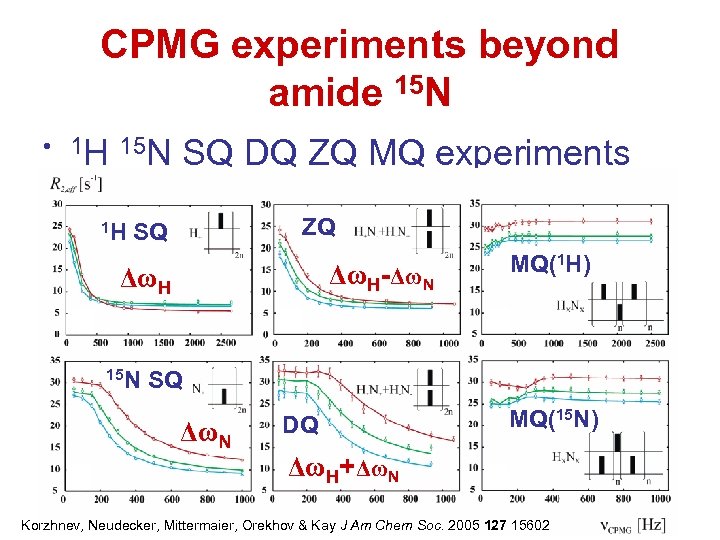 CPMG experiments beyond amide 15 N • 1 H 15 N 1 H SQ DQ ZQ MQ experiments ZQ SQ ΔωH-ΔωN ΔωH 15 N MQ(1 H) SQ ΔωN DQ MQ(15 N) ΔωH+ΔωN Korzhnev, Neudecker, Mittermaier, Orekhov & Kay J Am Chem Soc. 2005 127 15602
CPMG experiments beyond amide 15 N • 1 H 15 N 1 H SQ DQ ZQ MQ experiments ZQ SQ ΔωH-ΔωN ΔωH 15 N MQ(1 H) SQ ΔωN DQ MQ(15 N) ΔωH+ΔωN Korzhnev, Neudecker, Mittermaier, Orekhov & Kay J Am Chem Soc. 2005 127 15602
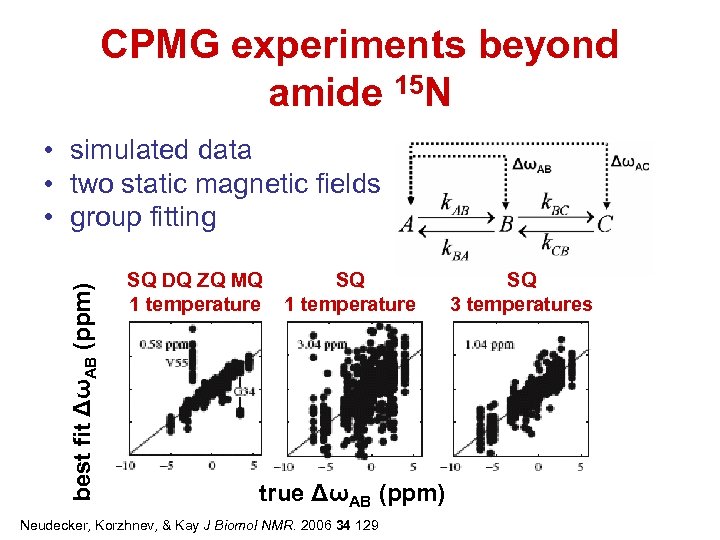 CPMG experiments beyond amide 15 N best fit ΔωAB (ppm) • simulated data • two static magnetic fields • group fitting SQ DQ ZQ MQ 1 temperature SQ 1 temperature true ΔωAB (ppm) Neudecker, Korzhnev, & Kay J Biomol NMR. 2006 34 129 SQ 3 temperatures
CPMG experiments beyond amide 15 N best fit ΔωAB (ppm) • simulated data • two static magnetic fields • group fitting SQ DQ ZQ MQ 1 temperature SQ 1 temperature true ΔωAB (ppm) Neudecker, Korzhnev, & Kay J Biomol NMR. 2006 34 129 SQ 3 temperatures
 CPMG experiments beyond amide 15 N • In general, dispersion profiles are well-fit by two-site model. • Even with 6 experiments, for singleresidue fits, 3 -site is better than 2 -site model for only 14 out of 40 residues. • Multi-site models explain inconsistencies between apparent two-site parameters for different residues.
CPMG experiments beyond amide 15 N • In general, dispersion profiles are well-fit by two-site model. • Even with 6 experiments, for singleresidue fits, 3 -site is better than 2 -site model for only 14 out of 40 residues. • Multi-site models explain inconsistencies between apparent two-site parameters for different residues.
 Characterizing minor states using CPMG chemical shift information
Characterizing minor states using CPMG chemical shift information
 Obtaining the signs of chemical shift differences 15 N ±Dw ? 1 H ppm
Obtaining the signs of chemical shift differences 15 N ±Dw ? 1 H ppm
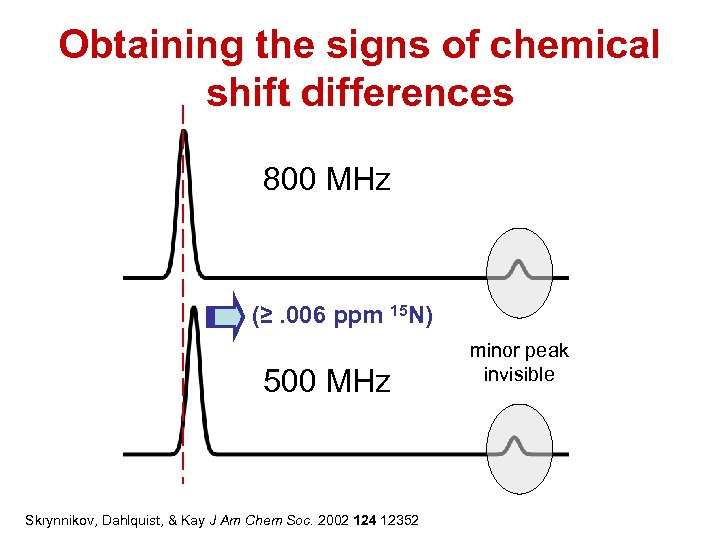 Obtaining the signs of chemical shift differences 800 MHz (≥. 006 ppm 15 N) 500 MHz Skrynnikov, Dahlquist, & Kay J Am Chem Soc. 2002 124 12352 minor peak invisible
Obtaining the signs of chemical shift differences 800 MHz (≥. 006 ppm 15 N) 500 MHz Skrynnikov, Dahlquist, & Kay J Am Chem Soc. 2002 124 12352 minor peak invisible
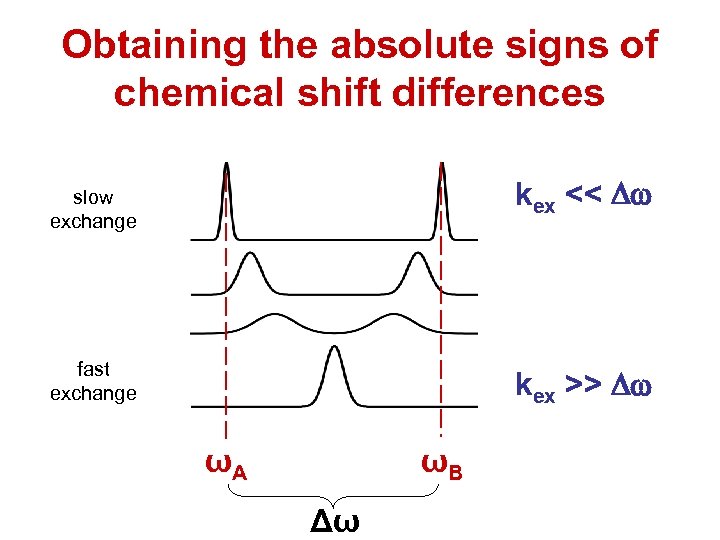 Obtaining the absolute signs of chemical shift differences kex << Dw slow exchange fast exchange kex >> Dw ωA ωB Δω
Obtaining the absolute signs of chemical shift differences kex << Dw slow exchange fast exchange kex >> Dw ωA ωB Δω
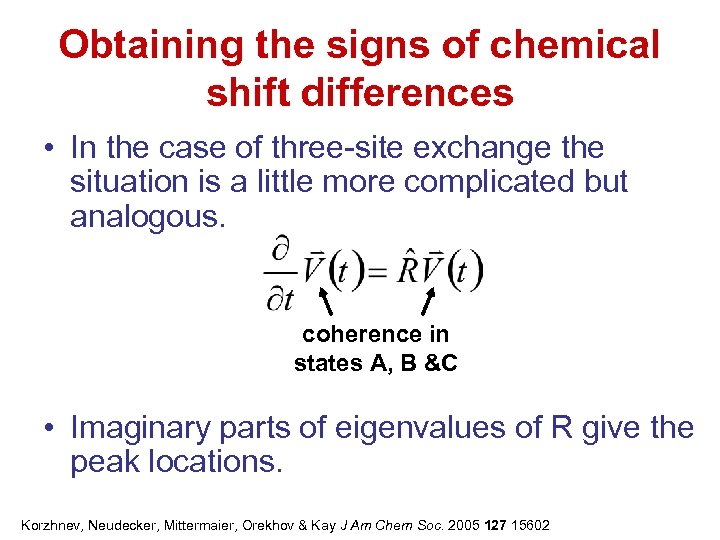 Obtaining the signs of chemical shift differences • In the case of three-site exchange the situation is a little more complicated but analogous. coherence in states A, B &C • Imaginary parts of eigenvalues of R give the peak locations. Korzhnev, Neudecker, Mittermaier, Orekhov & Kay J Am Chem Soc. 2005 127 15602
Obtaining the signs of chemical shift differences • In the case of three-site exchange the situation is a little more complicated but analogous. coherence in states A, B &C • Imaginary parts of eigenvalues of R give the peak locations. Korzhnev, Neudecker, Mittermaier, Orekhov & Kay J Am Chem Soc. 2005 127 15602
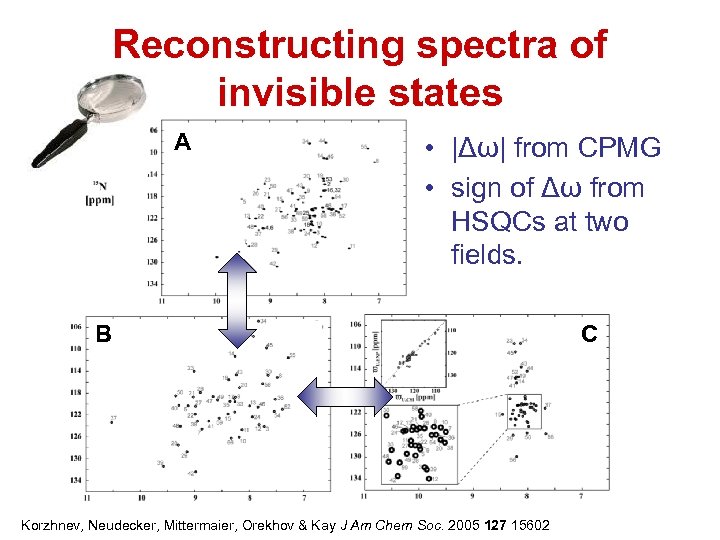 Reconstructing spectra of invisible states A • |Δω| from CPMG • sign of Δω from HSQCs at two fields. B Korzhnev, Neudecker, Mittermaier, Orekhov & Kay J Am Chem Soc. 2005 127 15602 C
Reconstructing spectra of invisible states A • |Δω| from CPMG • sign of Δω from HSQCs at two fields. B Korzhnev, Neudecker, Mittermaier, Orekhov & Kay J Am Chem Soc. 2005 127 15602 C
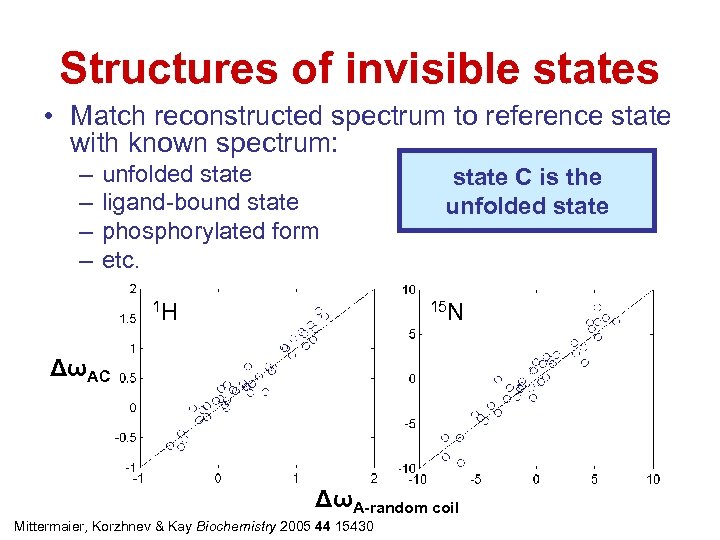 Structures of invisible states • Match reconstructed spectrum to reference state with known spectrum: – – unfolded state ligand-bound state phosphorylated form etc. 1 H state C is the unfolded state 15 N ΔωAC ΔωA-random coil Mittermaier, Korzhnev & Kay Biochemistry 2005 44 15430
Structures of invisible states • Match reconstructed spectrum to reference state with known spectrum: – – unfolded state ligand-bound state phosphorylated form etc. 1 H state C is the unfolded state 15 N ΔωAC ΔωA-random coil Mittermaier, Korzhnev & Kay Biochemistry 2005 44 15430
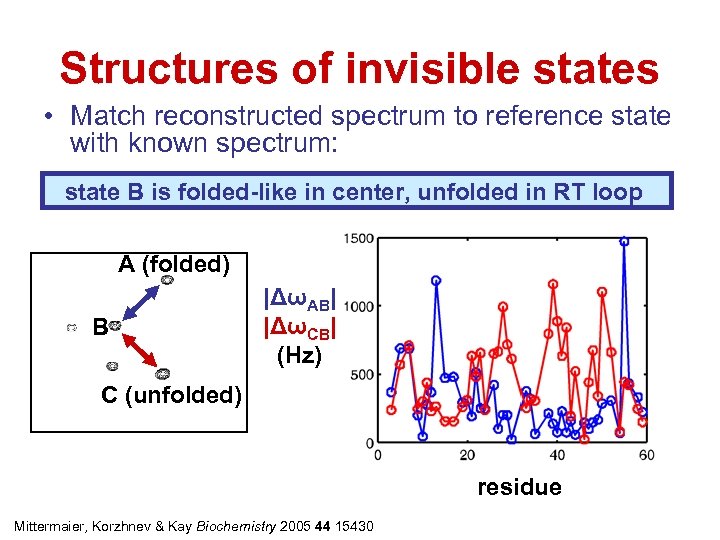 Structures of invisible states • Match reconstructed spectrum to reference state with known spectrum: state B is folded-like in center, unfolded in RT loop A (folded) B |ΔωAB| |ΔωCB| (Hz) C (unfolded) residue Mittermaier, Korzhnev & Kay Biochemistry 2005 44 15430
Structures of invisible states • Match reconstructed spectrum to reference state with known spectrum: state B is folded-like in center, unfolded in RT loop A (folded) B |ΔωAB| |ΔωCB| (Hz) C (unfolded) residue Mittermaier, Korzhnev & Kay Biochemistry 2005 44 15430
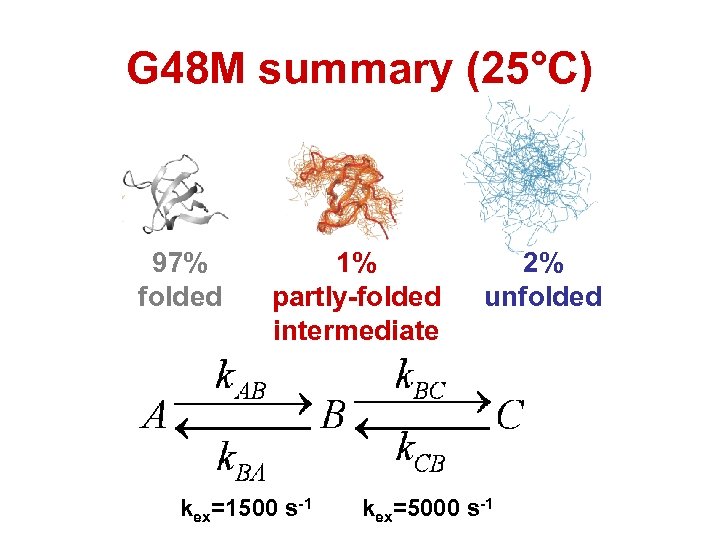 G 48 M summary (25°C) 97% folded 1% partly-folded intermediate kex=1500 s-1 2% unfolded kex=5000 s-1
G 48 M summary (25°C) 97% folded 1% partly-folded intermediate kex=1500 s-1 2% unfolded kex=5000 s-1
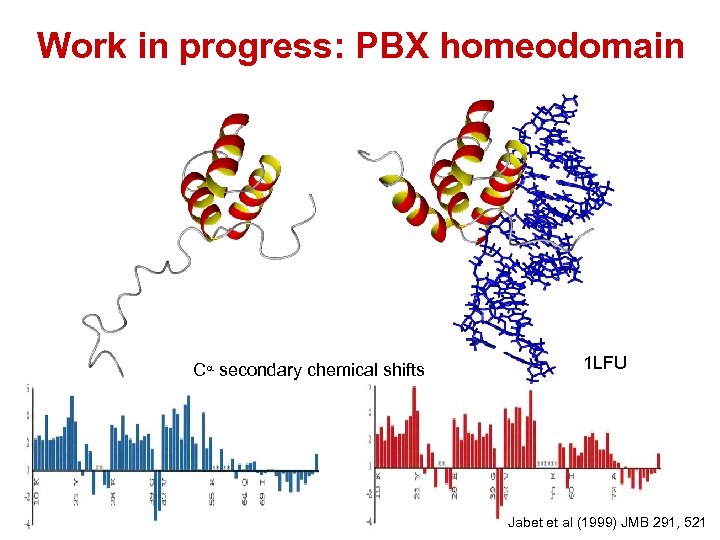 Work in progress: PBX homeodomain Ca secondary chemical shifts 1 LFU Jabet et al (1999) JMB 291, 521
Work in progress: PBX homeodomain Ca secondary chemical shifts 1 LFU Jabet et al (1999) JMB 291, 521
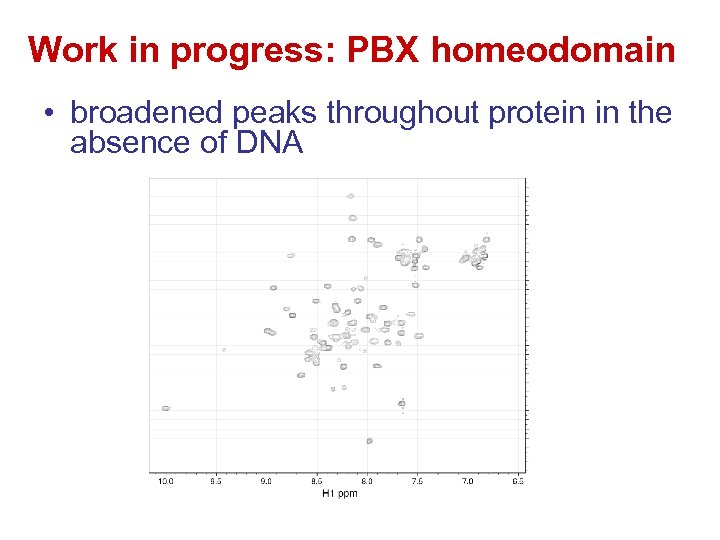 Work in progress: PBX homeodomain • broadened peaks throughout protein in the absence of DNA
Work in progress: PBX homeodomain • broadened peaks throughout protein in the absence of DNA
 Work in progress: PBX homeodomain ?
Work in progress: PBX homeodomain ?
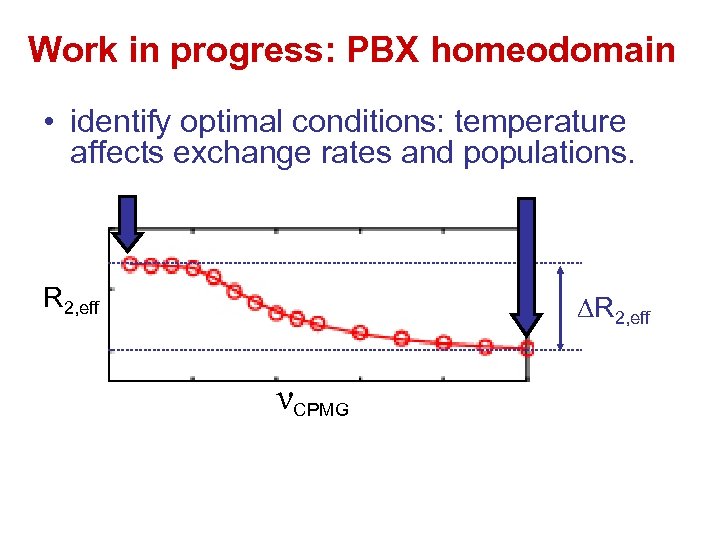 Work in progress: PBX homeodomain • identify optimal conditions: temperature affects exchange rates and populations. R 2, eff νCPMG
Work in progress: PBX homeodomain • identify optimal conditions: temperature affects exchange rates and populations. R 2, eff νCPMG
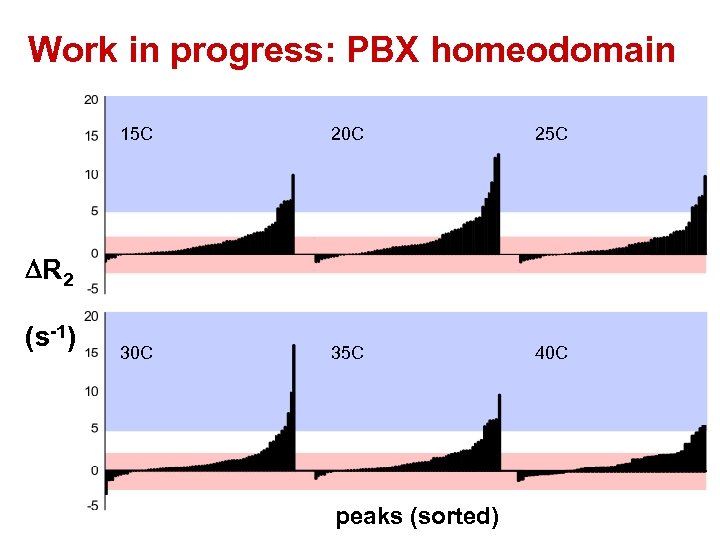 Work in progress: PBX homeodomain 15 C 20 C 25 C 30 C 35 C 40 C DR 2 (s-1) peaks (sorted)
Work in progress: PBX homeodomain 15 C 20 C 25 C 30 C 35 C 40 C DR 2 (s-1) peaks (sorted)
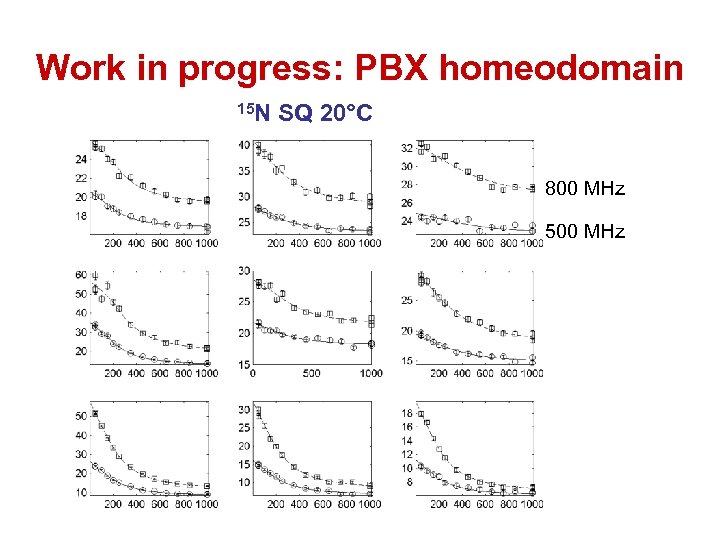 Work in progress: PBX homeodomain 15 N SQ 20°C 800 MHz 500 MHz
Work in progress: PBX homeodomain 15 N SQ 20°C 800 MHz 500 MHz
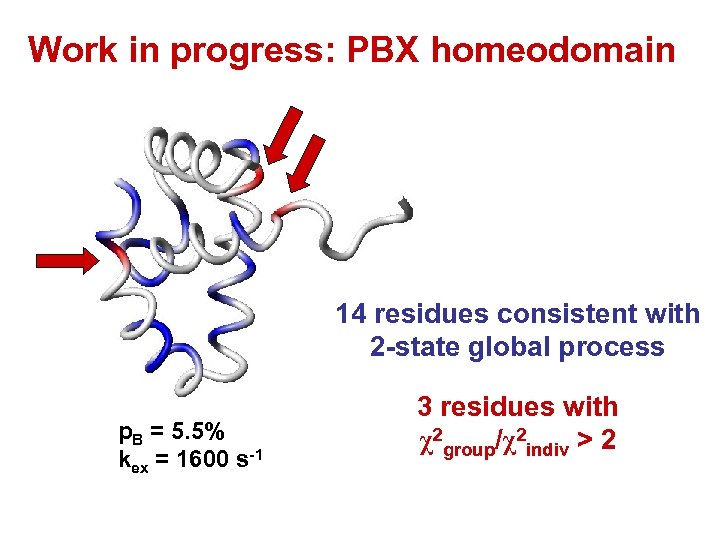 Work in progress: PBX homeodomain 14 residues consistent with 2 -state global process p. B = 5. 5% kex = 1600 s-1 3 residues with χ2 group/χ2 indiv > 2
Work in progress: PBX homeodomain 14 residues consistent with 2 -state global process p. B = 5. 5% kex = 1600 s-1 3 residues with χ2 group/χ2 indiv > 2
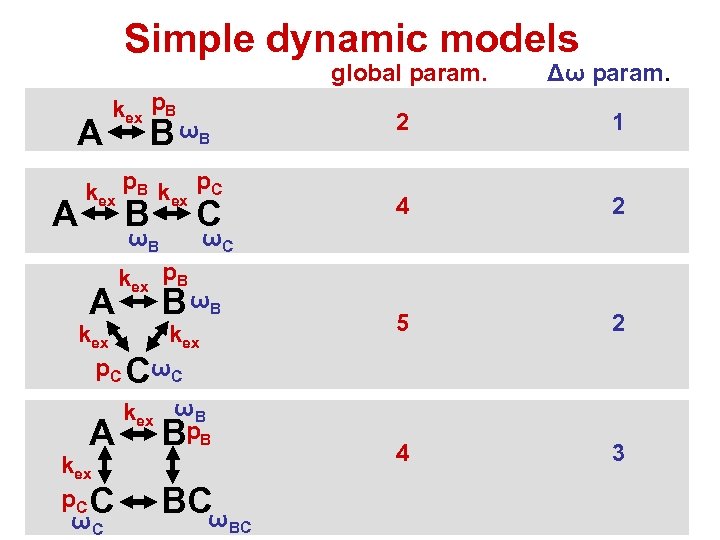 Simple dynamic models global param. A A kex p. B 2 ωB A kex p. C C ωC 3 ωC kex p. B B ωB kex p. C CωC A 2 4 C 2 5 kex p. B kex p. C 1 4 B ωB B Δω param. kex ωB Bp. B BCω BC
Simple dynamic models global param. A A kex p. B 2 ωB A kex p. C C ωC 3 ωC kex p. B B ωB kex p. C CωC A 2 4 C 2 5 kex p. B kex p. C 1 4 B ωB B Δω param. kex ωB Bp. B BCω BC





